Abstract
The levels of genetic relatedness of 139 Stenotrophomonas maltophilia strains recovered from 105 hospitalized non-cystic fibrosis patients (51% from medical wards, 35% from intensive care units, and 14% from surgical wards) and 7 environmental sources in the same hospital setting during a 4-year period were typed by the pulsed-field gel electrophoresis (PFGE) technique. A total of 99 well-defined distinct XbaI PFGE patterns were identified (Simpson's discrimination index, 0.996). The dendrogram showed a Dice similarity coefficient ranging from 28 to 80%. Two major clusters (I and II), three minor clusters (III, IV, and V), and two independent branches were observed when using a 36% Dice coefficient, indicating a high diversity of genetic relatedness. It is of note that 84% of strains were grouped within two major clonal lineages. No special cluster gathering was found among strains belonging to the same sample type specimen, patients' infection or colonization status, and ward of precedence. Despite this fact, three different clones (A, B, and C) recovered from respiratory samples from six, three, and two patients, respectively, and two clones, D and E, in two bacteremic patients each, were identified. Isolation of an S. maltophilia strain belonging to the clone A profile in a bronchoscope demonstrated a common source from this clone. This study revealed a high genetic diversity of S. maltophilia isolates despite their origin from a single hospital, which may be related to the wide environmental distribution of this pathogen. However, few clones could be transmitted among different patients, yielding outbreak situations.
Stenotrophomonas maltophilia is found in a great variety of environmental sources, including plant tissues, water, soil, sediments, and organic residues. Moreover, it has been identified in the hospital environment and has been recognized in human specimens (8). As a result of this adaptation to different habitats, great metabolic heterogeneity has been found (3, 15). During the last few years, S. maltophilia has risen to prominence, causing a wide spectrum of hospital-acquired infections, particularly pneumonia and bacteremia in severely debilitated or immunosuppressed patients with chronic underlying diseases admitted to intensive care units (ICUs) (8, 9, 11, 17, 19, 20). This situation may have been facilitated by its resistance to most of the currently available broad-spectrum antimicrobials (4, 30) and its ability to rapidly increase their multiresistance phenotype (1, 13).
On the other hand, information about the genomic relationship within this species and the degree of relationship among nosocomial strains remains scarce (3, 15). Nowadays, pulsed-field gel electrophoresis (PFGE) is considered to be the reference genotyping method for S. maltophilia and has been used not only to identify outbreaks and possible reservoirs and modes of transmission involving this species but also to understand population structure from isolates from cystic fibrosis patients (26). In addition, this technique provides a broad look at the whole chromosome of the microorganisms and is useful for determining the genetic relatedness among isolates of a given species by comparison of their macrorestriction profiles (2). In a previous work, Berg et al. (3) determined the diversity of 40 clinical and environmental S. maltophilia isolates and analyzed phenotypic profiles and molecular types by several molecular methods. In their study, the most discriminatory method was PFGE under DraI digestion. In our study, we have determined the genetic relatedness and epidemiological links among 139 isolates recovered in the same hospital over a long period by using profiles generated by PFGE under XbaI restriction. Moreover, detection of cross-transmissions among different patients is also presented.
MATERIALS AND METHODS
Bacterial strains.
One hundred thirty-nine S. maltophilia isolates (132 from 105 non-cystic fibrosis hospitalized patients and 7 from different hospital environments) were collected from 1995 to 1998 at Ramón y Cajal Hospital, a 1,200-bed university teaching hospital in northeast Madrid, Spain. Clinical isolates included all isolates from bacteremic episodes and 30% of isolates from nonblood samples recovered in our institution during the period studied. Moreover, all S. maltophilia isolates within a specific period and unit were included when an outbreak situation was suspected. The isolation sites of clinical isolates were the respiratory tract (n = 79), blood (n = 19), wounds (n = 15), urine (n = 5), rectal swabs (n = 2), peritoneal fluids (n = 2), ocular prosthesis (n = 2), and other sites (n = 8). All of these strains were previously analyzed for antimicrobial susceptibility profile (30). A control group including 12 S. maltophilia isolates was selected for molecular typing. This group included 10 clinical isolates (4 respiratory tract isolates, 3 blood isolates, and 1 isolate each from urine, ascitic fluid, and drainage) recovered in different Spanish hospitals in different geographic areas, one isolate from a tuna sample, and the S. maltophilia ATCC 13637 strain. Biochemical identification was performed with both the API-20NE (bioMerieux, La Balme Les Grottes, France) and WIDER (Fco. Soria Melguizo, Madrid, Spain) systems.
PFGE.
Chromosomal DNA was prepared in agarose plugs as described previously (31). After digestion with endonuclease XbaI (30 U; Roche Diagnostics, Barcelona, Spain), restriction fragments were resolved by PFGE with the CHEF-DRII system (Bio-Rad, Hemel Hempstead, United Kingdom). A second enzyme, SpeI (20 U; Roche), was used to confirm identical restriction XbaI profiles. The electrophoresis conditions were as follows: (i) for XbaI, pulse times were ramped from 10 to 60 s over 24 h at 5.4 V/cm with a second ramp from 5 to 20 s over 5 h at 5.4 V/cm at 12°C (30); and (ii) for SpeI, pulse times ranged from 25 to 45 s over 20 h at 6.0 V/cm at 12°C (22). Standard lambda ladders of 48.5-kbp concatemers (Roche) were run as molecular weight markers. Restriction fragments were visually compared and interpreted according to previous established criteria (27). Isolates with identical profiles were considered to represent a single strain.
Computer-monitored fingerprinting and discriminatory power analysis.
Computer analysis of the PFGE banding patterns was performed with Bio-Rad Molecular Analyst software. The images analyzed included two reference lanes representing concatemer phage lambda ladders. Bands were automatically assigned by the computer and were manually corrected after observation in the computer screen. Only fragments exceeding 97.0 kbp were included in the analysis. Fingerprinting profiles were scored for the presence and absence of bands at given molecular weights, and strains that differed by one band were assigned different PFGE profiles, codified by a number. Only one profile was represented in the dendrogram for the isolates with identical XbaI profiles. The Dice correlation coefficient was used to analyze the similarities of the banding patterns (21). Strains with an identical PFGE banding pattern (100% similarity coefficient) were considered isogenic strains. Moreover, as previously stated (5, 31), closely related strains within the same clone were those with a similarity coefficient ranging from 80 to 95%. Clustering was based on the unweighted pair group method with arithmetic averages (UPGMA). The tolerance position was 1%.
The discriminatory power of PFGE between isolates from Ramón y Cajal hospital and isolates from the control group was evaluated by using Simpson's index of diversity (16), which expresses the probability that two unrelated strains will be placed in two different typing groups.
Patient clinical data.
For all patients, demographic information and the presence of repetitive S. maltophilia-positive cultures and any other organism in the same positive culture were recorded. Upon identification of an outbreak situation, defined by indistinguishable PFGE profiles of isolates, the medical charts of patients colonized or infected with the outbreak strains were retrospectively reviewed. Hospital-acquired infections were classified according to the Centers for Disease Control and Prevention definitions (12). Clinical infection or colonization status by S. maltophilia was considered according to clinical judgment, high or moderate bacterial counts, tissue invasion, repetitive S. maltophilia isolation, monomicrobial culture, and antibiotic treatment response. Respiratory infection was considered when evidence of pulmonary infiltrates with X rays, fever (>38°C), cough, and respiratory function deterioration was observed (18, 20, 29). Nosocomial acquisition was considered as being 72 h after hospital admissions or within 30 days of a surgical patient's discharge (12).
RESULTS
S. maltophilia macrorestriction profile analysis.
The PFGE conditions under XbaI restriction resolved DNA fragment sizes ranging from <48 kbp to >1,000 kb. Approximately 10 bands (>97.0 to about <1,000.0 kbp) were identified, and these were used for the scoring and computer analysis of S. maltophilia strains. The control typing group, including 12 strains from outside our clinical setting, displayed a Simpson′s index of 1 (12 different profiles) with a genetic similarity coefficient ranging from 34 to 75%. Well-defined different profiles were obtained for 99 of the 139 isolates recovered in our hospital, and the Simpson's discrimination index value was 0.996.
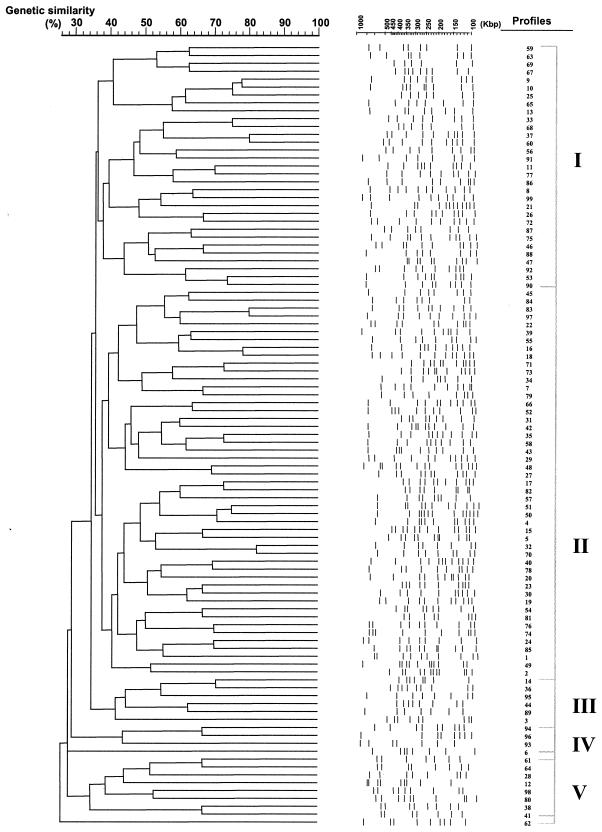
The dendrogram generated by computer-aided genotype analysis based on the unique PFGE patterns of all 139 S. maltophilia strains ranged from 28 to 80% similarity (Fig. 1). Two major clusters (designated clusters I and II, three minor clusters (III, IV, and V), and two independent branches (represented by profiles 6 and 62) were observed with a 36% similarity coefficient. The major clusters, I and II, included 37.7 and 38.5%, respectively, of the strains studied, whereas the minor clusters III, IV, and V only contained 3.6, 2.2, and 5.8%, respectively, of the strains studied. Profiles 6 and 62 did not fit into any of the five clusters even at similarity values of 28 and 26%, respectively. The origin of S. maltophilia clinical isolates for each defined XbaI profile is included in the legend to Fig. 1.
FIG. 1.
Phylogenetic analysis of digitized 99 PFGE XbaI profiles of 132 clinical and 7 environmental S. maltophilia isolates recovered in our hospital during a 4-year period. The dendrogram was constructed with PFGE data by similarity and clustering analysis by using the Dice coefficient and UPGMA with the Molecular Analysts software. A percent genetic similarity scale is shown above the dendrogram. Profile types are marked on the left in arabic numerals, and the clusters (cutoff value of 36% similarity) are marked on the right in roman numerals. The clinical origins of the isolates are as follows: respiratory specimens, profiles 1 to 51; blood, profiles 52 to 67; wounds, profiles 70 to 77; organic fluid, profiles 69 and 79; ocular specimens, profiles 68 and 78; urine, profiles 80 to 83; stool, profiles 84 and 85; environmental isolates, profiles 1, 86 to 91; and others, profiles 92 to 99.
Twenty-nine percent of isolates (40 of 139) showed an identical banding pattern to at least 1 other isolate. Repetitive isolates during the period studied were recovered in 18 patients: 14 patients presented two isolates each with identical profiles (100% similarity coefficient), and 4 patients presented three consecutive isolates each showing identical profiles (100% similarity coefficient). Persistence of S. maltophilia in these patients ranged from 2 to 25 days (8.5 ± 6.5 days).
It is of note that five isolates with different profiles (profiles 1, 4, 38, 52, and 61) were responsible for cross-transmission in 15 patients (see below). Profiles 1 (clone A), 4 (clone B), and 38 (clone C) were recovered from respiratory sources and were detected in six, three, and two patients, respectively. Moreover, profile 52 (clone D) was recovered from blood cultures in two patients, and profile 62 (clone E) was recovered from blood cultures in two patients. Strains belonging to these profiles represented different cross-infection situations among different patients and were associated with clusters II, II, V, and II and a nonclustered profile, respectively. These results were confirmed by SpeI restriction.
The degree of heterogeneity among control stains was also high. No new cluster or independent branches were observed when control strains were integrated in the dendrogram (data not shown).
Demographic data and patient characteristics.
One hundred thirty-two isolates were recovered from 105 patients. We had access to demographic data for 97 patients (61 male and 36 female). Thirty-five percent of patients (34 of 97) were hospitalized in different ICUs: Medical ICU (n = 15), Neurosurgical ICU (n = 6), Digestive Tract Surgical ICU (n = 7), Cardiovascular Surgery ICU (n = 3), and Pediatric ICU (n = 3). The other patients were from the Pulmonary Diseases (n = 12), Digestive Tract Surgery (n = 13), Infectious Diseases (n = 5), and Hematology (n = 4) Wards, while the rest of the patients were located in different units. S. maltophilia appeared as a monomicrobial culture in 13 of 19 (68.4%) blood cultures, 21 of 79 (26.6%) respiratory samples, 1 of 14 (7.1%) samples from wounds and cutaneous tissues, and 6 of 12 (50.0%) samples from other locations. In combination with other organisms, S. maltophilia appeared mainly associated with normal flora in respiratory samples (22%) and with gram-positive organisms (mainly coagulase-negative staphylococci) in blood (26%) and wound (61%) cultures.
Detection of cross-transmission.
A total of five episodes of cross-transmission were detected during the period studied: (i) three episodes involved three different S. maltophilia strains (clones A, B, and C) recovered from respiratory samples from 11 patients grouped as 6, 3, and 2 patients, and (ii) two episodes involved two different S. maltophilia strains (clones D and E) implicated in episodes of bacteremia in 4 patients. Risk factors, previous defined by others (7, 9, 11, 19) for S. maltophilia acquisition in all of these patients are shown in Table 1. Figure 2 shows progression of the respiratory outbreaks, the number of patients involved, the patients who underwent a bronchoscope procedure, and the patients' origin.
TABLE 1.
Comorbidities and potential risk factors for S. maltophilia infection and colonization in patients sharing similar clone typesa
| Patient (clone) | Ageb (gender) | Infection or colonization type or sitec | Ward | Central venous catheter | Mechanical ventilation | Bronchoscope procedure | Underlying illness | Previous hospitalization (< 6 mo) | Previous surgery (< 3 mo) | Previous (< 1 mo) antimicrobial therapy | Antimicrobial therapy | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (A) | 58 (M) | Respiratory (I) | ICU | Yes | Yes | Yes | Malignancy | Yes | Yes | AMX | CAZ + TOB | Death |
| 2 (A) | 82 (M) | Respiratory (I) | Thoracic Surgery | No | No | Yes | COPD | Yes | No | AMX | ERY | Recovered |
| 3 (A) | 24 (F) | Respiratory (C) | Pneumology | No | No | Yes | Tuberculosis | Yes | No | No | no | Recovered |
| 4 (A) | 76 (M) | Respiratory (C) | Outpatient | No | No | Yes | COPD | No | No | CRO | CIP | Recovered |
| 5 (A) | 76 (M) | Respiratory (C) | Outpatient | No | No | Yes | COPD, anemia | No | No | No | No | Death |
| 6 (A) | 60 (F) | Respiratory (I) | ICU | Yes | Yes | No | COPD, anemia neutropenia, hypertension | Yes | No | AMX + TOB | MER + VAN | Death |
| 7 (B) | 70 (M) | Respiratory (C) | Outpatient | No | No | Yes | Malignancy | No | No | No | No | Recovered |
| 8 (B) | 61 (F) | Respiratory (I) | ICU | Yes | Yes | Yes | Liver transplant | Yes | No | VAN + TOB + MER | CIP | Death |
| 9 (B) | 73 (M) | Respiratory (C) | ICU | No | No | Yes | COPD, traumatism | No | No | No | AMX | Recovered |
| 10 (C) | 75 (M) | Respiratory (I) | Vascular Surgery | No | Yes | No | Diabetes | Yes | Yes | CTX + TOB + MER | PIP-TAZ | Recovered |
| 11 (C) | 77 (M) | Respiratory (I) | ICU | Yes | No | No | COPD, heart failure | No | Yes | No | AMP + TOB | Death |
| 12 (D) | 18 (M) | Bacteremia (I) | ICU | Yes | No | No | Malignancy, neutropenia | No | No | VAN + FLZ | No | Recovered |
| 13 (D) | 56 (F) | Bacteremia (I) | Reumatology | Yes | No | No | Pleurocarditis, anemia | No | No | No | No | Recovered |
| 14 (E) | 60 (F) | Bacteremia (I) | ICU | Yes | Yes | No | Craniocerebral traumatism | Yes | No | No | No | Recovered |
| 15 (E) | 64 (F) | Bacteremia (I) | Hematology | Yes | Yes | No | Malignancy, neutropenia, diabetes | Yes | No | No | CAZ + VAN | Recovered |
M, male; F, female; AMX, amoxicillin-clavulanate; AMP, ampicillin; CAZ, ceftazidime; CIP, ciprofloxacin; CRO, ceftriaxone; CTX, cefotaxime; FLZ, fluconazol; MER, meropenem; PIP-TAZ, piperacillin-tazobactam; TOB, tobramycin; VAN, vancomycin; COPD, chronic obstructive pulmonary disease.
Ages are given in years.
Infection (I) or colonization (C) is indicated parenthetically.
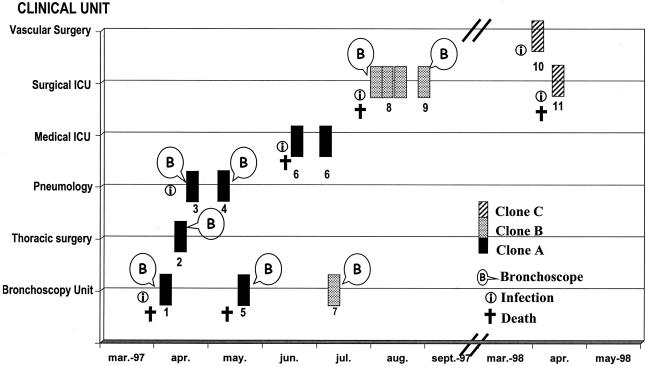
FIG. 2.
Progression of clones A, B, and C of S. maltophilia. Boxes represent one isolate each; the number below corresponds to the number of the patient. The time period, ward of precedence, bronchoscopy procedure, infection, and patient deaths are also indicated.
Seven isolates with an indistinguishable profile (clone A, profile 1) were recovered from respiratory sources from one outpatient (patient 5) and from five patients hospitalized in different wards (patients 1, 2, 3, 4, and 6) during a period of 84 days (Fig. 2). With the exception of patient 6, all patients had undergone a bronchoscope procedure. In patient 6, S. maltophilia was isolated twice during a period of 13 days. It is of note that as a result of an epidemiological investigation performed by our Hospital Infection Control Committee, an S. maltophilia strain with the same PFGE profile (profile 1) was cultured in a fiberbronchoscope in our Bronchoscopy Unit (Fig. 3). Cross-transmission of clone B was also demonstrated. Five strains with an indistinguishable pulse type (profile 4) were collected over a period of 47 days from an outpatient followed by the Pulmonary Diseases Unit (patient 7) and from two patients (patients 8 and 9) from the Digestive Tract Surgical ICU. S. maltophilia was isolated three times from a liver transplant recipient (patient 8) over an 11-day period as a monomicrobial culture. It is of note that a bronchoscope procedure was carried out in all patients, but epidemiological investigation failed to identify S. maltophilia in any of the fiberbronchoscopes used. Moreover, the same clone (clone C, profile 38) was observed in respiratory samples from two patients (patients 10 and 11) with a difference of 12 days in isolation. Neither of them had undergone a bronchoscope procedure, but both had undergone surgery. After the last patients positive for clones A, B and C, no further cases were detected.
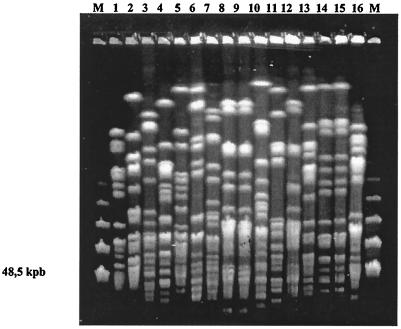
FIG. 3.
XbaI profiles obtained in S. maltophilia isolates from blood from 16 patients. Two episodes of cross-transmission were suspected (clone E, lanes 8 and 9, and clone D, lanes 14 and 15). Lane M, molecular size marker.
The same S. maltophilia clone (clone D, profile 52) was identified in two bacteremic patients (patients 12 and 13) (Table 1) hospitalized in the Pediatric ICU and in the Rheumatology Ward, respectively. Finally, the same clone (clone E, profile 61) was identified in blood cultures in two bacteremic patients (patients 14 and 15) (Table 1) hospitalized in the Neurosurgery ICU and the Hematology Ward.
DISCUSSION
Several multiresistant pathogens, including both gram-negative and gram-positive organisms, have been recognized during the last decades as nosocomial pathogens. Most of these organisms, such as Acinetobacter baumannii, Enterobacter spp., Klebsiella pneumoniae, or methicillin-resistant Staphylococus aureus, are generally found in epidemic or outbreak situations (10). In some institutions, these organisms have represented a clinical concern as a result of endemic isolation in hospitalized patients. Moreover, isolation of S. maltophilia in the nosocomial setting is not an infrequent situation, and different outbreaks have been identified (28). Until the present, few studies have focused on the genetic relatedness of S. maltophilia isolates, and most of them only included a small number of both environmental and nosocomial isolates (3, 7, 8, 20) or were focused on cystic fibrosis isolates (5, 31). In the present work, we studied a total of 139 S. maltophilia isolates from non-cystic fibrosis patients and recovered in the same hospital during a 4-year period and investigated their genetic relatedness.
The low interstrain variability of phenotypic typing methods in S. maltophilia (23, 34) makes molecular techniques, particularly PFGE, widely accepted methods for epidemiological typing of this species (3, 20, 26). As previously demonstrated in S. maltophilia, PFGE under XbaI restriction is appropriate for tracing isolates and generating stable profiles in long-term-colonized patients. In the present study, the discriminatory ability of the XbaI PFGE technique reached a Simpson coefficient value of 1.0, allowing us to establish the molecular relationships among clinical S. maltophilia isolates recovered in a single hospital. Despite being from the same institution, the majority of strains tested (71.7%) displayed different PFGE profiles, and only identical genotypic patterns were observed in isolates recovered from 15 patients (14.3%), suggesting a common source of these strains. Moreover, the PFGE fingerprinting analysis for the strains tested revealed a great discriminatory ability, resulting in a Simpson coefficient value of 0.996. Berg et al. (3), using PFGE under DraI restriction, also demonstrated a high intraspecific diversity in S. maltophilia clinical and environmental isolates from different locations and niches. A similar conclusion was obtained with arbitrarily primed PCR (32), ERIC-PCR (6), randomly amplified polymorphic DNA (35), and amplified-fragment length polymorphism (15) techniques in S. maltophilia isolates from different environments. However, the high intraspecific diversity decreased, because closely related strains could be detected in chronically colonized cystic fibrosis patients with this pathogen (5).
It is of note that unlike isolates from cystic fibrosis patients (5, 31), isolates serially recovered from the same patient showed identical PFGE profiles (100% similarity). In a previous work from our group (31), which included only isolates from cystic fibrosis patients, the presence of strains with a similarity coefficient ranging from 80 to 95% was observed within the same patient. This fact represents the dynamic situation of S. maltophilia isolates in the pulmonary environment in cystic fibrosis patients, which may persist for more than 6 years (31). On the contrary, nosocomial patients present a lower length of persistence of S. maltophilia isolates (8.5 ± 6.5 days in the present study).
On the basis of XbaI PFGE profiles, 99 distinct profiles were identified among 139 isolates studied. Isolates were grouped into five phylogenetic clusters when a cutoff of 36.0% in the genetic similarity scale was considered. The resolved profiles showed a great genetic distance, and the genetic diversity extended from low (28.0%) to high (80.0%) similarity. These results clearly demonstrated the low homogeneity level of S. maltophilia strains, irrespective of the time frame of collection in the same clinical setting. Interestingly, the major clonal lineages (I and II) were nearly grouped 84.0%% of the strains analyzed.
As previously noted, prolonged hospitalization and broad-spectrum antimicrobial therapy may facilitate the selection of S. maltophilia from respiratory or gastrointestinal locations (32, 33). In our study, the great number of distinct S. maltophilia profiles may reflect a wide environmental distribution of this species, allowing acquisition from different environmental sources. Moreover, S. maltophilia strains from different patients may have been acquired independently, discarding the presence of specific nosocomial clones. In S. maltophilia, the epidemiological relationship among different isolates needs to be analyzed, because unexpected results can be obtained. This was the case in a Croatian hospital in which, over a 4-month period (28), nine different profiles were observed in S. maltophilia isolates recovered from 20 patients. Six of these profiles were observed in different groups, including up to four hospitalized patients in the same or different units. Moreover, in an Italian university hospital, an epidemiological investigation of ICU patients revealed that although most patients were infected or colonized by different S. maltophilia clones, strains with identical genotypes were isolated, and two separate outbreaks were identified (7). Similarly, in our investigation, three consecutive respiratory outbreaks were detected in an approximately 1-year period. As previously noted, the acquisition of S. maltophilia isolates could be due to defective sterilization of the bronchoscope rather than dissemination of the organisms from other environmental sources or between patients. This hypothesis was reinforced when an S. maltophilia isolate with an indistinguishable profile from clone A was isolated in a fiberbronchoscope. However, in other clones, the sources and vehicles of infection could not be detected, as has occurred in most S. maltophilia outbreaks (8). Recently, an outbreak of Pseudomonas aeruginosa infections after bronchoscopic procedures was demonstrated (25), but to our knowledge, the involvement of these procedures as a potential source for S. maltophilia transmission has not been published. In contrast, faucet aerators and contaminated faucet water, electronic ventilator temperature sensors, and disinfectants have been recognized as contributing to disseminate epidemic S. maltophilia isolates (8).
In general, S. maltophilia is recovered from mixed cultures, particularly from respiratory tract and skin and soft tissue infections, which makes it difficult to establish an unequivocal role of this organism as a pathogen (8). In our study, S. maltophilia appeared as a single etiological agent in 26.6% of respiratory samples and 7.1% of wounds. This value reached 68.4% in blood cultures, similar to that found by Jang et al. (17). The pathogenicity of S. maltophilia is still controversial; however, in a systematic retrospective case control study excluding polymicrobial bacteremia, the mortality rate for S. maltophilia (26.7%) was similar to that observed for other nosocomial pathogens causing bloodstream infections (24). As in other clinical studies (14), most of our S. maltophilia isolates (79 of 139) were recovered from respiratory specimens. However, clinical evidence of true infection was difficult to determine. Previous risk factors for S. maltophilia pneumonia have been reported (29, 32, 33), including neutropenia, immunosuppression, use of H2 antagonists, previous antibiotic exposure, hospitalization and surgery, catheterization, mechanical ventilation support, prolonged hospitalization, and ICU stay (7-9, 11, 19). Most of these risk factors were observed in our 11 patients involved in respiratory cross-transmission (Table 1).
In conclusion, our study revealed a high genetic diversity among S. maltophilia isolates despite their origin in a single hospital. This result may be related to the high potential environmental distribution of this pathogen. However, a few clones could have been transmitted among different patients, producing outbreaks and epidemic situations. Transmission of S. maltophilia isolates was in fact demonstrated in six patients after the use of a bronchoscope device. Molecular typing investigations of S. maltophilia isolates are useful for control strategies to decrease infections due to this emerging pathogen.
Acknowledgments
This work was supported by a research grant from Comunidad de Madrid (2114/1998), Spain, and was partially supported by the Microbial Sciences Foundation, Madrid, Spain.
REFERENCES
- 1.Alonso, A., and J. L. Martínez. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1140-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field gel electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. [DOI] [PubMed] [Google Scholar]
- 3.Berg, G., N. Roskot, and K. Smalla. 1999. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 37:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betriu, C., A. Sánchez, M. L. Palau, M. Gómez, and J. J. Picazo. 2001. Antibiotic resistance surveillance of Stenotrophomonas maltophilia, 1993-1999. J. Antimicrob. Chemother. 48:152-154. [DOI] [PubMed] [Google Scholar]
- 5.Cantón, R., S. Valdezate, A. Vindel, B. Sánchez del Salz, L. Máiz, and F. Baquero. 2003. Antimicrobial susceptibility profile of molecular typed cystic fibrosis Stenotrophomonas maltophilia isolates and differences with noncystic fibrosis isolates. Pediatr. Pulmonol. 35:99-107. [DOI] [PubMed] [Google Scholar]
- 6.Chatelut, M., J. L. Dournes, G. Chabanon, and N. Marty. 1995. Epidemiological typing of Stenotrophomonas (Xanthomonas) maltophilia by PCR. J. Clin. Microbiol. 33:912-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crispino, M., M. C. Boccia, M. Bagattini, P. Villari, M. Triassi, and R. Zarrilli. 2002. Molecular epidemiology of Stenotrophomonas maltophilia in a university hospital. J. Hosp. Infect. 52:88-92. [DOI] [PubMed] [Google Scholar]
- 8.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11:57-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elting, L. S., N. Khardori, G. P. Bodey, and V. Fainstein. 1990. Nosocomial infection caused by Xanthomonas species and non-aeruginosa pseudomonas species: increasing incidence of catheter-related infections. Medicine 69:296-306. [DOI] [PubMed] [Google Scholar]
- 10.Fridkin, S. K. 2001. Increasing prevalence of antimicrobial resistance in intensive care units. Crit. Care Med. 29(Suppl. 4):N64-N68. [DOI] [PubMed] [Google Scholar]
- 11.Gales, A. C., R. N. Jones, K. R. Forward, J. Liñares, H. S. Sader, and J. Verhoef. 2001. Emerging importance of multidrug resistance Acinetobacter species and Stenotrophomonas maltophilia as pathogen in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY antimicrobial surveillance program (1997-1999). Clin. Infect. Dis. 32:S104-S113. [DOI] [PubMed] [Google Scholar]
- 12.Garner, J. S., R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections. Am. J. Infect. Control 16:128-140. [DOI] [PubMed] [Google Scholar]
- 13.Garrison, M. W., D. E. Anderson, D. M. Campbell, K. C. Carroll, C. L. Malone, J. D. Anderson, R. J. Hollis, and M. A. Pfaller. 1996. Stenotrophomonas maltophilia: emergence of multidrug-resistant strains during therapy and in an in vitro pharmacodynamic chamber model. Antimicrob. Agents Chemother. 40:2859-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gopalakrishnan, R., H. B. Hawley, J. S. Czachor, R. J. Markert, and J. M. Bernstein. 1999. Stenotrophomonas maltophilia infection and colonization in the intensive care units of two community hospitals: a study of 143 patients. Heart Lung 28:134-141. [DOI] [PubMed] [Google Scholar]
- 15.Hauben, L., L. Vauterin, E. R. B. Moore, B. Hoste, and J. Swings. 1999. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 49:1749-1760. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang, T. N., F. D. Wang, L. S. Wang, C. Y. Liu, and I. M. Liu. 1992. Xanthomonas maltophilia bacteremia: an analysis of 32 cases. J. Formos. Med. Assoc. 91:1170-1176. [PubMed] [Google Scholar]
- 18.Julve, R., E. Rovire, A. Belda, J. Prat, R. Escoms, A. Albert, and F. Gonzalvo. 1998. Clinical manifestations of Stenotrophomonas (Xanthomonas) maltophilia infections. An. Med. Interna 15:476-480. [PubMed] [Google Scholar]
- 19.Khardori, N., L. Elting, E. Wong, B. Schable, and G. P. Bodey. 1990. Nosocomial infections due to Xanthomonas maltophilia (Pseudomonas maltophilia) in patients with cancer. Rev. Infect. Dis. 12:997-1003. [DOI] [PubMed] [Google Scholar]
- 20.Laing, F. P. Y., K. Ramotar, R. R. Read, N. Alfieri, A. Kureishi, E. A. Henderson, and T. J. Louie. 1995. Molecular epidemiology of Xanthomonas maltophilia colonization and infection in the hospital environment. J. Clin. Microbiol. 33:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sader, H. S., A. C. Pignatari, R. Frei, R. J. Hollis, and R. N. Jones. 1994. Pulsed-field gel electrophoresis of restriction-digested genomic DNA and antimicrobial susceptibility of Xanthomonas maltophilia strains from Brazil, Switzerland and the USA. J. Antimicrob. Chemother. 33:615-618. [DOI] [PubMed] [Google Scholar]
- 23.Schable, B., D. L. Rhoden, W. R. Jarvis, and J. M. Miller. 1992. Prevalence of serotypes of Xanthomonas maltophilia from world-wide sources. Epidemiol. Infect. 108:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senol, E., J. DesJardin, P. C. Stark, L. Barefoot, and R. Snydman. 2002. Attributable mortality of Stenotrophomonas maltophilia bacteremia. Clin. Infect. Dis. 34:1653-1656. [DOI] [PubMed] [Google Scholar]
- 25.Srinivasan, A., L. L. Wolfenden, X. Song, K. Mackie, T. L. Hartsell, H. D. Jones, G. B. Diette, J. B. Orens, R. C. Yung, T. L. Ross, W. Merz, P. J. Scheel, E. F. Haponik, and T. M. Perl. 2003. An outbreak of Pseudomonas aeruginosa infections associated with flexible bronchoscopes. N. Engl. J. Med. 348:221-227. [DOI] [PubMed] [Google Scholar]
- 26.Talon, D., P. Bailly, R. Leprat, C. E. Deconnink, J. Y. Cahn, and Y. Michel-Briand. 1994. Typing of hospital strains of Xanthomonas maltophilia by pulsed-field gel electrophoresis. J. Hosp. Infect. 27:209-217. [DOI] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripkovic, V., M. Müller-Premru, S. Kalenic, V. Plecko, I. Jelic, B. Filipovic-Grcic, and M. Jandrlic. 2001. Clustering of infections caused by different PFGE types of Stenotrophomonas maltophilia occurring simultaneously in a university hospital. J. Hosp. Infect. 47:333-335. [DOI] [PubMed] [Google Scholar]
- 29.Ubeda, P., M. Salavert, S. Giner, I. Jarque, J. López-Alderguer, C. Pérez-Beller, and M. Gobernado. 1998. Bacteremia from Stenotrophomonas maltophilia: clinical epidemiological study and resistance profile. Rev. Esp. Quimioter. 11:206-215. [PubMed] [Google Scholar]
- 30.Valdezate, S., A. Vindel, E. Loza, F. Baquero, and R. Cantón. 2001. Antimicrobial susceptibility of unique Stenotrophomonas maltophilia clinical strains. Antimicrob. Agents Chemother. 45:1581-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdezate, S., A. Vindel, L. Máiz, F. Baquero, H. Escobar, and R. Cantón. 2001. Persistence and variability of Stenotrophomonas maltophilia in cystic fibrosis patients, Madrid, 1991-1998. Emerg. Infect. Dis. 7:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Couwenberghe, C. J., S. H. Cohen, Y. J. Tang, P. H. Gumerlock, and J. Silva. 1995. Genomic fingerprints of epidemic and endemic strains of Stenotrophomonas maltophilia (formerly Xanthomonas) by arbitrarily primed PCR. J. Clin. Microbiol. 33:1289-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villarino, M. E., L. E. Stevens, B. Schable, G. Mayers, J. M. Miller, J. P. Burke, and W. R. Jarvis. 1992. Risk factors for epidemic Xanthomonas maltophilia infection/colonization in intensive care unit patients. Infect. Control Hosp. Epidemiol. 13:201-206. [DOI] [PubMed] [Google Scholar]
- 34.Yang, P., L. Vauterin, M. Vancanneyt, J. Swings, and K. Kersters. 1993. Application of fatty acid methyl esters for the taxonomic analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 16:47-71. [Google Scholar]
- 35.Yao, J. D. C., J. M. Conly, and M. Krajden. 1995. Molecular typing of Stenotrophomonas (Xanthomonas) maltophilia by DNA macrorestriction analysis and random amplified polymorphic DNA analysis. J. Clin. Microbiol. 33:2195-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]