Abstract
We designed, optimized, and extensively tested several sensitive and specific real-time PCR assays for rapid detection of both smallpox and pan-orthopox virus DNAs. The assays are based on TaqMan 3′-minor groove binder chemistry and were performed on both the rapid-cycling Roche LightCycler and the Cepheid Smart Cycler platforms. The hemagglutinin (HA) J7R, B9R, and B10R genes were used as targets for the variola virus-specific assays, and the HA and DNA polymerase-E9L genes were used as targets for the pan-orthopox virus assays. The five orthopox virus assays were tested against a panel of orthopox virus DNAs (both genomic and cloned) at the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID). The results indicated that each assay was capable of detecting both the appropriate cloned gene and genomic DNA. The assays showed no cross-reactivity to the 78 DNAs in the USAMRIID bacterial cross-reactivity panel. The limit of detection (LOD) of each assay was determined to be between 12 and 25 copies of target DNA. The assays were also run against a blind panel of DNAs at the Centers for Disease Control and Prevention (CDC) on both the LightCycler and the Smart Cycler. The panel consisted of eight different variola virus isolates, five non-variola virus orthopox virus isolates, two varicella-zoster virus isolates, and one herpes simplex virus isolate. Each sample was tested in triplicate at 2.5 ng, 25 pg, 250 fg, and 2.5 fg, which represent 1.24 × 107, 1.24 × 105, 1.24 × 103, and 1.24 × 101 genome equivalents, respectively. The results indicated that each of the five assays was 100% specific (no false positives) when tested against both the USAMRIID panels and the CDC blind panel. With the CDC blind panel, the LightCycler was capable of detecting 96.2% of the orthopox virus DNAs and 93.8% of the variola virus DNAs. The Smart Cycler was capable of detecting 92.3% of the orthopox virus DNAs and between 75 and 93.8% of the variola virus DNAs. However, all five assays had nearly 100% sensitivity on both machines with samples above the LOD (>12 gene copies). These real-time PCR assays represent a battery of tests to screen for and confirm the presence of variola virus DNA. The early detection of a smallpox outbreak is crucial whether the incident is an act of bioterrorism or an accidental occurrence.
The orthopox viruses are large, enveloped viruses containing double-stranded DNA genomes of approximately 175,000 to 225,000 bp. Several viruses in this family are human pathogens, including variola major virus (the causative agent of smallpox) and variola minor virus (the causative agent of alastrim), monkeypox virus, cowpox virus, and vaccinia virus. In humans, orthopox viruses cause infections ranging from mild in the case of vaccinia and cowpox viruses and some strains of monkeypox virus to fatal in the case of smallpox. The 33rd World Health Assembly, at the end of a decade-long intensive surveillance and vaccination campaign, officially declared smallpox eradicated from the world in 1980. Member states of the World Health Organization (WHO) subsequently agreed to consolidate the variola virus stocks into two locations, i.e., the Centers for Disease Control and Prevention (CDC), Atlanta, Ga., and the Institute for Viral Preparations, Moscow, Russia, and to destroy the remaining laboratory samples of variola viruses. The Russian collection was subsequently moved to the State Research Center of Virology and Biotechnology, Novosibirsk, Russia, in the early 1990s. Those stocks in the WHO reference laboratories have been scheduled for destruction several times, but the World Health Assembly member states have since decided to retain them indefinitely in order to allow for the development of improved vaccines, antiviral drugs, and diagnostics (18, 25). The United States and WHO have significant concerns that undeclared stocks of smallpox have been obtained by terrorists or rogue governments and might be employed as a weapon of mass destruction (6, 7, 12, 23).
For many years, PCR analysis of orthopox virus DNA relied solely on the laborious method of determining amplicon size from an agarose gel after a 2- to 3-h PCR run (22, 26-29, 31). A revolutionary technique, the fluorogenic 5′ nuclease assay (now commonly called the TaqMan assay), was developed in 1991 (14) and subsequently improved by Lee et al. in 1993 (21). While TaqMan assays were initially run on the Applied Biosystems (ABI) 7700 nucleic acid detection system, more rapidly cycling systems have become available, including the Roche (Indianapolis, Ind.) LightCycler and the Cepheid (Sunnyvale, Calif.) Smart Cycler (5, 8-10, 33). Several papers have dealt with the general application of real-time TaqMan PCR technology for identifying biological agents (11, 13, 19), as well as specifically for orthopox viruses (8, 16, 17, 24, 31). The TaqMan chemistry has recently undergone a significant improvement by the addition of a minor groove binding protein (MGB) (1, 2, 20) and a nonfluorescent quencher (NFQ) to the 5′ end of the probe molecule. The objective of this study was to use TaqMan-MGB chemistry to improve our previous variola virus-specific TaqMan assay (17) and to design additional variola virus-specific and pan-orthopox virus TaqMan-MGB assays.
MATERIALS AND METHODS
Viruses and DNA preparation.
The orthopox viruses and controls used (see Tables 2 and 4) included eight isolates of variola virus and 10 different strains from camelpox virus, cowpox virus, herpes simplex virus, monkeypox virus, vaccinia virus, and varicella-zoster virus. The origin, propagation, and harvest procedures for these viruses are documented elsewhere (3, 31). Our studies also included 11 monkeypox-human virus isolates obtained from clinical samples during the 1996 outbreak in the Congo (unpublished data) and four cidofovir-resistant orthopox virus DNAs (32). Variola virus infectious titers were determined by the plaque assay and were between 108 and 1010 PFU per ml. DNA was extracted from virus-infected cells, virions, and crusts by using the AquaPure DNA kit, using a procedure modified slightly from that of the manufacturer (Bio-Rad, Hercules, Calif.). Briefly, 100 μl of cell lysate or crust suspension was mixed with 500 μl of AquaPure lysis buffer, incubated at 55°C for 8 h, cooled to 37°C, and removed from the CDC Maximum Containment Laboratory after appropriate safety tests. Five microliters (12 μg) of AquaPure RNase solution was added to the mixture and incubated at 37°C for 5 min. Two hundred microliters of AquaPure protein precipitation solution was added and mixed by vortexing. The samples were centrifuged at 13,000 × g for 20 min, the supernatants were transferred into sterile microcentrifuge tubes, 600 μl of isopropanol was added, and the DNA was precipitated by centrifugation at 13,000 × g for 5 min. The DNA was washed once with 70% alcohol, suspended in 50 μl of AquaPure DNA hydration buffer, and stored at −20°C until used.
TABLE 2.
USAMRIID orthopox virus panel DNAs
| DNA species | Dilution (pg/μl) | Assay resulta
|
||||
|---|---|---|---|---|---|---|
| pan-HA | pan-E9L3 | Variola virus HA | Variola virus B9R | Variola virus B10R | ||
| Human genomic DNA | 1,000 | − | − | − | − | − |
| Vaccinia virus E9L gene (cloned) | 1 | − | + | − | − | − |
| Variola virus India 7124 HA gene (cloned) | 1 | + | − | + | − | − |
| Variola virus India 7124 B9R-B10R (cloned) | 1 | − | − | − | + | + |
| Variola major virus Nepal v73-175 (E9L cloned) | 1 | ND | + | ND | ND | ND |
| Variola major virus India 7124 (E9L cloned) | 1 | ND | + | ND | ND | ND |
| Variola major virus Bangladesh 1975 (v75-550) (E9L cloned) | 1 | ND | + | ND | ND | ND |
| Variola major virus Horn (E9L cloned) | 1 | ND | + | ND | ND | ND |
| Variola major virus Somalia v77-1605 (E9L cloned) | 1 | ND | + | ND | ND | ND |
| Variola major virus Congo (v70-46) (E9L cloned) | 1 | ND | + | ND | ND | ND |
| Variola major virus Afgan Variolator 4 (E9L cloned) | 1 | ND | + | ND | ND | ND |
| Variola major virus Nepal v73-175 (B9R-B10R cloned) | 1 | ND | ND | ND | + | + |
| Variola major virus India 7124 (B9R-B10R cloned) | 1 | ND | ND | ND | + | + |
| Variola major virus Bangladesh 1975 (v75-550) (B9R-B10R cloned) | 1 | ND | ND | ND | + | + |
| Variola major virus Horn (B9R-B10R cloned) | 1 | ND | ND | ND | + | + |
| Variola major virus Somalia v77-1605 (B9R-B10R cloned) | 1 | ND | ND | ND | + | + |
| Variola major virus Congo (v70-46) (B9R-B10R cloned) | 1 | ND | ND | ND | + | + |
| Variola major virus Afgan Variolator 4 (B9R-B10R cloned) | 1 | ND | ND | ND | + | + |
| Vaccinia virus Western Reserve | 100 | + | + | − | − | − |
| Modified vaccinia virus Ankara | 100 | + | + | − | − | − |
| Camelpox virus Somalia | 100 | + | + | − | − | − |
| Cidofovir-resistant camelpox virus somalia | 100 | + | + | − | − | − |
| Cowpox virus Brighton Red | 100 | + | + | − | − | − |
| Cidofovir-resistant cowpox virus Brighton Red | 100 | + | + | − | − | − |
| MPXb | 100 | + | + | − | − | − |
| Cidofovir-resistant MPX | 100 | + | + | − | − | − |
| Vaccinia virus Copenhagen | 100 | + | + | − | − | − |
| Cidofovir-resistant vaccinia virus Copenhagen | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-04) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-16) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-03) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-0035) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-07) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-05) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-12) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-17) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-11) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-06) | 100 | + | + | − | − | − |
| MPX-human isolate (CDC V79-I-08) | 100 | + | + | − | − | − |
+, detected; −, not detected; ND, not determined.
MPX, monkeypox virus Zaire.
TABLE 4.
Results of OHS-IWG CDC panel testing for all five orthopox virus-MGB assays
| Organism | Specimen material | Sample | Resulta with the following amt (gene copy no.)b of DNA:
|
||||
|---|---|---|---|---|---|---|---|
| Predc | 2.5 ng (1.24 × 107) | 25 pg (1.24 × 105) | 250 fg (1.24 × 103) | 2.5 fg (1.24 × 101) | |||
| Variola virus | Purified virus | Bangladesh | 12345 | 12345 | 12345 | 1234d5d | 123d45 |
| Congo | 12345 | 12345 | 12345 | 12345 | 123d4d 5 | ||
| India 7124 | 12345 | 12345 | 12345 | 12345 | 123d45 | ||
| Nepal (v73-175) | 12345 | 12345 | 12345 | 12345 | 123d4d 5 | ||
| Afgan (var4) | 12345 | 12345 | 12345 | 12345 | 123d4d 5 | ||
| Horn | 12345 | 12345 | 12345 | 12345 | 123d45 | ||
| Virus-cell lysate | Botswana (v73-225) | 12345 | 12345 | 12345 | 12345 | ----5e | |
| Primary clinical crust | Parvin | 12345 | 12345 | 12345 | 12345 | ----- | |
| Vaccinia virus | Purified virus | Dryvax | 12--- | 12--- | 12--- | 12--- | 12--- |
| IHDJ | 12--- | 12--- | 12--- | 12--- | 12--- | ||
| Cowpox virus | Purified virus | CP58 | 12--- | 12--- | 12--- | 12--- | 1d2--- |
| Monkeypox virus | Purified virus | v79-I-005 | 12--- | 12--- | 12--- | 12--- | 1d2d--- |
| Camelpox virus | Purified virus | v78-2379 | 12--- | 12--- | 12--- | 12--- | 12--- |
| Varicella-zoster virus | Purified virus | Varicella OKA | ----- | ----- | ----- | ----- | ----- |
| Varicella WEB | ----- | ----- | ----- | ----- | ----- | ||
| Herpes simplex virus | Purified virus | HSV-1 Justin | ----- | ----- | ----- | ----- | ----- |
| Positive control | Assay genomic DNA | PTC | 12345 | NDf | ND | 12345 | ND |
| Blank (H2O) | Negative control | NTC | ----- | ----- | ----- | ----- | ----- |
The presence of a number indicates that the sample was detected as follows: 1, pan-Orthopox virus HA-MGB assay; 2, pan-Orthopox virus E9L3-MGB assay; 3, Variola virus HA-MGB assay; 4, Variola virus B9R-MGB assay; 5, Variola virus B10R-MGB assay. A dash indicates that the sample was not detected by that assay on either the LightCycler or the Smart Cycler.
Total amount of unknown genomic DNA and approximate gene copy number added to each assay.
Predicted assay outcome based on sample DNA origin.
Detected only on the LightCycler.
Detected only on the Smart Cycler.
ND, not determined.
Cloning and sequencing of HA, E9L, B9R, and B10R genes.
Several variola virus and non-variola virus gene targets, i.e., hemagglutinin (HA) (J7R), E9L (DNA polymerase), and B9R-B10R fragments, were cloned (see Table 2). The variola major virus B9R protein was 74 amino acids, while the B10R was 65 amino acids; the exact functions of both are unknown. A fragment of 942 bp which contained the HA (J7R) gene was PCR amplified from viral genomic DNA by using the forward primer 5′-ATG ACA CGA TTG TCA ATA CTT TTG T-3′ and reverse primer 5′-CTA GAC TTT GTT TTC TGT TTT GTA T-3′. A fragment of 3,015 bp which contained the E9L gene was PCR amplified from viral genomic DNA by using the forward primer 5′-ATG GAT GTT CGG TGT ATT AAT TGG T-3′ and reverse primer 5′-TTA TGC TTC GTA AAA TGT AGG TCT TG-3′. A fragment of 504 bp (variola major virus) or 1,138 bp (variola minor virus) which contained the B9R and B10R genes was PCR amplified from the appropriate viral genomic DNA by using the forward primer 5′-ATG GAC ATT TCT TAT GTT ATT AAT G-3′ and reverse primer 5′-TCA AAA CGT GTA TCT CAT ATA TAC T-3′. The fragments were cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, Calif.) according to the manufacturer's protocol. Briefly, target genes were amplified by high-fidelity PCR from genomic DNA. DNA fragments were isolated on 0.8% agarose gels, and 2 μl of the isolated PCR product was added to 2 μl of pCR2.1-TOPO cloning vector. Mixtures were incubated for 5 min at room temperature, and 2 μl of each was used to transform competent DH5α (LacI+) Escherichia coli bacteria that were plated on Luria-Bertani agar containing ampicillin (100 μg/ml) (Sigma, St. Louis, Mo.), IPTG (isopropyl-β-d-thiogalactopyranoside) (0.5 mM) (Sigma), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml) (Sigma). Positive clones were detected by restriction enzyme digestion, PCR amplification of the inserted DNA, and sequencing. Plasmid DNA was isolated from bacterial cultures by using a commercially available kit (Qiagen, Valencia, Calif.) and measured by optical density at 260 nm.
DNA sequencing.
The pCR2.1 clones containing HA and B9R-B10R inserts were sequenced on an ABI (Foster City, Calif.) Prism 3100 genetic analyzer. Approximately 100 ng of plasmid DNA was used as the template in BigDye terminator cycle ready reactions (Applied Biosystems) according to the manufacturer's instructions. Clones containing HA gene inserts were sequenced with pCR2.1 vector-specific T7 promoter (5′-TAA TAC GAC TCA CTA TAG GG-3′) and M13 reverse (5′-CAG GAA ACA GCT ATG AC-3′) primers and HA gene-specific forward (5′-AGT GAC GTC TTG TAT TTT GAT-3′) and reverse (5′-TCT GTT TTG TAT TTA CGT G-3′) sequencing primers. Clones containing variola major and minor virus B9R-B10R gene inserts were sequenced with vector-specific T7 promoter and M13 reverse primers. In addition, variola minor virus B9R-B10R genes were sequenced with B9R-B10R-specific forward (5′-TCT GAT TTG GAA GCG TAT TT-3′) and reverse (5′-AGT AGC GGA GGT AGT CGT CT-3′) sequencing primers. All sequence data were assembled and analyzed with SeqMan II software (DNASTAR, Inc., Madison, Wis.).
PCR primers, target sequences, and fluorogenic probes.
The real-time PCR assay primers and TaqMan-MGB probe sequences are listed in Table 1. The HA sequence was selected from the variola virus HA J7R gene (GenBank accession number L22579) and from the report of Ibrahim et al. (17). We also cloned and sequenced several new HA, E9L (DNA polymerase), B9R, and B10R gene fragments (Table 2) and used this new sequence information (data not shown) to perform our gene alignments and TaqMan-MGB assay design. All new sequences will be submitted to GenBank. In all, 28 HA genes (including 12 variola virus, 7 vaccinia virus, and 11 non-variola virus genes), 48 E9L DNA polymerase genes (including 39 variola virus genes, 1 camelpox virus gene, 1 cowpox virus gene, 1 monkeypox virus gene, 1 vaccinia virus gene, 3 herpes simplex virus genes, and 1 varicella-zoster virus gene), and 11 B9R-B10 fragments (including 8 variola major virus genes and 2 variola minor virus genes) were used for alignments. All alignments were done by using OMIGA 2.0 (Accelrys, San Diego, Calif.). Regions of homology were used as target sequences for the pan-orthopox virus assays, and regions of nonhomology were used as targets for the variola virus-specific assays. The specific primer and TaqMan-MGB sequences were designed by using Primer Express version 2.0 for Windows (Applied Biosystems). All primers were synthesized by using standard phosphoramidite chemistry with an ABI 394 DNA-RNA synthesizer. The TaqMan-MGB probes were also synthesized by ABI and contained 6-carboxyfluorescein (FAM) at the 5′ end. An NFQ and the MGB were added to the 3′ end.
TABLE 1.
Primer and TaqMan-MGB sequences
| Organism | Gene target or probe | Amplicon size (bp) | Primer or probe
|
MgCl2 concn (mM) | ||
|---|---|---|---|---|---|---|
| Name | Sequence | Final concn (μM) | ||||
| pan-Orthopox virus | HA (J7R) | 130 | OPHA-F89 | 5′-GAT GAT GCA ACT CTA TCA TGT A-3′a | 0.5 | 5 |
| OPHA-R219 | 5′-GTA TAA TTA TCA AAA TAC AAG ACG TC-3′ | 0.5 | ||||
| TaqMan-MGB probe | OPHA-p143S-MGB | 6′FAM-AGT GCT TGG TAT AAG GAG-MGBNFQ-3′ | 0.1 | |||
| DNA polymerase (E9L) | 177 | OPE9L-F1880 | 5′-GAA CAT TTT TGG CAG AGA GAG CC-3′ | 0.5 | 5 | |
| OPE9L-R2057 | 5′-CAA CTC TTA GCC GAA GCG TAT GAG-3′ | 0.5 | ||||
| TaqMan-MGB probe | OPE9L-p1924S-MGB | 6′FAM-CAG GCT ACC AGT TCA A-MGBNFQ-3′ | 0.1 | |||
| Variola virus | HA (J7R) | 139 | VARHA-F601 | 5′-TCA TCT GGA GAA TCC ACA ACA-3′ | 0.4 | 7 |
| VARHA-R740 | 5′-CAT CAT TGG CGG TTG ATT TA-3′ | 0.4 | ||||
| TaqMan-MGB Probe | VARHA-p625S-MGB | 6′FAM-AAG ACG TCG GGA CCA AT-MGBNFQ-3′b | 0.1 | |||
| B9R | 87 | VARB9R-F103 | 5′-CAG ATA GCG GTT GTC AGA ATA CCA-3′ | 0.5 | 5 | |
| VAR9R-R189 | 5′-ATA CGC TTC CAA ATC AGA TCC TG-3′c | 0.5 | ||||
| TaqMan-MGB Probe | VARB9R-p143S-MGB | 6′FAM-CAA TGG AAC AAT TAG GTA TTA C-MGBNFQ-3′ | 0.1 | |||
| B10R | 88 | VARB10R-F66 | 5′-CAA AAT GCA GGG TAC AAC AAA CA-3′ | 0.5 | 5 | |
| VARB10R-R153 | 5′-CAA TGA ATC CTT AGT ATT GCC AAC G-3′ | 0.5 | ||||
| TaqMan-MGB Probe | VARB10R-p90S-MGB | 6′FAM-TAA TGA CGG AAG TAA ACG-MGBNFQ-3′ | 0.05 | |||
The primer has a single internal base mismatch with monkeypox virus isolate 3945 (base 13, C→A). However, the primer also has a single internal base mismatch with vaccinia virus strain Ankara (base 13, C→T) and is easily detected (see Table 2).
We have also recently become aware that there are at least two potential mismatched bases with our variola virus HA-MGB probe. This probe is the MGB version of the probe originally reported by Ibrahim et al. (17). There are five strains of variola virus (var-sln, accession no. AF375142; var-nig, accession no. 375138; var-cm6, accession no. AF375130; var-but, accession no. AF375129; and var-gar, accession no. Y16780) with a G→T change at base 6 in the MGB probe. There are two strains of variola virus (var-rat, accession no. AF375141; and var-mad, accession no. AF375137) with a G→C change at base 9 in the MGB probe. We therefore recommend that the variola virus HA-MGB assay always be used in conjunction with the B9R and B10R assays.
The primer has a single internal base mismatch with variola virus India-1967 strain (base 23, G→A).
5′ nuclease PCR (TaqMan-MGB) assays.
Following the Primer Express version 2.0 software design of potential orthopox virus TaqMan-MGB assays (pan-orthopox virus HA-MGB and E9L3-MGB and variola virus-specific HA-MGB, B9R-MGB, and B10R-MGB), we optimized each assay according to a standard protocol instituted by the Diagnostic Systems Division at the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID). Briefly, potential primer pairs were initially tested in the LightCycler with the fluorescent dye SYBR Green I (Roche Biochemicals). The optimum primer pair was selected based on specificity (a single, appropriately sized amplicon) and efficiency of amplification (lowest Ct value, which is defined as the real-time PCR cycle at which the LightCycler software determines the reaction to be positive). The selected primer pair was then optimized to the final concentration (0.1 to 1.0 μM) with the lowest Ct value and had the highest fluorescent signal. Next, several potential TaqMan-MGB probes were tested with the optimized primer pair by varying the probe and MgCl2 concentrations. The final assay was that with the primer-probe pair concentrations and reaction conditions that combined the lowest level of detection (LOD) (the gene copy number which was detected by the assay 100% of the time), lowest Ct value, and highest fluorescent signal-to-noise ratio. The LODs of the assays were determined from serial dilutions of both the cloned variola virus genes (HA, E9L, B9R, and B10R) and genomic DNA purified from variola virus (India 7125) virions.
All assays were carried out in 20-μl volumes for the LightCycler and 25-μl volumes for the Smart Cycler. Each reaction mixture contained PCR buffer (50 mM Tris [pH 8.3], 25 μg of bovine serum albumin per ml, and 0.2 mM deoxynucleoside triphosphate mix [Idaho Technology, Salt Lake City, Utah]). Platinum Taq DNA polymerase (Invitrogen) at 0.8 U for the LightCycler and 1 U for the Smart Cycler was added to each reaction mixture. The final MgCl2, primer, and probe concentrations for each assay are listed in Table 1. Each Smart Cycler reaction mixture also contained 1× Smart Cycler additive reagent (0.2 mM Tris [pH 8.0], 0.2 mg of bovine serum albumin per ml, 150 mM trehalose, and 0.2% Tween 20). Five microliters of control or template DNA was added to each reaction mixture. Thermal cycling for the LightCycler and the Smart Cycler was performed as follows: one cycle at 95°C for 2 min, followed by 45 cycles of 95°C for 1 s and 60°C for 20 s. A fluorescence reading was taken at the end of each 60°C step. For the LightCycler, each reaction capillary tube was read in channel 1 (F1) at a gain setting of 16, with data being analyzed by using the LightCycler data analysis software (version 3.5.3). Sample curves were analyzed by using the second derivative maximum with the baseline adjustment set to arithmetic. For the Smart Cycler, data were analyzed by using the Cepheid Smart Cycler software (version 1.2d). The Smart Cycler settings consisted of a primary curve analysis with a manual threshold setting of 10, background subtraction turned on, boxcar average set to five cycles, background minimum cycle set to 5, and background maximum cycle set to 45.
Extended assay evaluation.
All five assays were extensively evaluated, first against various cloned and genomic orthopox virus DNAs available at USAMRIID (Table 2), second against various strains and isolates from the USAMRIID cross-reactivity bacterial DNA panel (1 ng of each DNA) (Table 3), and finally against a blind panel of orthopox virus DNAs prepared by the CDC under the auspices of the Office of Homeland Security Interagency Working Group (OHS-IWG). The OHS-IWG CDC blind panel consisted of the 16 DNAs listed in Table 4. The DNAs represent a cross section of different orthopox viruses from various sources along with three non-orthopox virus DNAs. Each sample (20 μl total) was initially diluted 1:2 with sterile water to have sufficient volume to run the sample in triplicate with all five assays on both the LightCycler and the Smart Cycler. Five microliters of each 1:2 dilution was added to the LightCycler master mix (15 μl) and to the Smart Cycler master mix (260 μl). Therefore, after the 1:2 dilutions, the actual amount of DNA loaded into each assay mixture was 1 ng/μl (2.5 ng), 10 pg/μl (25 pg), 100 fg/μl (250 fg), and 1 fg/μl (2.5 fg). The genome equivalents for each assay (based on an average orthopox virus genome size of 186,000 bp and a GC content of 33%) were 62.5 ng = 1.24 × 107 genomes, 25 pg = 1.24 × 105 genomes, 250 fg = 1.24 × 103 genomes, and 2.5 fg = 1.24 × 101 genomes. For the OHS-IWG CDC blind DNA panel, each sample (68 in all) was tested in triplicate and any questionable sample results were rerun in triplicate. All reactions performed during testing of the coded samples at CDC included at least one positive control that contained 1.24 × 103 copies (250 fg) of purified variola virus genomic DNA (India 7125) and two no-template controls (NTC): reagent NTC and sample NTC. The CDC testing took place over 3 days (16 to 18 December 2002). The unblinded OHS-IWG CDC results are also shown in Table 4. Calculations of sensitivity and specificity for the CDC panel (Table 5) were determined as follows: percent sensitivity = [TP/(TP + FN)] × 100 and percent specificity = [TN/(TN + FP)] × 100, where TP is the number of true-positive samples, FN is the number of false-negative samples, TN is the number of true-negative samples, and FP is the number of false-positive samples.
TABLE 3.
USAMRIID cross-reactivity panel DNAs
| Organism |
|---|
| Acinetobacter baumanni |
| Alcaligenes xylosoxydans |
| Bacillus anthracis |
| BA0068 |
| Ames |
| Sterne |
| SPS 97.13.213 |
| Bacillus cereus |
| Bacillus coagulans |
| Bacillus licheniformis |
| Bacillus macerans |
| Bacillus megaterium |
| Bacillus polymyxa |
| Bacillus sphaericus |
| Bacillus stearothermophilus |
| Bacillus subtilis subsp. niger |
| Bacillus thuringiensis |
| Bacillus popilliae |
| Bacteroides distasonis |
| Bordetella bronchiseptica |
| Budvicia aquatica |
| Burkholderia cepacia |
| Burkholderia pseudomallei |
| Clostridium perfringens |
| Clostridium sporogenes |
| Comanonas acidovorans |
| Enterococcus durans |
| Enterococcus faecalis |
| Escherichia coli |
| Francisella tularensis |
| Haemophilus influenzae |
| Klebsiella pneumoniae |
| Listeria monocytogenes |
| Moraxella cattaharalis |
| Neisseria lactamica |
| Proteus mirabilis |
| Proteus vulgaris |
| Providencia stuartii |
| Pseudomonas aeruginosa |
| Ralstonia pickettii |
| Salmonella choleraesuis |
| Serratia odorifera |
| Shigella flexneri |
| Shigella sonnei |
| Staphylococcus aureus |
| Staphylococcus hominis |
| Stenotrophomonas maltophilia |
| Streptococcus pneumoniae |
| Streptococcus pyogenes |
| Yersinia enterocolitica |
| Yersinia frederiksenii |
| Yersinia kristensii |
| Yersinia pestis |
| Yersinia pseudotuberculosis |
| Yersinia ruckeri |
TABLE 5.
Sensitivities and specificities of orthopox virus-MGB assays with the IWG CDC blind panel
| Instrument | TaqMan-MGB assay | Sensitivitya (%) with:
|
Specificity (%) (all samples) | ||
|---|---|---|---|---|---|
| 250 fg (1.24 × 103 genome equivalents) | 2.5 fg (12.4 genome equivalents) | All samplesb | |||
| LightCycler | pan-Orthopox virus HA | 100 | 85 | 96 | 100 |
| pan-Orthopox virus E9L3 | 100 | 85 | 96 | 100 | |
| Variola virus HA-MGB | 100 | 75 | 94 | 100 | |
| Variola virus B9R-MGB | 100 | 75 | 94 | 100 | |
| Variola virus B10R-MGB | 100 | 75 | 94 | 100 | |
| Smart Cycler | pan-Orthopox virus HA | 100 | 69 | 92 | 100 |
| pan-Orthopox virus E9L3 | 100 | 77 | 92 | 100 | |
| Variola virus HA-MGB | 100 | 0 | 75 | 100 | |
| Variola virus B9R-MGB | 88 (100) | 38 | 81 (84) | 100 | |
| Variola virus B10R-MGB | 88 (100) | 88 | 94 (97) | 100 | |
Values in parentheses are those when the result for aberrant sample 839 (Bangladesh DNA at 250 fg) is not included in the calculations.
Includes all sample amounts: 2.5 ng, 25 pg, 250 fg, and 2.5 fg.
All five assays were also tested for their ability to be quantitative on the LightCycler. A dilution series of both cloned target DNA and genomic DNA was made, and three readings of optical density at 260 nm were done to determine the concentration of each stock DNA sample. Each stock sample was then prepared as a 10-fold serial dilution from 5 × 106 to 5 gene copies per 5 μl. The dilutions were run in triplicate (cloned DNA) or duplicate (genomic DNA) with each of the five assays on the LightCycler with the cycling profile previously described. The LightCycler analysis software version 3.5.3 was used to generate linear regression curves and accompanying attributes (slope, intercept, error, and r value) for each assay. Additional data for both the pan-orthopox virus standard TaqMan and TaqMan-MGB HA assays will be published elsewhere (C. A. Whitehouse, D. A. Kulesh, M. S. Ibrahim, J. Paragas, and J. W. Huggins, submitted for publication).
RESULTS
Development of assays.
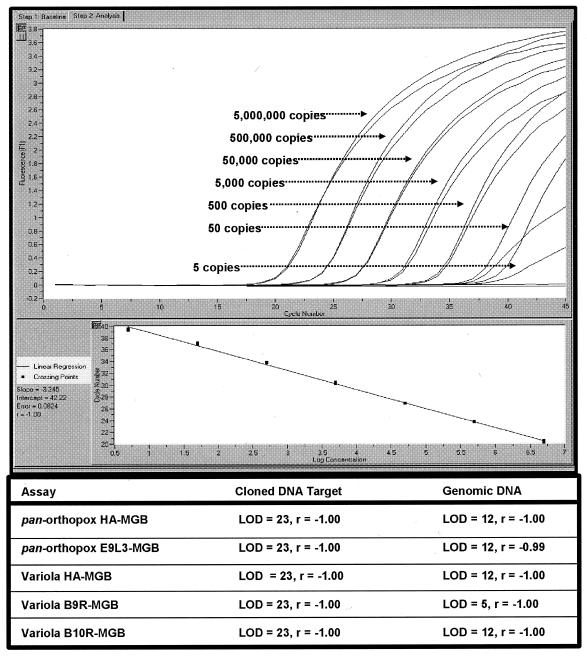
The final primer sequences, TaqMan-MGB probe sequences, and reaction conditions for each assay are shown in Table 1. The assays reproducibly detected 0.1 to 0.2 fg of plasmid DNA and 5 to 10 fg of genomic DNA, which represented approximately 12 to 25 copies of each gene, respectively. Figure 1 shows both a graph from a representative run using the B9R-MGB assay with variola virus genomic DNA samples in duplicate and each assay's LOD and dynamic range as obtained with cloned genes and purified variola virus genomic DNA on the LightCycler. The data from both plasmid and genomic DNA LOD experiments showed linear correlation with a dynamic range of six orders of magnitude, representing approximately 25 to 2,500,000 copies.
FIG. 1.
LODs and r values of linear curves for all five orthopox virus-MGB assays. The graph shown represents the results from a representative run using the B9R-MGB assay with variola virus genomic DNA samples in duplicate.
USAMRIID DNA panel evaluations.
Each assay was tested against two USAMRIID DNA panels: (i) an orthopox virus DNA panel and (ii) a DNA cross-reactivity panel. The results in Table 2 indicate that the pan-orthopox virus assays (pan-HA-MGB and pan-E9L3-MGB) were capable of detecting both the cloned and genomic DNAs of the orthopox virus species available in the USAMRIID panel, including DNA of the vaccine candidate vaccinia virus strain Ankara (which has a single internal base mismatch with the forward primer of the pan-HA-MGB assay). The results for the variola virus-specific assays (HA-MGB, B9R-MGB, and B10R-MGB) indicate that the assays were capable of differentially detecting the cloned genes of various variola virus species. Non-variola virus DNAs were not detected by the variola virus-specific assays. All five assays were negative (DNA was not detected) for various strains and isolates (78 total) in the USAMRIID bacterial cross-reactivity panel (Table 3).
OHS-IWG CDC blind DNA panel. (i) LightCycler.
Both pan-orthopox virus assays (HA-MGB and E9L3-MGB) detected every potential positive except Botswana (v73-225) and Parvin at the lowest amount (2.5 fg). All three variola virus-specific assays (HA-MGB, B9R-MGB, and B10R-MGB) also detected every potential positive except Botswana (v73-225) and Parvin at the lowest amount (2.5 fg). There were no false positives among the 15 non-orthopox virus DNA samples for any of the five assays on the LightCycler. There also were no false positives for the variola virus-specific assays with the 25 non-variola virus orthopox virus DNAs.
(ii) Smart Cycler.
For the pan-orthopox virus HA-MGB assay, the SmartCycler detected every potential positive except Botswana (v73-225), Parvin, cowpox virus (CP58), and monkeypox virus (v79-I-005), all at the lowest amount (2.5 fg). For the pan-orthopox virus E9L3-MGB assay, the Smart Cycler detected every potential positive except Botswana (v73-225), Parvin, and monkeypox virus (v79-I-005), all at the lowest amount (2.5 fg). For the variola virus-specific HA-MGB assay, no sample was detected at the 2.5-fg level; for the B9R-MGB assays, all DNAs except Parvin, Afgan (var4), Nepal (v73-225), Congo, and Botswana (v73-225) (each at 2.5 fg) and Bangladesh (250 fg) were detected. For B10R-MGB, all DNAs were detected except Parvin (2.5 fg) and Bangladesh (250 fg). Surprisingly, both the B9R-MGB and B10R-MGB assays detected the Bangladesh DNA at the 2.5-fg level. There were no false positives among the 15 non-orthopox virus DNA samples for any of the five assays on the Smart Cycler. There also were no false positives for the variola virus-specific assays with the 25 non-variola virus orthopox virus DNAs. Overall, the variola virus HA-MGB assay (and to a lesser extent the variola virus B9R-MGB assay) was significantly less sensitive on the Smart Cycler than on the LightCycler (Table 5).
Assay quantification.
Each assay was tested for its ability to predict the orthopox virus genome equivalence on the LightCycler. Results also verified the LOD for each assay for both cloned DNA and genomic DNA. The data (Fig. 1) from both the plasmid and genomic DNA experiments showed a linear correlation with a dynamic range of six orders of magnitude, representing 5,000,000 to 25 copies. In addition, several assays showed an LOD of 12 gene copies, and one assay, B9R-MGB, was able to detect 5 gene copies with genomic variola virus DNA. Similar results for cloned DNA (LODs of 12 to 25 gene copies) were obtained for the Smart Cycler (data not shown).
DISCUSSION
Smallpox vaccinations were discontinued after the eradication of the smallpox virus in the United States and elsewhere in 1979. However, recent events in the world have prompted the U.S. military to begin vaccinating troops against smallpox. The overriding fear is that even though smallpox has been eradicated for nearly two decades, the potential for a bioterrorism attack has increased substantially following the events of 11 September 2001. It is conceivable that an individual or possibly a rogue government has used their technical expertise to mass produce infectious smallpox virus (or a genetically engineered variant). It is therefore crucial that fast, reliable, yet simple diagnostic molecular tests be developed with the latest detection and identification technology available. We present the development and extended evaluation of five real-time PCR assays to either screen for orthopox virus DNA (pan-orthopox virus assays) or confirm smallpox DNA (variola virus-specific assays) on both the LightCycler and the Smart Cycler. Each assay is either an improvement on a previous real-time PCR assay (variola virus HA-MGB) (17) or an entirely new assay (pan-orthopox virus HA-MGB, pan-orthopox virus E9L3-MGB, variola virus HA-MGB, variola virus B9R-MGB, and variola virus B10R-MGB) incorporating various orthopox virus gene target sequences (4, 15, 29, 30). In order to ensure the development of the most reliable assays possible, we cloned and sequenced the appropriate genes from various species of orthopox and variola viruses. Also, because these assays were to be used in rapid-cycling machines (one cycle per 15 to 20 s), the primers were specifically designed for amplicon sizes of less than 180 bp. We chose primer-TaqMan-MGB probe pairs that exhibited the maximum efficiency of amplicon synthesis, the lowest Ct value, and the maximum LOD. We opted to use TaqMan-MGB probe technology because it possesses significantly improved hybridization properties (20). TaqMan-MGB probes are more stable, display increased mismatch discrimination, and have an improved signal-to-noise ratio due to the use of an NFQ instead of the fluorescent quencher dye TAMRA (2). In addition, the MGB stabilizes AT-rich duplexes, resulting in an increased probe Tm (that temperature at which 50% of an oligonucleotide is annealed to its complement strand). Therefore, the MGB probes simplified assay design for the orthopox viruses, which have a high A/T ratio (∼66%) (1). We also improved the original variola virus HA assay (17) by redesigning the standard TaqMan probe as a TaqMan-MGB probe, thereby reducing the probe sequence from 26 nucleotides (Tm = 60.4°C) to 18 nucleotides (Tm = 69.2°C). The four other assays were designed based on TaqMan-MGB chemistry. Each assay was designed to be as broad (pan-orthopox virus MGB) or specific (variola virus MGB) as possible, but several primer-probe mismatches have been noted (see footnotes to Table 1).
In this study, the LOD for each TaqMan-MGB assay was evaluated on the LightCycler with both the appropriately cloned gene target fragments and purified variola virus genomic DNA. The LOD with cloned gene targets was 10 to 100 atg (12 to 23 gene copies), and that with variola virus genomic DNA was 2.5 fg (12 copies). Each assay was also highly quantitative over a range of 1 ng (5 × 106) to 2.4 pg (12 copies) when tested in the LightCycler. Initial testing against DNAs (cloned genes and genomic DNA) available at USAMRIID increased our confidence in the overall specificity of each assay. While each assay detected only the appropriate DNA samples in the USAMRIID orthopox virus DNA panel, none of the assays detected any DNAs in the cross-reactivity panel. These data showed that all five of our orthopox virus TaqMan-MGB assays were both highly specific and exceptionally sensitive. In addition, it is our experience that a single internal base mismatch in a primer (as in the forward primer of the pan-HA-MGB assay with vaccinia virus strain Ankara) is tolerated more easily than any mismatches in a TaqMan-MGB probe. Therefore, the variola virus HA-MGB assay should be used with caution and in conjunction with the B9R-MGB and B10R-MGB assays when used for smallpox virus DNA verification.
At the CDC, the assays were run against a blind panel of a mixture of 68 different samples (16 different types of DNAs at four different concentrations and water). The results indicate that our assays were not only highly specific (there were no false positives with varicella-zoster virus or herpes simplex virus DNA) but also highly sensitive (most of the 2.5 fg [12 gene copies] was detected by the LightCycler). It is noteworthy that the two samples missed by all five assays on the LightCycler were the 2.5-fg (12 gene copies) amounts of a virus-cell lysate and a primary clinical crust (Parvin). Each of these samples was derived from either a cell culture lysate or a variola virus-infected tissue; thus, the sample material contained an estimated amount (probably less than 2.5 fg) of viral DNA mixed with background cellular DNA. In the Smart Cycler, the assay results were more variable (no sample was detected at 2.5 fg by the variola virus HA-MGB assay, while the other assays exhibited less sensitivity for some low-level samples [2.5 fg]). The observation that the Smart Cycler missed sample 839 (Bangladesh at 250 fg; 1,240 gene copies) but detected the lower levels of the same sample (2.5 fg; 12 gene copies) by both the B9R-MGB and B10R-MGB assays is at present unexplained. These results also confirm our previous results (unpublished data) that using a TaqMan-MGB probe instead of the standard TaqMan probe did not significantly change the sensitivity of an assay on either the LightCycler or the Smart Cycler. However, the use of TaqMan-MGB probes does increase the robustness of the signal (more fluorescence) on the LightCycler, probably because the NFQ is a better signal quencher than the TAMRA found on the standard TaqMan probes. This effect is seen only on the LightCycler. Overall, the specificity of the five assays on both platforms with all of the OHS-IWG CDC blind samples was 100%. The sensitivity for all CDC samples with the five assays on the LightCycler ranged from 93.8 to 96.2%, and that on the Smart Cycler ranged from 75 to 93.8%. The biggest difference in sensitivity was with the variola virus HA-MGB assays on the Smart Cycler. To avoid false negatives, this assay should not be run in its present configuration on the Smart Cycler if a dilute sample is suspected.
Recent world events have focused attention on the need for a smallpox (variola) animal model for testing new and improved vaccines and antiviral compounds. Jahrling et al. (unpublished data) recently developed a monkey model for studying the progression of smallpox infections. In these experiments, the quantitative pan-orthopox virus HA-MGB assay presented here has been used to monitor the viral load in monkey blood and tissues after infection with smallpox. Future studies on the preserved monkey tissues with the other four quantitative assays are planned. Further, monkeypox infections in monkeys are an important surrogate model for human smallpox infections (34). The pan-orthopox virus HA-MGB assay has been used to monitor, in real time, the viral loads in both placebo- and cidofovir-treated monkeys challenged with monkeypox virus (unpublished data).
In addition, during Operation Noble Eagle (11 September 2001 to 15 May 2002), more than 12,500 similar non-MGB pan-orthopox virus HA assays (primers identical to those in Table 1 with a slightly longer, “standard” TaqMan probe [6′FAM-AGT GCT TGG TAT AAG GAG CCC AAT TC-TAMRA-3′]) were run on numerous environmental samples in the Ruggedized Advanced Pathogen Identification Device (RAPID) (Idaho Technology) at USAMRIID. The RAPID is the improved version of the LightCycler with slightly different software. There were approximately 50 false positives (99.6% specificity), all of which were negative for orthopox virus DNA upon rescreening. The assay also detected 98.5% of all vaccinia virus DNA “concentration-positive” controls that were prepared for each batch of samples processed for real-time PCR. In addition, from 16 May to 24 September 2002, Midwest Research Institute (Rockville, Md.) assayed over 15,000 samples for pan-orthopox virus-HA PCR on the RAPID and found only two false positives (which were negative upon rescreening) (unpublished data).
In conclusion, this report demonstrates the reliable and specific identification of orthopox virus DNAs from environmental, clinical, and virus-infected cell cultures by TaqMan-MGB real-time PCR on both the LightCycler and Smart Cycler. By using the assays as a battery of PCR tests, one can easily establish the presence of orthopox virus DNA in a sample and then quickly and accurately determine whether the DNA is from the smallpox virus by using three PCR confirmatory assays.
Acknowledgments
We thank John P. Kondig for his excellent technical assistance with the sequencing work and Jeffery Garrison for his technical assistance in optimizing and testing the orthopox virus assays. We also thank Erik Henchal, Randal Schoepp, Melanie Ulrich, and Katheryn Kenyon (USAMRIID, Fort Detrick, Md.) for reviewing the manuscript, Sofi Ibrahim for access to his orthopox virus HA gene sequences, and Inger Damon, Russ Regnery, and the rest of their Pox Group at CDC for preparing the orthopox and non-orthopox virus DNA samples for the OHS-IWG assay extended evaluation.
The opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
The research described here was sponsored by the DTO CB.26.J00, U.S. Army Medical Research and Materiel Command (Plan/Contract no. DTO1-18)
REFERENCES
- 1.Afonina, I., M. Zivarts, I. Kutyavin, E. Lukhtanov, H. Gamper, and R. B. Meyer. 1997. Efficient priming of PCR with short oligonucleotides conjugated to a minor groove binder. Nucleic Acids Res. 25:2657-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonina, I. A., M. W. Reed, E. Lusby, I. G. Shishkina, and Y. S. Belousov. 2002. Minor groove binder-conjugated DNA probes for quantitative DNA detection by hybridization-triggered fluorescence. BioTechniques 32:940-949. [DOI] [PubMed] [Google Scholar]
- 3.Baker, R. O., M. Bray, and J. W. Huggins. 2003. Potential antiviral therapeutics for smallpox, monkeypox, and other orthopoxvirus infections. Antiviral Res. 57:13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaud, G. 1995. Vaccinia virus DNA replication: a short review. Biochimie 77:774-779. [DOI] [PubMed] [Google Scholar]
- 5.Belanger, S. D., M. Boissinot, C. Menard, F. J. Picard, and M. G. Bergeron. 2002. Rapid detection of Shiga toxin-producing bacteria in feces by multiplex PCR with molecular beacons on the Smart Cycler. J. Clin. Microbiol. 40:1436-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berche, P. 2001. The threat of smallpox and bioterrorism. Trends Microbiol. 9:15-18. [DOI] [PubMed] [Google Scholar]
- 7.Breman, J. G., and D. A. Henderson. 2002. Diagnosis and management of smallpox. N. Engl. J. Med. 346:1300-1308. [DOI] [PubMed] [Google Scholar]
- 8.Espy, M. J., J. R. Uhl, P. S. Mitchell, J. N. Thorvilson, K. A. Svien, A. D. Wold, and T. F. Smith. 2000. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espy, M. J., J. R. Uhl, L. M. Sloan, J. E. Rosenblatt, F. R. Cockerill III, and T. F. Smith. 2002. Detection of vaccinia virus, herpes simplex virus, varicella-zoster virus, and Bacillus anthracis DNA by LightCycler polymerase chain reaction after autoclaving: implications for biosafety of bioterrorism agents. Mayo Clin. Proc. 77:624-628. [DOI] [PubMed] [Google Scholar]
- 10.Hearps, A., Z. Zhang, and S. Alexandersen. 2002. Evaluation of the portable Cepheid SmartCycler real-time PCR machine for the rapid diagnosis of foot-and-mouth disease. Vet. Rec. 150:625-628. [DOI] [PubMed] [Google Scholar]
- 11.Henchal, E. A., and M. S. Ibrahim. 2003. Evaluation of polymerase chain reaction assays for the identifying biological agents, p. 239-249. In P. J. Stoppa and M. Bartoszcze (ed.), Rapid methods for analysis of biological materials in the environment. Kluwer Academic Publishing, Amsterdam, The Netherlands.
- 12.Henderson, D. A., T. V. Inglesby, J. G. Bartlett, M. S. Ascher, E. Eitzen, P. B. Jahrling, J. Hauer, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. Perl, P. K. Russell, K. Tonat, et al. 1999. Smallpox as a biological weapon: medical and public health management. JAMA 281:2127-2137. [DOI] [PubMed] [Google Scholar]
- 13.Higgins, J. A., M. S. Ibrahim, F. K. Knauert, G. V. Ludwig, T. M. Kijek, J. W. Ezzell, B. C. Courtney, and E. A. Henchal. 1999. Sensitive and rapid identification of biological threat agents. Ann. N.Y. Acad. Sci. 894:130-148. [DOI] [PubMed] [Google Scholar]
- 14.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard, S. T., Y. S. Chan, and G. L. Smith. 1991. Vaccinia virus homologues of the Shope fibroma virus inverted terminal repeat proteins and a discontinuous ORF related to the tumor necrosis factor receptor family. Virology 180:633-647. [DOI] [PubMed] [Google Scholar]
- 16.Ibrahim, M. S., J. J. Esposito, P. B. Jahrling, and R. S. Lofts. 1997. The potential of 5′ nuclease PCR for detecting a single-base polymorphism in Orthopoxvirus. Mol. Cell Probes. 11:143-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibrahim, M. S., D. A. Kulesh, S. S. Saleh, I. K. Damon, J. J. Esposito, A. A. Schmaljohn, and P. B. Jahrling. 2003. Real-time PCR assay to detect smallpox virus. J. Clin. Microbiol. 41:3835-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joklik, W. K., B. Moss, B. N. Fields, D. H. Bishop, and L. S. Sandakhchiev. 1993. Why the smallpox virus stocks should not be destroyed. Science 262:1225-1226. [DOI] [PubMed] [Google Scholar]
- 19.Krafft, A. E., and D. A. Kulesh. 2001. Applying molecular biological techniques to detecting biological agents. Clin. Lab Med. 21:631-660. [PubMed] [Google Scholar]
- 20.Kutyavin, I. V., I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov, A. A. Gall, R. Dempcy, M. W. Reed, R. B. Meyer, and J. Hedgpeth. 2000. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, L. G., C. R. Connell, and W. Bloch. 1993. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 21:3761-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loparev, V. N., R. F. Massung, J. J. Esposito, and H. Meyer. 2001. Detection and differentiation of Old World orthopoxviruses: restriction fragment length polymorphism of the crmB gene region. J. Clin. Microbiol. 39:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutwick, L. I., B. Goozner, and E. Bourke. 2002. Bioterrrorism: a primer for 2002. J. Assoc. Acad. Minor. Phys. 13:9-13. [PubMed] [Google Scholar]
- 24.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahy, B. W., J. W. Almond, K. I. Berns, R. M. Chanock, D. K. Lvov, R. F. Pettersson, H. G. Schatzmayr, and F. Fenner. 1993. The remaining stocks of smallpox virus should be destroyed. Science 262:1223-1224. [DOI] [PubMed] [Google Scholar]
- 26.Meyer, H., H. Neubauer, and M. Pfeffer. 2002. Amplification of ‘variola virus-specific’ sequences in German cowpox virus isolates. J. Vet. Med. B 49:17-19. [DOI] [PubMed] [Google Scholar]
- 27.Meyer, H., M. Pfeffer, and H. J. Rziha. 1994. Sequence alterations within and downstream of the A-type inclusion protein genes allow differentiation of Orthopoxvirus species by polymerase chain reaction. J. Gen. Virol. 75:1975-1981. [DOI] [PubMed] [Google Scholar]
- 28.Neubauer, H., M. Pfeffer, and H. Meyer. 1997. Specific detection of mousepox virus by polymerase chain reaction. Lab Anim. 31:201-205. [DOI] [PubMed] [Google Scholar]
- 29.Neubauer, H., U. Reischl, S. Ropp, J. J. Esposito, H. Wolf, and H. Meyer. 1998. Specific detection of monkeypox virus by polymerase chain reaction. J. Virol. Methods 74:201-207. [DOI] [PubMed] [Google Scholar]
- 30.Price, N., D. C. Tscharke, and G. L. Smith. 2002. The vaccinia virus B9R protein is a 6 kDa intracellular protein that is non-essential for virus replication and virulence. J. Gen. Virol. 83:873-878. [DOI] [PubMed] [Google Scholar]
- 31.Ropp, S. L., Q. Jin, J. C. Knight, R. F. Massung, and J. J. Esposito. 1995. PCR strategy for identification and differentiation of smallpox and other orthopoxviruses. J. Clin. Microbiol. 33:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smee, D. F., R. W. Sidwell, D. Kefauver, M. Bray, and J. W. Huggins. 2002. Characterization of wild-type and cidofovir-resistant strains of camelpox, cowpox, monkeypox, and vaccinia viruses. Antimicrob. Agents Chemother. 46:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhl, J. R., C. A. Bell, L. M. Sloan, M. J. Espy, T. F. Smith, J. E. Rosenblatt, and F. R. Cockerill III. 2002. Application of rapid-cycle real-time polymerase chain reaction for the detection of microbial pathogens: the Mayo-Roche rapid anthrax test. Mayo Clin. Proc. 77:673-680. [DOI] [PubMed] [Google Scholar]
- 34.Zaucha, G. M., P. B. Jahrling, T. W. Geisbert, J. R. Swearengen, and L. Hensley. 2001. The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis). Lab Investig. 81:1581-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]