Abstract
Inflammatory responses in many cell types are coordinately regulated by the opposing actions of NF-κB and the glucocorticoid receptor (GR). The human glucocorticoid receptor (hGR) gene encodes two protein isoforms: a cytoplasmic alpha form (GRα), which binds hormone, translocates to the nucleus, and regulates gene transcription, and a nuclear localized beta isoform (GRβ), which does not bind known ligands and attenuates GRα action. We report here the identification of a tumor necrosis factor (TNF)-responsive NF-κB DNA binding site 5′ to the hGR promoter that leads to a 1.5-fold increase in GRα mRNA and a 2.0-fold increase in GRβ mRNA in HeLaS3 cells, which endogenously express both GR isoforms. However, TNF-α treatment disproportionately increased the steady-state levels of the GRβ protein isoform over GRα, making GRβ the predominant endogenous receptor isoform. Similar results were observed following treatment of human CEMC7 lymphoid cells with TNF-α or IL-1. The increase in GRβ protein expression correlated with the development of glucocorticoid resistance.
Inflammation is a multifaceted immune response that generates tissue damage. Proinflammatory cytokines such as tumor necrosis factor (TNF)-α and IL-1 initiate this cascade in part by activating the transcription factor NF-κB, which in turn stimulates proinflammatory genes (1–3). In contrast, corticosteroids are potent antiinflammatory agents. Recently, the mechanisms underlying the antiinflammatory actions of corticosteroids have indicated that ligand-bound glucocorticoid receptor (GR) interacts with nuclear localized NF-κB and alters its ability to promote transcription (4). The antagonism between NF-κB and GR is mutual, such that NF-κB can also abolish the transactivation of glucocorticoid-responsive genes (5, 6). Additionally, glucocorticoids can also increase the expression of I-κB (7, 8). Thus, these two opposing regulatory systems appear inherently coupled in the control of the inflammatory response.
Glucocorticoid resistance can arise whereby the antiinflammatory effects of the corticosteroids are ablated, and several mechanisms have been suggested. For example, following chronic exposure to glucocorticoid, there is a down-regulation of the glucocorticoid receptor (GRα) at both the mRNA and protein level (9, 10). The phosphorylation status of the receptor also influences sensitivity and selectivity to glucocorticoids (11). Cell type-specific expression of several GR coactivators and/or corepressors could also contribute to glucocorticoid resistance.
When the human glucocorticoid receptor was first isolated and cloned, two distinct isoforms were described (12). The alpha isoform binds hormone, translocates to the nucleus, and activates transcription of hormone-sensitive genes. The beta isoform does not activate known hormone-sensitive genes (13). Whereas many studies have elucidated how GRα alters gene expression, little effort has been devoted to understanding the function of GRβ. GRβ does not bind typical corticosteroids or antagonists, is predominantly located in the nucleus, and can attenuate the ligand-mediated transactivation of hormone-sensitive genes by GRα (13–15). This dominant negative property of the GRβ isoform makes it an attractive candidate to explain glucocorticoid resistance (16–18).
We report here that a consensus NF-κB sequence is located in the previously published (19) 5′-flanking sequences of the hGR gene, which is identical to that identified in the lymphotoxin (TNF-β) gene (20). Based on the relationship between NF-κB and glucocorticoid signaling, we examined the potential for proinflammatory cytokines such as TNF-α and IL-1 to regulate hGR isoform expression. We show that TNF-α leads to the selective accumulation of GRβ protein and the development of a state of glucocorticoid resistance.
Materials and Methods
Dexamethasone (9α-fluoro-16α-methyl-11β, 17α, 21-trihydroxypregn-1, 4-diene-3, 20-dione) was purchased from Steraloids (Wilton, NH). The TNF-α and IL-1 were purchased from R & D systems. The [14C]chloramphenicol (40–60 Ci/mmol) was from DuPont/NEN. Biotrans nylon membranes were purchased from ICN. Protran nitrocellulose BA85 was purchased from Schleicher & Schuell, and 20 × 20-cm TLC Silica gel 60 sheets were purchased from EM Separation Technology (Gibstown, NJ). The antipeptide polyclonal antibodies AShGR and BShGR (14, 21) were used for both immunohistochemistry and Western analysis.
Cell Culture and Transfection.
HeLaS3 and COS1 cells were grown as previously described (10, 22). CEMC7 cells were grown in RPMI-1640 containing 100 units/ml penicillin, 100 μg/ml streptomycin and supplemented with 2 mM glutamine and 10% FCS. Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 (HeLaS3 and COS1) or 7% CO2 (CEMC7). Subconfluent monolayers of COS1 cells were transfected by using DMRIE-C reagent (GIBCO/BRL).
Reverse Transcription (RT)–PCR.
Total RNA from HeLaS3 cells treated with TNF-α or vehicle was isolated by using TriZol Reagent (Life Technologies, Grand Island, NY). cDNA was prepared and amplified by using the hGRα and hGRβ primers previously described (13). Amplified DNA fragments were electrophoretically fractionated on 1.75% agarose gels. Restriction enzyme analysis of the RT-PCR products amplified by the hGRα- and hGRβ-specific primers confirmed that these fragments contained the appropriate sequences.
Semiquantitative RT-PCR.
RNA was reverse transcribed, and the resulting cDNA was amplified as described. Five microliters of the PCR reaction was removed at 2-cycle intervals and electrophoresed on 1.75% agarose gels stained with ethidium bromide. The intensity of the ethidium bromide fluorescence was measured densitometrically and plotted as a function of cycle number to generate amplification curves for the GRα and GRβ PCR fragments. The amplification efficiencies for the GRα and GRβ fragments were nearly identical.
Immunocytochemistry.
HeLaS3 cells were placed in two-chamber glass slides in JMEM supplemented with 4% dextran-coated charcoal-treated serum. The cells were incubated for 24 h with either TNF-α (1 ng/ml) or vehicle and subsequently treated with 100 nM dexamethasone (DEX) for 1 h. Cells were fixed and processed as previously described (14, 21, 23, 24). Image analysis was performed by using the video image analysis software NIH Image 1.56 for the Power PC (National Institutes of Health, Bethesda, MD).
Western Blot Analysis.
Extracts from HeLaS3 or CEMC7 cells, which were treated with TNF-α or IL-1 as stated or vehicle, were prepared as described (25–28). Membranes were then incubated with epitope-purified polyclonal anti-hGRα (AShGR) or anti-hGRβ (BShGR) at a dilution of 1:500 or anti-hGR (no. 57) antibody at a dilution of 1:1,000. Immunoreactivity was detected by using enhanced chemiluminescence (ECL, Amersham Pharmacia). For the homologous down-regulation studies, COS1 cells were transfected with either hGRα or hGRβ as described. Eighteen hours posttransfection, the cells were treated with 100 nM DEX or vehicle for 24 h, and extracts were prepared for Western blotting.
Half-Life Studies of hGR mRNA and Protein.
Turnover of GRα and GRβ mRNA was determined by semiquantitative RT-PCR of GRα and GRβ mRNA obtained from actinomycin D-treated HeLaS3 cells. Twenty-four hours after treatment with TNF-α (1 ng/ml), the cells were treated with actinomycin D (1 μg/ml). Total RNA was isolated at time points from 0 to 9 h. Western analysis was used to measure receptor protein half-life. Twenty-four hours after treatment with TNF-α (1 ng/ml), the cells were treated with cycloheximide (1 μM). Whole cell lysates were prepared at various time points from 0 to 48 h.
Luciferase Assays.
Cells were transfected as described, and luciferase assays were performed according to the manufacturer's instructions (Analytical Luminescence Laboratory, San Diego). Luciferase activity was reported in relative light units with a coefficient of variance calculated for triplicate samples.
Chloramphenicol Acetyltransferase (CAT) Assays.
HeLaS3 cells were transfected with the GRE2-TATA-CAT reporter plasmid that contains a tandem repeat of a consensus glucocorticoid response element (GRE) (22, 29). After transfection, cells were treated with TNF-α, DEX, or vehicle and allowed to incubate for 24 h. Cell extracts were prepared and analyzed as previously described (30).
Alkaline Phosphatase Assay.
To determine the effect TNF-α had on an endogenous glucocorticoid-responsive gene, alkaline phosphatase activity was measured in HeLaS3 cells incubated for 24 h with either TNF-α (1 ng/ml) or vehicle and subsequently treated with 100 nM dexamethasone for 16 h (31).
Results
NF-κB-Mediated Regulation of GRα and GRβ Expression.
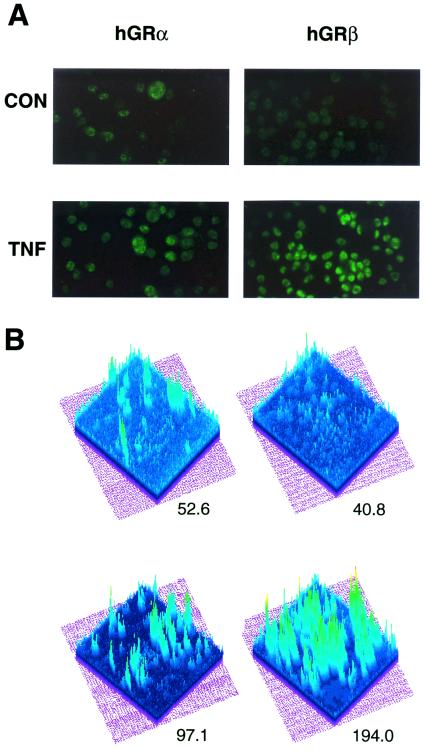
NF-κB represses the ability of GRα to activate glucocorticoid-responsive promoters (4, 6) via a physical interaction between NF-κB and GRα in the nucleus; however, whether NF-κB-mediated changes in GRα and/or GRβ expression contributes to this antagonism is unknown. To address this question, we treated HeLaS3 cells, which endogenously express both GR isoforms, for 24 h with or without the cytokine TNF-α. Semiquantitative RT-PCR was then performed on the isolated RNA with primers specific for hGRα mRNA, hGRβ mRNA, and β-actin mRNA. Representative amplification curves are shown (Fig. 1). From the exponential phase of the amplification curve, we measured a 1.5-fold increase in GRα mRNA and a slightly greater 2.0-fold increase in GRβ mRNA following TNF-α treatment. This small change in cellular GR mRNA levels is typical of the response of this promoter to other agents (32). We next evaluated whether TNF-α treatment altered the GRα and GRβ protein levels by immunocytochemistry. HeLaS3 cells were treated with or without TNF-α for 24 h by using well characterized antipeptide antibodies specific for GRα or GRβ. DEX was added during the last hour of treatment to promote the nuclear localization of GRα and facilitate its comparison with the constitutively nuclear GRβ isoform. As shown in Fig. 2A, both receptor isoforms increased in response to TNF-α; however, the increase in GRβ was greater than that observed for GRα. Quantitation of these data (Fig. 2B) indicates the intensity for GRα staining was 52.6 in the control cells and 97.1 in the TNF-α treated cells, representing a 1.8-fold increase in GRα protein. In contrast, the mean intensity for GRβ staining was 40.8 in control cells and 194 in TNF-α treated cells, representing a 4.8-fold increase in GRβ protein.
Figure 1.
GRβ-specific mRNA increases after cytokine treatment. Total RNA from HeLaS3 cells was isolated, and RT-PCR was performed on 0.5 μg total RNA per treatment group by using hGRα- and hGRβ-specific primers. Primers specific for actin were used as an internal control.
Figure 2.
Immunostaining of the GR isoforms by using isoform-specific epitope-purified polyclonal antibodies. (A) HeLaS3 cells were treated with 1 ng/ml TNF-α or vehicle for 24 h followed by treatment with 100 nM dexamethasone for one hour (TNF) or vehicle (CON). Cells were fixed and processed as described (14, 21, 23, 24). (B) Image analysis of the fluorescent staining was performed by using NIH Image 1.56. Mean peak intensities were calculated for each isoform with and without exposure to TNF.
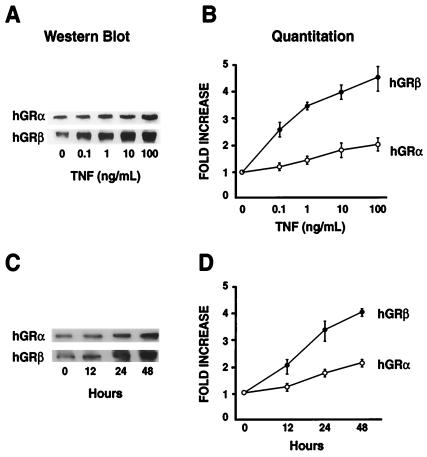
The effects of TNF-α on GRα and GRβ protein expression were further examined by Western blot analysis. We observed a modest 1.5-fold increase in GRα protein levels following a 24-h treatment with TNF-α (1 ng/ml), whereas GRβ increased by 3.5-fold. The preferential increase in GRβ was both dose and time dependent. The highest concentration of TNF-α tested produced a 2-fold increase in GRα and a 4.5-fold increase in GRβ (Fig. 3A). The longest duration of TNF-α treatment produced a 2-fold increase in GRα and a 4.0-fold increase in GRβ (Fig. 3B). Therefore, consistent with the immunocytochemical analysis, both GR isoforms increased following TNF-α treatment of HeLaS3 cells; however, GRβ accumulated in cells to a greater extent than GRα.
Figure 3.
Western analysis of GR isoform levels after cytokine treatment. (A) HeLaS3 cells were treated with increasing concentrations of TNF-α (0.1 to 100 ng/ml) for 24 h. The alpha-specific AShGR and beta-specific BShGR antibodies were used for Western analysis. (B) Quantitation of the Western blots showing the average (SEM) of three separate experiments. (C) HeLaS3 cells were treated with 1 ng/ml TNF-α for 0, 12, 24, or 48 h. Whole cell extracts were prepared and analyzed as described. (D) Quantitation of the results given as a mean (SEM) of three experiments.
Immune cells are often the primary mediators of inflammatory response and, as such, are subject to glucocorticoid regulation. Therefore, we evaluated whether the preferential increase in GRβ was also induced by TNF-α in cells of lymphoid origin. We used CEMC7 cells, a human lymphoid cell line known to express both GRα and GRβ (33). Immunoblots were performed with the AShGR and BShGR specific antibodies on CEMC7 cells treated with or without TNF-α (Fig. 4A). Similar to our findings in HeLaS3 cells, TNF-α treatment of the CEMC7 cells produced a 1.5-fold increase in GRα but a 4.0-fold increase in GRβ. To determine whether other activators of NF-κB also preferentially increased GRβ, we treated both HeLaS3 and CEMC7 cells with IL-1 (34) and evaluated the expression of GRα and GRβ. As shown in Fig. 4A, IL-1 treatment produced a modest 1.5-fold increase in GRα and a greater 4.0–5.5-fold induction in GRβ. Therefore, the preferential increase in GRβ is mediated by two different activators of NF-κB and occurs in cells of both epithelial and lymphoid origin.
Figure 4.
Preferential up-regulation of GRβ occurs in other cell types and with other cytokines. (A) HeLaS3 cells and CEMC7 cells were left untreated or treated with TNF-α (1 ng/ml) or IL-1 (10 ng/ml) for 24 h. Whole cell extracts were prepared and subjected to Western analysis by using AShGR and BShGR antibodies. (B) HeLaS3 cells were treated with TNF-α for 24 or 48 h or vehicle. Whole cell extracts were prepared and analyzed by Western blot with the anti-hGR antibody (no. 57) directed against an epitope common to both GRα and GRβ.
Because different antibodies were used to evaluate the relative changes in the levels of GRα and GRβ, it is impossible to determine the precise ratio of accumulated GRα and GRβ protein in these cells following proinflammatory insult by TNF-α. To address this issue, we evaluated the relative ratios of GRα and GRβ in HeLaS3 cells treated for 24 and 48 h with or without TNF-α and then immunoblotted by using an antibody directed toward a common epitope in each receptor isoform. The data in Fig. 4B show that the ratio of GRα to GRβ in untreated cells (CON) is ≈4:1, whereas after a 24-h treatment with TNF-α (24H TNF), the ratio is ≈1:1, and after treatment with TNF-α for 48 h (48H TNF), the ratio of GRα to GRβ is 1:2. These data conclusively show that TNF-α treatment of HeLa S3 cells results in a condition where GRβ becomes the predominant cellular receptor.
Mechanism of NF-κB-Mediated Regulation of GRα and GRβ Expression.
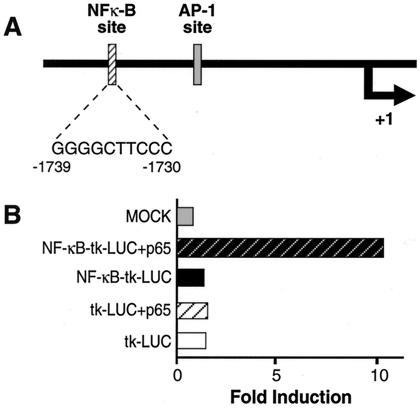
Activation of NF-κB leads to an increase in both GR isoforms; however, GRβ increases to a greater extent than GRα. To elucidate the mechanism(s) responsible for these changes in GR expression, we first searched the human GR promoter for potential NF-κB binding sites. An NF-κB consensus sequence, GGGGCTTCCC, was found between −1739 and −1730 relative to the transcription start-site (19). Evaluation of this putative NF-κB binding site was performed by placing it upstream of a thymidine kinase promoter driving the expression of a luciferase reporter. Coexpression of the p65 subunit of NF-κB resulted in a 6-fold increase in luciferase activity over that measured in cells expressing the pNF-κB-tk-LUC reporter alone (Fig. 5B). These results suggest that the NF-κB binding site found in the GR promoter is probably functional in vivo and indicates that the cytokine-mediated increase in both GRα and GRβ mRNA may be because of an NF-κB induced increase in transcription of the GR gene.
Figure 5.
Location of an NF-κB consensus sequence in the human glucocorticoid receptor promoter. (A) Location of the NF-κB consensus sequence relative to the transcription start site (+1) of the hGR promoter (2). The AP-1 site is also designated. Sequence of the NF-κB site is also shown. (B) COS1 cells were transfected with either pBR322 (MOCK) or the NF-κB consensus sequence linked to a heterologous thymidine kinase promoter driving expression of a luciferase reporter (NF-κB-tk-LUC) and p65 or NF-κB-tk-LUC alone or the tk-LUC plus p65 or tk-LUC alone. Luciferase assays represent the mean of three transfections.
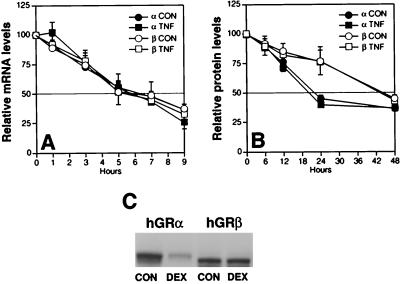
However, the effect of NF-κB on the GR promoter would be expected to increase both isoforms similarly, and although our data clearly show slightly more GRβ mRNA accumulating in cells, this small increase in GRβ mRNA over GRα mRNA probably cannot account for the increase in GRβ. This disproportionate increase in GRβ protein suggests that other components of the GRα and GRβ expression and catabolism pathways are involved. Therefore, we tested whether cytokine treatment differentially altered the stability of the GRα and GRβ mRNA and/or the GRα and GRβ protein. As shown in Fig. 6A, TNF-α treatment had no apparent effect on the stability of either the GRα or the GRβ mRNA. The mRNA half-life was ≈6 h for both GR isoforms in the absence and presence of TNF-α. Similarly, TNF-α treatment did not appear to effect the stability of the GRα and GRβ proteins (Fig. 6B). Interestingly, the half-life of the GRα protein at 24 h was consistent with previous findings in transfected cells (11), whereas the half-life of the GRβ protein was 48 h in the absence and presence of TNF-α. One property that determines the level of GRα is ligand-dependent down-regulation, which is known to reduce the half-life of GRα protein to 10 h (11). The ability of GRβ to undergo ligand-dependent down-regulation is unknown as it does not bind ligand. Comparison of the ability of transfected GRα and GRβ to undergo ligand-induced down-regulation in Fig. 6C clearly shows that GRα down-regulates in response to glucocorticoid (DEX), whereas GRβ does not.
Figure 6.
Half-life studies of GRα and GRβ mRNA and protein after TNF-α treatment, and down-regulation of receptor protein in response to glucocorticoids. (A) HeLaS3 cells were treated for 24 h with 1 ng/ml TNF-α then treated with actinomycin D for 1 h. mRNA was obtained at various time intervals, and RT-PCR by using alpha-isoform or beta-isoform specific primers was performed. (B) HeLaS3 cells were treated for 24 h with 1 ng/ml TNF-α then treated with cyclohexamide for 1 h. Whole cell extracts were obtained at various time intervals, and Western analysis was performed by using AShGR or BShGR antibodies. (C) COS1 cells were transfected with either hGRα or hGRβ. Sixteen hours posttransfection, the cells were treated with 100 nM DEX or vehicle (CON) for 24 h. Cell extracts were prepared, and GR protein levels were evaluated by Western analysis by using no. 57 antibody.
NF-κB-Mediated Regulation of GRα and GRβ Expression Produces Glucocorticoid Resistance.
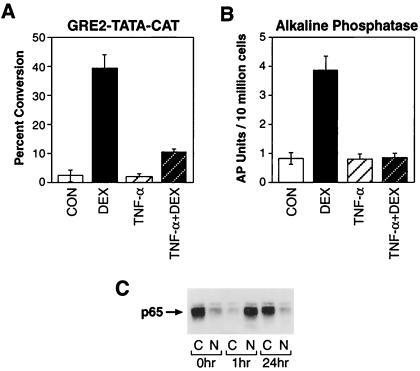
GRβ has been shown to act as a dominant negative by repressing the transcriptional activity of GRα (15). It has also been reported to be preferentially increased over GRα in patients suffering from both generalized and tissue-specific glucocorticoid resistances (18, 35–37). Therefore, we next evaluated whether the cytokine-mediated selective up-regulation of GRβ resulted in altered sensitivity to glucocorticoids. HeLaS3 cells were transfected with a glucocorticoid-responsive reporter plasmid. Treatment with DEX produced a 20-fold induction in CAT activity over that measured in untreated control cells (Fig. 7A) but only a 4-fold increase in CAT activity in cells pretreated with TNF-α. We also analyzed the effect of TNF-α on GRα-mediated activation of the endogenous glucocorticoid-responsive alkaline phosphatase activity. This endogenous response was also completely abolished in cells pretreated with TNF-α. Direct protein interaction and antagonism between p65 and GRα are unlikely to account for the inhibition of GRα activity because the nuclear level of p65 in TNF-α treated cells had returned to that found in control cells (Fig. 7C). These findings suggest that prolonged exposure to cytokines can produce glucocorticoid resistance in cells by mechanisms involving the cellular accumulation of the dominant negative GRβ protein. Unlike GRα, this receptor isoform is not susceptible to homologous down-regulation in the presence of hormone and persists in cells despite glucocorticoid administration.
Figure 7.
Hormone-mediated transactivation is abrogated after pretreatment with cytokine. (A) HeLaS3 cells were transiently transfected with the glucocorticoid-inducible reporter plasmid GRE2-TATA-CAT. Cells were treated for 24 h with vehicle alone (CON), 100 nM DEX, 1 ng/ml TNF-α, or 1 ng/ml TNF-α for 24 h plus 100 nM dexamethasone for 1 h (TNF-α + DEX). Cell extracts were prepared and assayed for CAT activity. (B) Endogenous alkaline phosphatase activity was analyzed by using the same treatment scheme as above. (C) Western analysis of nuclear and cytoplasmic levels of p65 after 0, 1, and 24 h of TNF-α treatment.
Discussion
The molecular processes whereby inflammatory responses are abrogated by glucocorticoids are poorly understood. There is evidence, however, that a primary signal for the inflammatory process to occur is mediated by the transcription factor NF-κB. The glucocorticoid receptor and NF-κB family members antagonize each other's transactivation functions in the nuclear compartment of the cell (4–6). Alternatively, it is possible that NF-κB-mediated changes in the levels of GRα and/or GRβ play a role in this mutual antagonism. We report here the discovery of an NF-κB consensus-binding element located in the 5′-flanking sequences of the hGR promoter and show that this element is functional. We examined whether activation of NF-κB by the proinflammatory cytokine TNF-α would have any direct effect on GR levels in a human cell line and found that there was a disproportionate increase in GRβ over GRα. The selective increase in the levels of the beta isoform was both dose and time dependent and occurred in both epithelial and lymphoid cells. Additionally, another proinflammatory cytokine (IL-1β), which also activates NF-κB, also disproportionately increased GRβ. To determine whether activation of NF-κB by proinflammatory cytokine signals would lead to an alteration in the levels of GRα- and GRβ-specific mRNAs, semiquantitative RT-PCR was used. Treatment with TNF-α resulted in a slight increase in GRβ-specific mRNA over GRα-specific mRNA that could only partially account for the large differences observed between the two receptor protein isoforms. Cytokines may regulate mRNA stability leading to a state whereby the alpha-specific message is destabilized or the beta-specific message is more stable after cytokine treatment; however, our studies do not support this possibility, as there was no apparent alteration in either isoform mRNA half-life when TNF-α was present. When we examined the protein half-life of each isoform, we again saw no differences between the half-life of each isoform when treated with cytokine. Interestingly, the beta isoform of the receptor had a much longer protein half-life than the alpha isoform, and, unlike GRα, GRβ was not subject to ligand-dependent homologous down-regulation. However, the mechanism responsible for the selective increase in GRβ protein remains to be established.
The most striking result was that the pretreatment of cells with a proinflammatory cytokine resulted in the abolition of hormone-mediated transactivation of a glucocorticoid-responsive gene by ligand. Expression from both an endogenous and a transiently transfected reporter were completely repressed after cytokine treatment when GRβ accumulated in the cells. Although we cannot completely rule out that a component of this resistance is mediated by a direct antagonism of GRα with NF-κB, it is clear that nuclear p65 had returned to control levels before our analysis. Previous studies (13–15) have shown in transient transfection systems that GRβ acts as a dominant negative regulator of GRα gene transactivation. In these studies, much higher levels of GRβ plasmid were required to achieve this dominant negative effect. However, it was impossible to discern the precise cellular ratio of GRα and GRβ. Our new data indicate that much lower levels of GRβ protein can achieve the dominant negative function previously observed. Recently, Pariante et al. (38) also demonstrated that another proinflammatory cytokine, IL-1α, was able to moderately attenuate dexamethasone action in mouse L929 cells. This observation suggests that proinflammatory cytokines can also affect glucocorticoid-mediated functions. Although some of the repression they observed might be because of direct physical antagonism between GRα and NF-κB, the increase in GRβ cannot be discounted because its action parallels the late effect glucocorticoids have on NF-κB by up-regulating the IκB inhibitor (7, 8). In this situation, both transcription families repress each other's function by both direct interactions and by increasing the availability of cognate repressors.
Chronic inflammation is a consequence of numerous disease states such as rheumatoid arthritis, systemic lupus erythematosus, and arteriosclerosis, which all progress with inflammatory components (39–41). Corticosteroids have long been used in treatment regimes for these diseases (41–46) as well as Alzheimer's disease in which an inflammatory response has been characterized (47–49). However, states of steroid resistance often develop after prolonged treatment with glucocorticoids (50–53). Unfortunately, the molecular mechanisms underlying the development of glucocorticoid resistance are poorly understood.
Our studies identify a possible molecular mechanism by which glucocorticoid resistance is achieved because of cumulative levels of a dominant negative GRβ isoform. Prolonged exposure to proinflammatory cytokines allows for large increases in the steady-state levels of the dominant negative beta isoform of the glucocorticoid receptor thwarting the action of GRα. This preferential increase in the beta isoform of the human glucocorticoid receptor has also been seen in disease states resistant to glucocorticoid therapy. Recently, there have been reports (17, 36, 54) on increases in GRβ levels in T cells in the airway, peripheral blood mononuclear cells, and in tuberculin-induced inflammatory lesions in glucocorticoid-insensitive asthmatics. In another report, higher levels of GRβ were found in 10 of 12 patients with glucocorticoid-resistant colitis (37). Additionally, Hauk et al. (55) have demonstrated that isolated peripheral blood mononuclear cells, when stimulated with various superantigens, became insensitive to glucocorticoids, and this insensitivity is believed to be the result of increased GRβ. These studies underscore the importance of the beta isoform in human disease states and suggest that a strong correlation exists between inflammation, the expression of GRβ, and resistance to glucocorticoids.
Abbreviations
- GR
glucocorticoid receptor
- hGR
human glucocorticoid receptor
- TNF
tumor necrosis factor
- RT
reverse transcription
- DEX
dexamethasone
- CAT
chloramphenicol acetyltransferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Baeuerle P A. Curr Biol. 1998;8:R19–R22. doi: 10.1016/s0960-9822(98)70010-7. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A S. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Sen R, Baltimore D. Cell. 1986;46:706–716. [Google Scholar]
- 4.Scheinman R I, Gualberto A, Jewell C M, Cidlowski J A, Baldwin A S. Mol Cell Biol. 1995;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldenhoven E, Liden J, Wissink S, Van de Stolpe A, Raaijimakers J, Koenderman L, Okret S, Gustafsson J A, Van der Saag P T. Mol Endocrinol. 1995;9:401–412. doi: 10.1210/mend.9.4.7659084. [DOI] [PubMed] [Google Scholar]
- 6.McKay L I, Cidlowski J A. Mol Endocrinol. 1998;12:45–56. doi: 10.1210/mend.12.1.0044. [DOI] [PubMed] [Google Scholar]
- 7.Auphan N, DiDonato J A, Rosette C, Helmberg A, Karin M. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 8.Scheinman R I, Cogswell P C, Lofquist A K, Baldwin A S. Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- 9.Burnstein K L, Jewell C M, Cidlowski J A. J Biol Chem. 1990;265:7284–7291. [PubMed] [Google Scholar]
- 10.Burnstein K L, Jewell C M, Sar M, Cidlowski J A. Mol Endocrinol. 1994;8:1764–1773. doi: 10.1210/mend.8.12.7708063. [DOI] [PubMed] [Google Scholar]
- 11.Webster J C, Jewell C M, Sar M, Bodwell J E, Munck A, Cidlowski J A. J Biol Chem. 1997;272:9287–9293. doi: 10.1074/jbc.272.14.9287. [DOI] [PubMed] [Google Scholar]
- 12.Hollenberg S M, Weinberger C, Ong E S, Cerelli G, Oro A, Lebo R, Thompson E B, Rosenfeld M G, Evans R M. Nature (London) 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oakley R H, Sar M, Cidlowski J A. J Biol Chem. 1996;271:9550–9559. doi: 10.1074/jbc.271.16.9550. [DOI] [PubMed] [Google Scholar]
- 14.Oakley R H, Webster J C, Sar M, Parker C R, Cidlowski J A. Endocrinology. 1997;138:5028–5038. doi: 10.1210/endo.138.11.5501. [DOI] [PubMed] [Google Scholar]
- 15.Oakley R H, Jewell C M, Yudt M R, Bofetiado D M, Cidlowski J A. J Biol Chem. 1999;274:27857–27866. doi: 10.1074/jbc.274.39.27857. [DOI] [PubMed] [Google Scholar]
- 16.Chrousos G P, de Castro M, Leung D Y, Webster E, Kino T, Bamberger C, Elliot S, Strakis C, Karl M. Am J Respir Crit Care Med. 1996;154:S39–S40. doi: 10.1164/ajrccm/154.2_Pt_2.S39. [DOI] [PubMed] [Google Scholar]
- 17.de Castro M, Elliot S, Kino T, Bamberger C, Karl M, Webster E, Chrousos G P. Mol Med. 1996;2:597–607. [PMC free article] [PubMed] [Google Scholar]
- 18.Leung D Y M, Hamid Q, Vottero A, Szefler S J, Surs W, Minshall E, Chrousos G P, Klemm D J. J Exp Med. 1997;186:1567–1574. doi: 10.1084/jem.186.9.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zong J, Ashraf J, Thompson E B. Mol Cell Biol. 1990;10:5580–5585. doi: 10.1128/mcb.10.10.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zabel U, Schreckand R, Baeuerle P A. J Biol Chem. 1991;266:252–260. [PubMed] [Google Scholar]
- 21.Oakley R H, Webster J C, Jewell C M, Sar M, Cidlowski J A. Steroids. 1999;64:742–751. doi: 10.1016/s0039-128x(99)00059-8. [DOI] [PubMed] [Google Scholar]
- 22.Allgood V E, Powell-Oliver F E, Cidlowski J A. J Biol Chem. 1990;265:12424–12433. [PubMed] [Google Scholar]
- 23.Jewell C M, Webster J C, Burnstein K L, Sar M, Bodwell J E, Cidlowski J A. J Steroid Biochem Mol Biol. 1995;55:135–146. doi: 10.1016/0960-0760(95)00174-x. [DOI] [PubMed] [Google Scholar]
- 24.Webster J C, Jewell C M, Sar M, Cidlowski J A. Endocrine J. 1994;2:967–969. [Google Scholar]
- 25.Tully D B, Cidlowski J A. Biochemistry. 1989;28:1968–1975. doi: 10.1021/bi00431a003. [DOI] [PubMed] [Google Scholar]
- 26.Fairbanks G, Steck T L, Wallach D F H. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 27.Silva C M, Tully D B, Petch L A, Jewell C M, Cidlowski J A. Proc Natl Acad Sci USA. 1987;84:1744–1748. doi: 10.1073/pnas.84.7.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;79:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allgood V E, Oakley R H, Cidlowski J A. J Biol Chem. 1993;268:20870–20876. [PubMed] [Google Scholar]
- 30.Gorman C. In: DNA Cloning: A Practical Approach. Glover D M, editor. III. Oxford: IRL; 1985. pp. 143–190. [Google Scholar]
- 31.Littlefield B A, Cidlowski J A. Endocrinology. 1984;114:566–575. doi: 10.1210/endo-114-2-566. [DOI] [PubMed] [Google Scholar]
- 32.Vig E, Barrett T J, Vadeckis W V. Mol Endocrinol. 1994;8:1336–1346. doi: 10.1210/mend.8.10.7854351. [DOI] [PubMed] [Google Scholar]
- 33.Harmon J M, Thompson E B. Mol Cell Biol. 1981;1:512–521. doi: 10.1128/mcb.1.6.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baeuerle P A, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 35.Shahidi H, Vottero A, Stratakis C A, Taymans M K, Longui C A, Chrousos G P, Daughaday W H, Gregory S A, Plate J M D. Biochem Biophys Res Commun. 1999;254:559–565. doi: 10.1006/bbrc.1998.9980. [DOI] [PubMed] [Google Scholar]
- 36.Sousa A R, Lane S J, Cidlowski J A, Staynov D Z, Lee T H. J Allergy Clin Immunol. 2000;105:943–950. doi: 10.1067/mai.2000.106486. [DOI] [PubMed] [Google Scholar]
- 37.Honda M, Orii F, Ayabe T, Imai S, Ashida T, Obara T, Kohgo Y. Gastroenterology. 2000;118:859–866. doi: 10.1016/s0016-5085(00)70172-7. [DOI] [PubMed] [Google Scholar]
- 38.Pariante C M, Pearce B D, Pisell T L, Sanchez C I, Po C, Su C, Miller A H. Endocrinology. 1999;140:4359–4366. doi: 10.1210/endo.140.9.6986. [DOI] [PubMed] [Google Scholar]
- 39.Hagihara H, Nomoto A, Mutoh S, Yamaguchi I, Ono T. Atherosclerosis. 1991;91:107–116. doi: 10.1016/0021-9150(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 40.Matsuda Y, Yamanaka H, Higami K, Kashiwazaki S. J Rheumatol. 1998;25:427–432. [PubMed] [Google Scholar]
- 41.Redford T W, Small R E. Am J Health Syst Pharm. 1995;52:2686–2695. doi: 10.1093/ajhp/52.23.2686. [DOI] [PubMed] [Google Scholar]
- 42.Asai K, Funaki C, Hayashi T, Yamada K, Naito M, Kuzuya M, Yoshida F, Yoshimine N, Kuzuya F. Arterioscler Thromb. 1993;13:892–899. doi: 10.1161/01.atv.13.6.892. [DOI] [PubMed] [Google Scholar]
- 43.Heytman M, Ahern M J, Smith M D, Roberts-Thomson PJ. J Rheumatol. 1994;21:435–441. [PubMed] [Google Scholar]
- 44.Macanovic M, Sinicropi D, Shak S, Baughman S, Thiru S, Lachmann P J. Clin Exp Immunol. 1996;106:243–252. doi: 10.1046/j.1365-2249.1996.d01-839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voisard R, Seitzer U, Baur R, Dartsch P C, Osterhues H, Hoher M, Hombach V. Int J Cardiol. 1994;43:257–267. doi: 10.1016/0167-5273(94)90206-2. [DOI] [PubMed] [Google Scholar]
- 46.Weisman M H. Curr Opin Rheumatol. 1995;7:183–190. doi: 10.1097/00002281-199505000-00005. [DOI] [PubMed] [Google Scholar]
- 47.Eikelenboom P, Zhan S S, van Gool W A, Allsop D. Trends Pharmacol Sci. 1994;15:447–450. doi: 10.1016/0165-6147(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 48.Kalaria R N, Harshbarger-Kelly M, Cohen D L, Premkumar D R. Neurobiol Aging. 1996;17:687–693. doi: 10.1016/0197-4580(96)00114-5. [DOI] [PubMed] [Google Scholar]
- 49.Breitner J C. Annu Rev Med. 1996;47:401–411. doi: 10.1146/annurev.med.47.1.401. [DOI] [PubMed] [Google Scholar]
- 50.Nijhuis E, Hinloopen B, van Duijn C, Hofman A, Rozing J, Nagelkerken L. Clin Immunol Immunopathol. 1994;73:45–52. doi: 10.1006/clin.1994.1168. [DOI] [PubMed] [Google Scholar]
- 51.Barnes P J. N Engl J Med. 1995;332:868–875. doi: 10.1056/NEJM199503303321307. [DOI] [PubMed] [Google Scholar]
- 52.Barnes P J, Greening A P, Crompton G K. Am J Respir Crit Care Med. 1995;152:S125–S142. doi: 10.1164/ajrccm/152.6_Pt_2.S125. [DOI] [PubMed] [Google Scholar]
- 53.Spahn J D, Leung D Y, Szefler S J. J Asthma. 1997;34:177–194. doi: 10.3109/02770909709068188. [DOI] [PubMed] [Google Scholar]
- 54.Hamid Q A, Wenzel S E, Hauk P J, Tsicopoulos A, Wallaert B, Lafitte J J, Chrousos G P, Szefler S J, Leung D Y. Am J Respir Crit Care Med. 1999;159:1600–1604. doi: 10.1164/ajrccm.159.5.9804131. [DOI] [PubMed] [Google Scholar]
- 55.Hauk P J, Hamid Q A, Chrousos G P, Leung D Y. J Allergy Clin Immunol. 2000;105:782–787. doi: 10.1067/mai.2000.105807. [DOI] [PubMed] [Google Scholar]