Abstract
Background
Evidence indicates that CYP1A1 MspI polymorphism might be a possible risk factor for several malignancies. A growing body of literature has been devoted to the association of CYP1A1 MspI polymorphism with acute myeloid leukemia (AML). However, the results remain conflicting. The aim of the present study was to derive a more precise estimation of the relationship.
Methods
Meta-analyses assessing the association of CYP1A1 MspI variation with AML were conducted and subgroup analyses on ethnicity and age groups were further performed. Eligible studies were identified for the period up to May 2012.
Results
A total of ten case–control studies including 1330 cases and 3688 controls were selected for analysis. The overall data failed to indicate a significant association of CYP1A1 MspI polymorphism with AML risk (C vs T: OR = 1.13; 95%CI = 0.87-1.48; CC vs TT: OR = 1.72; 95%CI = 0.99-3.01; CC + TC vs TT: OR = 1.16; 95%CI = 0.86-1.55). In subgroup analysis stratified by ethnicity, significant AML risk was shown among Asians (CC + TC vs TT: OR = 1.33; 95%CI = 1.09-1.62) but not Caucasians or mixed races. In subgroup analysis regarding age groups, no associations were observed in either the childhood AML or the adult AML subgroups.
Conclusion
The results of the present study suggested that CYP1A1 MspI polymorphism might be a risk factor for AML among Asians. Further investigations are needed to confirm the conclusions.
Keywords: CYP1A1 MspI, Acute myeloid leukemia, Malignancy, Susceptibility, Meta-analysis, Polymorphism
Introduction
Acute myeloid leukemia (AML), also known as acute nonlymphocytic leukemia (ANLL), is the most common acute leukemia mostly affecting adults, characterized by the rapid growth of abnormal white blood cells in the bone marrow and impaired production of normal blood cells. The mechanisms for AML genesis are still rarely understood. Evidence suggests that radiation, smoking, obesity and exposure to chemical carcinogens are considered as its possible risk factors [1]. Nevertheless, AML only develops in a small proportion of people exposed to these environmental and lifestyle risk factors, indicating that the host genetic background might play a critical role in its genesis.
Several genetic polymorphisms have been determined as possible risk factors for leukemia by meta-analyses. Variations of GSTM1, GSTT1, MTHFR C677T and XRCC1 Arg399Gln have been indicated to raise leukemia susceptibility [2-4]. Nevertheless, polymorphic MTR A2756G has been shown to decrease acute leukemia risk [5]. Therefore, different genetic polymorphisms might exert different effects on leukemia risk. Nevertheless, only a few gene polymorphisms associated with leukemia susceptibility have been identified to date.
Recent evidence indicates that carcinogen-metabolizing genes might play critical roles in determining individual susceptibility to cancers [6]. Susceptibility to cancer is determined by the activation of enzymes involved in carcinogen activation or deactivation. Polymorphisms in these genes encoding the enzymes, possibly by altering their functions, might increase or decrease carcinogen activation/detoxification and modulate DNA repair process. Cytochrome P450 (CYP) enzymes catalyze Phase I metabolism reaction. Cytochrome P450 1A1 (CYP1A1) is a member of the CYP family that participates in the metabolism of xenobiotics and endogenous compounds, particularly polycyclic aromatic hydrocarbons (PAHs) such as benzo[a]pyrene in smoke [7]. A commonly studied single nucleotide polymorphism (SNP) in the CYP1A1 gene has been indicated to associate with cancer susceptibility. The SNP locates at nucleotide 3801 in the 3’ non-coding region containing a single T to C base substitution that results in a polymorphic restriction site for the MspI enzyme (MspI or CYP1A1*2A polymorphism, rs4646903). The MspI restriction site polymorphism results in three genotypes: a predominant homozygous m1 allele without the MspI site (type A, TT), the heterozygote (type B, TC) and a homozygous rare m2 allele with the MspI site (type C, CC) [8].
Published studies devoted to the relationship between CYP1A1 MspI polymorphism and AML risk have generated controversial results. The issue of whether CYP1A1 MspI polymorphism is a risk factor for AML remains uncertain. Therefore, in this study, we aimed to perform a quantitative meta-analysis that increased statistical power to generate more confidential results.
Materials and methods
Literature search strategy
We carried out a search in the Medline, EMBASE, OVID, Sciencedirect, and Chinese National Knowledge Infrastructure (CNKI) without a language limitation, covering all publications published up to May 2012, with a combination of the following keywords: Cytochrome P450 1A1, CYP1A1, T3801C, MspI, acute myeloid leukemia, acute nonlymphocytic leukemia, hematology,malignancy, neoplasm, cancer, variation and polymorphism. All searched studies were retrieved and the bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand searched to find additional eligible studies.
Inclusion and exclusion criteria
The following criteria were used for the literature selection: first, studies should concern the association of CYP1A1 MspI polymorphism with AML risk; second, studies must be observational studies (Case—control or cohort); third, papers must offer the size of the sample, odds ratios (ORs) and their 95% confidence intervals (CIs), the genetic distribution or the information that can help infer the results. Accordingly, the following criteria for exclusion were also utilized: first, the design and the definition of the experiments were obviously different from those of the selected articles; second, the source of cases and controls and other essential information were not offered; third, reviews and duplicated publications. After deliberate searching, we reviewed all papers in accordance with the criteria defined above for further analysis.
Data extraction
Data were carefully extracted from all eligible publications independently by two of the authors according to the inclusion criteria mentioned above. For conflicting evaluations, an agreement was reached following a discussion. If a consensus could not be reached, another author was consulted to resolve the dispute and then a final decision was made by the majority of the votes. The extracted information was entered into a database.
Statistical analysis
The odds ratio (OR) of CYP1A1 MspI polymorphisms and AML risk was estimated for each study. The pooled ORs were performed for an allelic contrast (C allele versus T allele), a homozygote comparison (CC versus TT) and a dominant model (CC + TC versus TT). For detection of any possible sample size biases, the OR and its 95% confidence interval (CI) to each study was plotted against the number of participants respectively. A Chi-square based Q statistic test was performed to assess heterogeneity. If the result of the Q-test was P >0.1, ORs were pooled according to the fixed-effect model (Mantel-Haenszel); otherwise, the random-effect model (DerSimonian and laird) was used. The significance of the pooled ORs was determined by Z-test. The Hardy-Weinberg equilibrium (HWE) was assessed by Fisher’s exact test. Publication bias was assessed by visual inspection of funnel plots [9], in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot indicates a possible publication bias. The symmetry of the funnel plot was further evaluated by Egger’s linear regression test [10]. Statistical analysis was undertaken using the program STATA 11.0 software (Stata Corporation, Texas).
Results
Study characteristics
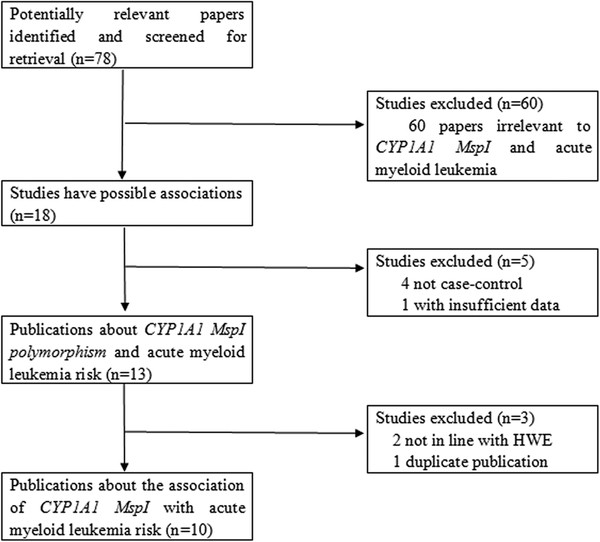
Relevant publications were retrieved and screened originally. A total of seventy-eight publications were identified, of which sixty irrelevant papers were excluded. As shown in Figure1, eighteen publications were preliminary eligible, of which four publications not being case–control studies [11-14] and one article not presenting sufficient information [15] were discarded. Next, two studies [16,17] whose genetic distributions of the control groups exhibited evident deviation from HWE were excluded. Then, one duplicate publication [18] which concerned the same research with one of the included studies [19] was further excluded. Lastly, ten case–control studies were selected for data extraction [19-28].
Figure 1 .
The flow diagram of included/excluded studies.
Of the selected publications, one was written in Chinese [24] while the remaining nine were in English. The relevant information was listed in Table1. According to this table, the first author and the number and characteristics of cases and controls for each study as well as other necessary information are presented.
Table 1.
Characteristics of studies included in the meta-analysis
| First Author | Publication Year | Number of Leukemia Cases (male/female) | Number of Controls (male/female) | Number of AML cases | Type of controls | Median (or mean) age, (range) year (Cases/Controls) | Racial decent | Country |
|---|---|---|---|---|---|---|---|---|
| Balta |
2003 |
33 (19/14) |
185 (120/65) |
33 AML |
Healthy controls (PB) |
8.7(1–17)/7.4(0.58-17) |
Mixed |
Turkey |
| D'Alo |
2004 |
193 (107/86) |
273(147/126) |
193 AML |
Healthy controls (PB) |
62(19–87)/60(19–90) |
Caucasian |
Italy |
| Clavel |
2005 |
219 (129/90) |
105 (57/48) |
28 AML |
Non-cancer controls (age,- gender-, hospital-, ethnicity-matched; HB) |
NA(0–15)/NA(0–15) |
Mixed |
France |
| Aydin-Sayitoglu |
2006 |
249 (143/106) |
140 (73/67) |
50 adult AML; 44 pediatric AML |
Healthy controls (PB) |
Adult:33(19–75); pediatric: 7.8(2–18)/28.7(16–59) |
Caucasian |
Turkey |
| Bolufer |
2007 |
443 (223/190) |
454 (223/231) |
302 AML |
Healthy controls (PB) |
39.48(0.8-84)/38.38(1–85) |
Caucasian |
Spain |
| Jiang |
2008 |
98 (NA) |
120 (NA) |
98 AML |
Healthy controls (age-, sex-matched; PB) |
NA(16–68)/NA(16–68) |
Asian |
China |
| Majumdar |
2008 |
110 (70/40) |
126 (54/72) |
110 AML |
Healthy controls (PB) |
35(4–81)/30(8–73) |
Asian |
India |
| Yamaguti |
2009 |
133 (70/63) |
133 (70/63) |
133 AML |
Healthy controls (PB) |
47(11–89)/53(25–60) |
Mixed |
Brazil |
| Bonaventure |
2012 |
493 (266/227) |
549 (292/257) |
51 AML |
Healthy controls (age-, gender-matched; PB) |
NA(0–15)/NA(0–15) |
Caucasian |
France |
| Kim | 2012 | 415 (223/192) | 1700 (821/879) | 415 AML | Healthy controls (PB) | 50.5(15–86)/52.2(20–74) | Asian | Korea |
NA: not available; AML, acute myeloid leukemia; PB: population-based; HB: hospital-based.
There were four groups of Caucasians [19,21,25,26], three of Asians [20,23,24] and three of mixed races [22,27,28] in this meta-analysis. As for age groups, there were seven groups of adult AML [19,20,22-26] and four groups of childhood AML [21,25,27,28] in this study. Noticeably, the study conducted by Aydin-Sayitoglu et al… [25] involved two subgroups regarding adult AML and childhood AML, respectively.
The distributions of CYP1A1 MspI genotype as well as the genotyping methods of the included studies are presented in Table2. The genetic distributions of the control groups in all included studies were consistent with HWE.
Table 2.
Distribution of CYP1A1 MspI genotypes among acute myeloid leukemia cases and controls included in the meta-analysis
|
First Author |
Year |
Genotyping method |
Cases |
Controls |
HWE (control) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | TC | TT | CC | TC | TT | Chi-squre | P | |||
| Balta |
2003 |
PCR-RFLP |
0 |
6 |
20 |
7 |
35 |
103 |
2.862 |
> 0.05 |
| D'Alo |
2004 |
PCR-RFLP |
0 |
17 |
161 |
0 |
42 |
226 |
1.937 |
> 0.05 |
| Clavel |
2005 |
PCR-RFLP |
0 |
5 |
22 |
0 |
24 |
81 |
1.748 |
> 0.05 |
| Aydin-Sayitoglu |
2006 |
PCR-RFLP |
5 |
24 |
65 |
4 |
30 |
106 |
1.049 |
> 0.05 |
| Bolufer |
2007 |
Real-time PCR |
0 |
31 |
168 |
2 |
84 |
317 |
2.062 |
> 0.05 |
| Jiang |
2008 |
PCR-RFLP |
19 |
50 |
29 |
26 |
50 |
44 |
2.610 |
> 0.05 |
| Majumdar |
2008 |
PCR-RFLP |
30 |
39 |
41 |
9 |
51 |
66 |
0.040 |
> 0.05 |
| Yamaguti |
2009 |
PCR-RFLP |
9 |
59 |
65 |
6 |
32 |
95 |
2.199 |
> 0.05 |
| Bonaventure |
2012 |
Infinium platform |
2 |
7 |
41 |
7 |
87 |
454 |
1.435 |
> 0.05 |
| Kim | 2012 | PCR-RFLP | 61 | 219 | 135 | 263 | 801 | 636 | 0.170 | > 0.05 |
Test of heterogeneity
As shown in Table3, we analyzed the heterogeneity for the allelic contrast (C allele versus T allele), homozygote comparison (CC versus TT) and dominant model (CC + TC versus TT), respectively. Evident heterogeneities were observed for the overall data in the three genetic comparisons (C allele versus T allele: P = 0.000 for Q-test; CC versus TT: P = 0.026 for Q-test; CC + TC versus TT: P = 0.002 for Q-test). Additionally, I-square value is another index for the heterogeneity test [29], with value less than 25% indicating low, 25% to 50% indicating moderate, and greater than 50% indicating high heterogeneity. The I-square values were 71.7%, 55.9% and 65.5 for the overall data of the allelic contrast, homozygote comparison and dominant model, respectively, indicating marked heterogeneities between the studies. Hence, the random-effect models were utilized. However, when subgroup analyses regarding ethnicity and age groups were further conducted, we found loss of heterogeneities in the subgroups regarding Caucasians and childhood AML, respectively.
Table 3.
Main results of the pooled data in the meta-analysis
| |
No. (cases/controls) |
C allele vs T allele |
CC vs TT |
(CC + TC) vs TT |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | P (OR) | P (Q-test) | OR (95%CI) | P (OR) | P (Q-test) | OR (95%CI) | P (OR) | P (Q-test) | ||
| Total |
1330/3688 |
1.13 (0.87-.1.48) |
0.349 |
0.000 |
1.72 (0.99-3.01) |
0.055 |
0.026 |
1.16 (0.86-1.55) |
0.326 |
0.002 |
| Ethnicity |
|
|
|
|
|
|
|
|
||
| Caucasian |
521/1359 |
0.90 (0.59-.1.37) |
0.629 |
0.062 |
2.02 (0.76-5.36) |
0.160 |
0.462 |
0.85 (0.57-1.26) |
0.415 |
0.127 |
| Asian |
623/1946 |
1.35 (0.90-2.02) |
0.150 |
0.004 |
1.77 (0.72-4.35) |
0.214 |
0.002 |
1.33 (1.09-1.62) |
0.004 |
0.382 |
| Mixed |
186/383 |
1.11 (0.48-2.55) |
0.807 |
0.029 |
1.40 (0.28-6.90) |
0.681 |
0.227 |
1.24 (0.48-3.22) |
0.654 |
0.021 |
| Age groups |
|
|
|
|
|
|
|
|
||
| Adult AML |
1183/2890 |
1.21 (0.88-1.66) |
0.244 |
0.000 |
1.76 (0.94-3.30) |
0.078 |
0.015 |
1.26 (0.88-1.81) |
0.213 |
0.000 |
| Childhood AML | 147/938 | 1.02 (0.69-1.49) | 0.938 | 0.620 | 1.78 (0.60-5.32) | 0.299 | 0.376 | 0.97 (0.63-1.49) | 0.877 | 0.856 |
AML, acute myeloid leukemia.
Meta-analysis results
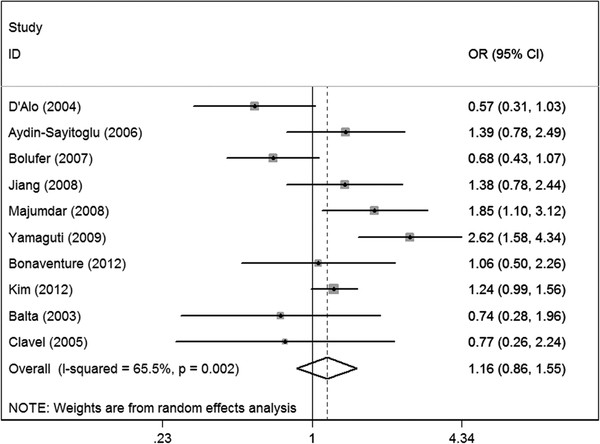
The main results of the meta-analysis were listed in Table3. For the overall data containing 1330 cases and 3688 controls, the pooled ORs for the allelic contrast, homozygote comparison and dominant model were 1.13 (95%CI = 0.87-1.48), 1.72 (95%CI = 0.99-3.01) and 1.16 (95%CI = 0.86-1.55), respectively, indicating that CYP1A1 MspI polymorphism might not have a correlation with AML risk (Figure2).
Figure 2 .
Meta-analysis for the association of acute myeloid leukemia risk with CYP1A1 MspI polymorphism for the overall data (CC + TC versus TT).
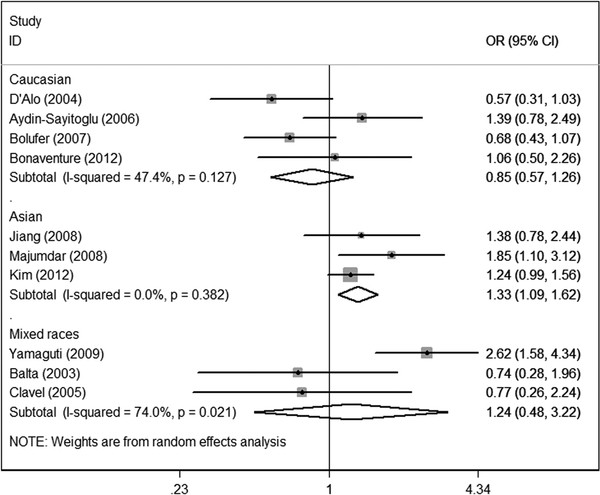
However, in subgroup analysis according to ethnicity, increased risk was shown among Asians (OR = 1.33; 95%CI = 1.09-1.62; P = 0.382 for heterogeneity) under the dominant model, but not the allele contrast or homozygote comparison models. No increased risk could be observed among Caucasians or mixed races under the three genetic models. The data indicated that Asians who carry variant C allele might have increased AML risk relative to those who harbor wild type TT alleles. (Figure3).
Figure 3 .
Meta-analysis for the association of acute myeloid leukemia risk with CYP1A1 MspI polymorphism (CC + TC versus TT; stratified by ethnicity).
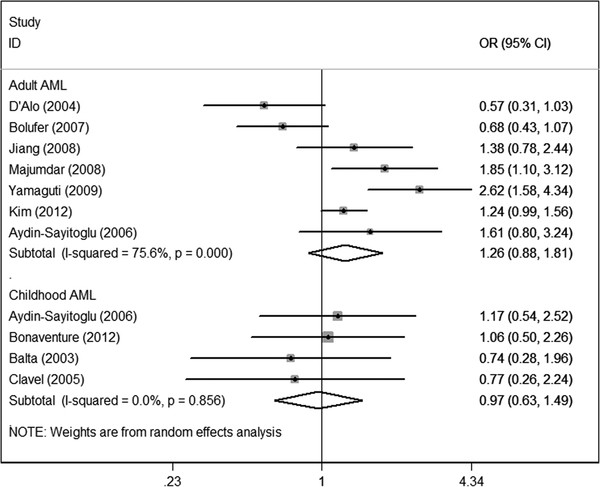
In subgroup analyses regarding age groups, no increased risk was found among either the childhood AML subgroup or the adult AML subgroup under the three genetic comparisons (Figure4).
Figure 4 .
Meta-analysis for the association of acute myeloid leukemia risk with CYP1A1 MspI polymorphism stratified by age groups (CC + TC versus TT). AML, acute myeloid leukemia.
Sensitivity analysis
When the effect-models were changed, the significance of the overall data for the two comparisons, respectively, was not statistically altered (data not shown). Then, one-way sensitivity analysis [30] was carried out to assess the stability of the meta-analysis. The statistical significance of the results was not changed when any single study was omitted (data not shown), indicating the credibility of the results.
Bias diagnostics
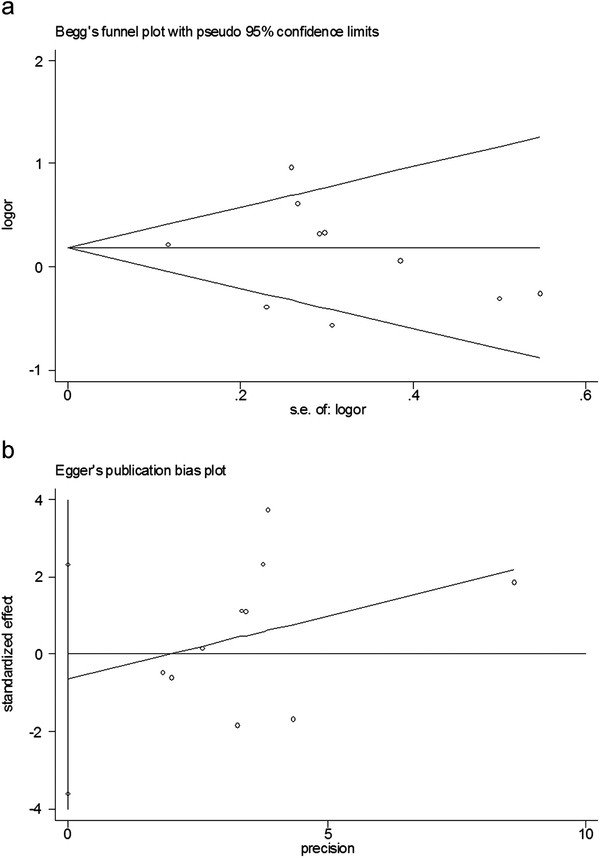
Funnel plots were created to detect possible publication bias. Then, Egger’s linear regression tests were used to assess the symmetries of the plots. The funnel plots appeared to be symmetrical for the overall data (Figure5a). Moreover, results of the Egger’s tests also indicated that the potential publication bias was not evident (Figure5b) (C allele versus T allele: t = −0.20, P > 0.05; CC versus TT: t = 0.66, P >0.05; CC + TC versus TT: t = −0.50, P >0.05).
Figure 5 .
Publication bias tests for the overall data (CC + TC versus TT). (a): Funnel plot; (b) Egger’s linear regression test.
Discussion
For the overall data, the results showed that CYP1A1 MspI polymorphism might not have a significant correlation with AML risk. Moreover, in subgroup analyses stratified by ethnicity, the data suggested an excess AML risk among Asians but not Caucasians or mixed races.
Previously, several meta-analyses have been devoted to the association of CYP1A1 MspI polymorphism with other cancer risk. Nevertheless, the results were conflicting. CYP1A1 MspI genetic variations have been indicated to raise risk for lung cancer, cervical cancer, prostate cancer and laryngeal cancer [31-34]. However, negligible relations between polymorphic CYP1A1 MspI and gastric cancer, colorectal cancer, breast cancer and esophageal cancer risks have been found [35-38]. Therefore, polymorphism of CYP1A1 MspI might play different roles in different cancers.
As for leukemia, a recent meta-analysis by Zhang et al… [39] regarding the relations of CYP1A1 MspI polymorphism with childhood acute leukemia failed to suggest a significant association regarding childhood ANLL (AML), in line with the present study. However, in the study by Zhang et al. [39], only two studies regarding childhood AML were selected [27,28]. Another two important studies that met the inclusion criteria were ignored [21,25]. In the present meta-analysis, a total of ten studies concerning childhood AML as well as adult AML were included, which statistically increased power to assess the associations.
In subgroup analysis according to ethnicity, significant increased risk was found among Asians, but not Caucasians and mixed races. Notably, this association could be only observed in the dominant model but not the allele contrast and homozygote comparison models, indicating that Asians who bear variant C allele of CYP1A1 MspI polymorphism might have an excess AML risk compared with those who carry wild type TT alleles. Possible racial differences in presentation, treatment patterns and survival with respect to AML might exist [40]. The difference might be owing to a possible role of ethnic differences in genetic backgrounds and the environment they lived in. However, the differences might be due to chance because the limited number of included studies and small sample sizes might give rise to insufficient statistical power for detection of a minor effect. Thus, the results should be interpreted with caution because undulated risk estimation might be obtained. Further studies regarding different ethnicities with large sample sizes are needed to clarify this issue.
In the subgroup analysis stratified by age groups, no increased risk was shown among either the childhood AML or the adult AML subgroups. Evidence indicates that the etiologies of childhood AML and adult AML might be different [41]. Moreover, host genetic differences between the two age groups might exist [42]. Therefore, the possible differences regarding CYP1A1 MspI polymorphism between the two age groups should be noted in further investigations. However, the data indicated that the potential difference was not evident in the present meta-analysis.
The overall data were not stratified by source of controls because all studies concerned the population-based controls, except for one study with limited sample sizes [28]. Hospital-based controls might not be always truly representative of the general population. In addition, the population-based controls in several studies were not strictly matched to the cases. Thus, any selection bias might exist. Future studies using proper control participants with strict matching criteria and large sample sizes are important for reducing such selection bias.
In the present meta-analysis, evident between-study heterogeneities for the overall data were observed for the three comparisons, respectively, and thus, the random-effect models were utilized. In the subgroup analyses, loss of heterogeneities was also found in the subgroups regarding Caucasian and childhood AML, respectively. Though we tried to minimize the possibility of encountering heterogeneity problems by conducting a careful search of the literature and using rigorous criteria for data pooling, evident heterogeneities still existed in some of the comparisons. Therefore, heterogeneities might be multifactoral. In addition to ethnicity and age groups, other factors such as gender, source of controls, histological types and prevalence of lifestyle factors might also yield the heterogeneities.
Several limitations should be concerned in the present meta-analysis. First, the primary articles only provided data about Caucasians, Asians and mixed races. Detailed information regarding other ethnicities such as African should be concerned. Second, subgroup analyses regarding gender and other factors such as smoking, drinking and radiation exposure have not been conducted in the present study because relevant information was insufficient in the primary articles. Third, only studies written in English and Chinese were included in this meta-analysis. Any selection bias should be noted. Furthermore, although the meta-analysis in this study is suggestive, high heterogeneity and lack of significant association in any genetic model among Caucasian and Mixed subgroups or age subgroups observed in this study could also originate from the nature of AML as a genetically heterogeneous disease and further assessment on the relationship between CYP1A1 MspI polymorphism and risk of AML subtypes might provide more instructive information. Additionally, gene-gene and gene-environment interactions should also be considered in the further investigations.
In summary, the results of the present meta-analysis suggest that variant C allele of CYP1A1 MspI polymorphism might have an association with increased AML risk among Asians. Further investigations are needed to confirm the conclusions.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
WZand ZC conceived of the study, and carried out the analysis of the literatures and drafted the manuscript. LZ and YW carried out the collection of the literatures. BZ helped with the statistical analysis and manuscript drafting. ZC and WZ conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Wenlei Zhuo, Email: zhuowenlei@yahoo.com.cn.
Liang Zhang, Email: zhangliangsyd@163.com.
Yan Wang, Email: wangyanflower@163.com.
Bo Zhu, Email: sydzhubo@163.com.
Zhengtang Chen, Email: cztsyd@yahoo.com.
Acknowledgements
None declared.
References
- Ilhan G, Karakus S, Andic N. Risk factors and primary prevention of acute leukemia. Asian Pacific journal of cancer prevention : APJCP. 2006;7:515–517. [PubMed] [Google Scholar]
- Yan J, Yin M, Dreyer ZE, Scheurer ME, Kamdar K, Wei Q, Okcu MF. A meta-analysis of MTHFR C677T and A1298C polymorphisms and risk of acute lymphoblastic leukemia in children. Pediatric blood & cancer. 2012;58:513–518. doi: 10.1002/pbc.23137. [DOI] [PubMed] [Google Scholar]
- Ye Z, Song H. Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. European journal of cancer (Oxford, England : 1990) 2005;41:980–989. doi: 10.1016/j.ejca.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Wang L, Yin F, Xu X, Hu X, Zhao D. X-Ray Repair Cross-Complementing Group 1 (XRCC1) Genetic Polymorphisms and Risk of Childhood Acute Lymphoblastic Leukemia: A Meta-Analysis. PloS one. 2012;7:e34897. doi: 10.1371/journal.pone.0034897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Zhang J, Dou C, Gu S, Xie Y, Mao Y, Ji C. Methionine synthase A2756G polymorphism and cancer risk: a meta-analysis. European journal of human genetics : EJHG. 2010;18:370–378. doi: 10.1038/ejhg.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffetta P. Biomarkers in cancer epidemiology: an integrative approach. Carcinogenesis. 2010;31:121–126. doi: 10.1093/carcin/bgp269. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mutation research. 1998;400:201–213. doi: 10.1016/S0027-5107(98)00037-2. [DOI] [PubMed] [Google Scholar]
- Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug metabolism reviews. 2009;41:89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry research. 2004;129:39–44. doi: 10.1016/j.psychres.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Tian Y, Jin X, Yan C, Jiang F, Zhang Y, Tang J, Shen X. A case-only study of interactions between metabolic enzyme polymorphisms and industrial pollution in childhood acute leukemia. Environmental toxicology and pharmacology. 2009;28:161–166. doi: 10.1016/j.etap.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Pelloso LA, Da Silva ID, De Souza NC, Yamamoto M, Botelho CA, Chauffaille Mde L. CYP1A1 polymorphisms modify overall survival in acute myeloid leukemia patients. Leukemia & lymphoma. 2007;48:1211–1215. doi: 10.1080/10428190701332431. [DOI] [PubMed] [Google Scholar]
- Barragan E, Collado M, Cervera J, Martin G, Bolufer P, Roman J, Sanz MA. The GST deletions and NQO1*2 polymorphism confers interindividual variability of response to treatment in patients with acute myeloid leukemia. Leukemia research. 2007;31:947–953. doi: 10.1016/j.leukres.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Voso MT, D'Alo F, Gumiero D, Guidi F, Hohaus S, Leone G. The CYP1A1*2a allele is an independent prognostic factor for acute myeloid leukemia. Haematologica. 2005;90:982–984. [PubMed] [Google Scholar]
- Infante-Rivard C, Krajinovic M, Labuda D, Sinnett D. Parental smoking, CYP1A1 genetic polymorphisms and childhood leukemia (Quebec, Canada) Cancer causes & control : CCC. 2000;11:547–553. doi: 10.1023/A:1008976116512. [DOI] [PubMed] [Google Scholar]
- Liu QX, Chen HC, Liu XF, Cao YF, Zhang J, Liu J. Study on the relationship between polymorphisms of Cyp1A1, GSTM1, GSTT1 genes and the susceptibility to acute leukemia in the general population of Hunan province. Zhonghua liu xing bing xue za zhi. 2005;26:975–979. [PubMed] [Google Scholar]
- Chen HC, Hu WX, Liu QX, Li WK, Chen FZ, Rao ZZ, Liu XF, Luo YP, Cao YF. Genetic polymorphisms of metabolic enzymes CYP1A1, CYP2D6, GSTM1 and GSTT1 and leukemia susceptibility. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2008;17:251–258. doi: 10.1097/CEJ.0b013e3282b72093. [DOI] [PubMed] [Google Scholar]
- Bolufer P, Collado M, Barragan E, Calasanz MJ, Colomer D, Tormo M, Gonzalez M, Brunet S, Batlle M, Cervera J, Sanz MA. Profile of polymorphisms of drug-metabolising enzymes and the risk of therapy-related leukaemia. British journal of haematology. 2007;136:590–596. doi: 10.1111/j.1365-2141.2006.06469.x. [DOI] [PubMed] [Google Scholar]
- Bolufer P, Collado M, Barragan E, Cervera J, Calasanz MJ, Colomer D, Roman-Gomez J, Sanz MA. The potential effect of gender in combination with common genetic polymorphisms of drug-metabolizing enzymes on the risk of developing acute leukemia. Haematologica. 2007;92:308–314. doi: 10.3324/haematol.10752. [DOI] [PubMed] [Google Scholar]
- Kim HN, Kim NY, Yu L, Tran HT, Kim YK, Lee IK, Shin MH, Park KS, Choi JS, Kim HJ. Association of GSTT1 polymorphism with acute myeloid leukemia risk is dependent on smoking status. Leukemia & lymphoma. 2012;53:681–687. doi: 10.3109/10428194.2011.625576. [DOI] [PubMed] [Google Scholar]
- Bonaventure A, Goujon-Bellec S, Rudant J, Orsi L, Leverger G, Baruchel A, Bertrand Y, Nelken B, Pasquet M, Michel G. et al. Maternal smoking during pregnancy, genetic polymorphisms of metabolic enzymes, and childhood acute leukemia: the ESCALE study (SFCE) Cancer causes & control : CCC. 2012;23:329–345. doi: 10.1007/s10552-011-9882-9. [DOI] [PubMed] [Google Scholar]
- Yamaguti GG, Lourenco GJ, Costa FF, Lima CS. High risk of 'de novo' acute myeloid leukaemia in individuals with cytochrome P450 A1 (CYP1A1) and NAD(P)H:quinone oxidoreductase 1 (NQO1) gene defects. European journal of haematology. 2009;83:270–272. doi: 10.1111/j.1600-0609.2009.01272.x. [DOI] [PubMed] [Google Scholar]
- Majumdar S, Mondal BC, Ghosh M, Dey S, Mukhopadhyay A, Chandra S, Dasgupta UB. Association of cytochrome P450, glutathione S-transferase and N-acetyl transferase 2 gene polymorphisms with incidence of acute myeloid leukemia. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2008;17:125–132. doi: 10.1097/CEJ.0b013e3282b6fd68. [DOI] [PubMed] [Google Scholar]
- Jiang L, Chen M, Qin G. Association between the polymorphisms of cytochrome P4501A1 and glutathione S-transferase M1, T1 Genes and acute myeloid leukemia in Guangxi. Guangxi Medical Journal. 2008;30:464–466. [Google Scholar]
- Aydin-Sayitoglu M, Hatirnaz O, Erensoy N, Ozbek U. Role of CYP2D6, CYP1A1, CYP2E1, GSTT1, and GSTM1 genes in the susceptibility to acute leukemias. American journal of hematology. 2006;81:162–170. doi: 10.1002/ajh.20434. [DOI] [PubMed] [Google Scholar]
- D'Alo F, Voso MT, Guidi F, Massini G, Scardocci A, Sica S, Pagano L, Hohaus S, Leone G. Polymorphisms of CYP1A1 and glutathione S-transferase and susceptibility to adult acute myeloid leukemia. Haematologica. 2004;89:664–670. [PubMed] [Google Scholar]
- Balta G, Yuksek N, Ozyurek E, Ertem U, Hicsonmez G, Altay C, Gurgey A. Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes in childhood acute leukemia. American journal of hematology. 2003;73:154–160. doi: 10.1002/ajh.10339. [DOI] [PubMed] [Google Scholar]
- Clavel J, Bellec S, Rebouissou S, Menegaux F, Feunteun J, Bonaiti-Pellie C, Baruchel A, Kebaili K, Lambilliotte A, Leverger G. et al. Childhood leukaemia, polymorphisms of metabolism enzyme genes, and interactions with maternal tobacco, coffee and alcohol consumption during pregnancy. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2005;14:531–540. doi: 10.1097/00008469-200512000-00007. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Techn Bull. 1999;8:15–17. [Google Scholar]
- Zhuo WL, Wang Y, Zhuo XL, Zhu B, Zhu Y, Chen ZT. Polymorphisms of CYP1A1 and GSTM1 and laryngeal cancer risk: evidence-based meta-analyses. Journal of cancer research and clinical oncology. 2009;135:1081–1090. doi: 10.1007/s00432-009-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik AP, Jamil K, Das P. CYP1A1 polymorphisms and risk of prostate cancer: a meta-analysis. Urology journal. 2009;6:78–86. [PubMed] [Google Scholar]
- Zhan P, Wang Q, Qian Q, Wei SZ, Yu LK. CYP1A1 MspI and exon7 gene polymorphisms and lung cancer risk: an updated meta-analysis and review. Journal of experimental & clinical cancer research : CR. 2011;30:99. doi: 10.1186/1756-9966-30-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergentanis TN, Economopoulos KP, Choussein S, Vlahos NF. Cytochrome P450 1A1 (CYP1A1) gene polymorphisms and cervical cancer risk: a meta-analysis. Molecular biology reports. 2012. [DOI] [PubMed]
- Zhuo WL, Zhang YS, Wang Y, Zhuo XL, Zhu B, Cai L, Chen ZT. Association studies of CYP1A1 and GSTM1 polymorphisms with esophageal cancer risk: evidence-based meta-analyses. Archives of medical research. 2009;40:169–179. doi: 10.1016/j.arcmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Sergentanis TN, Economopoulos KP. Four polymorphisms in cytochrome P450 1A1 (CYP1A1) gene and breast cancer risk: a meta-analysis. Breast cancer research and treatment. 2010;122:459–469. doi: 10.1007/s10549-009-0694-5. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wang JJ, Sun L, Li HL. Association between CYP1A1 polymorphism and colorectal cancer risk: a meta-analysis. Molecular biology reports. 2012;39:3533–3540. doi: 10.1007/s11033-011-1126-2. [DOI] [PubMed] [Google Scholar]
- Guo R, Guo X. Quantitative assessment of the associations between CYP1A1 polymorphisms and gastric cancer risk. Tumour biology. the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012. [DOI] [PubMed]
- Zhang YD, Tan LN, Zhang XL, Wei HY, Xiong H, Hu Q. Meta-analysis of cytochrome P4501A1 MspI gene polymorphism and childhood acute leukemia. Biomedical and environmental sciences : BES. 2011;24:683–687. doi: 10.3967/0895-3988.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Bierenbaum J, Davidoff AJ, Ning Y, Tidwell ML, Gojo I, Baer MR. Racial differences in presentation, referral and treatment patterns and survival in adult patients with acute myeloid leukemia: a single-institution experience. Leukemia research. 2012;36:140–145. doi: 10.1016/j.leukres.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larfors G, Hallbook H, Simonsson B. Parental age, family size, and offspring's risk of childhood and adult acute leukemia. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012. [DOI] [PubMed]
- Juhl-Christensen C, Ommen HB, Aggerholm A, Lausen B, Kjeldsen E, Hasle H, Hokland P. Genetic and epigenetic similarities and differences between childhood and adult AML. Peditric blood & cancer. 2012;58:525–531. doi: 10.1002/pbc.23397. [DOI] [PubMed] [Google Scholar]