Abstract
Background
Nicotinic acetylcholine receptors (nAChRs) play an important role as excitatory neurotransmitters in vertebrate and invertebrate species. In insects, nAChRs are the site of action of commercially important insecticides and, as a consequence, there is considerable interest in examining their functional properties. However, problems have been encountered in the successful functional expression of insect nAChRs, although a number of strategies have been developed in an attempt to overcome such difficulties. Ten nAChR subunits have been identified in the model insect Drosophila melanogaster (Dα1-Dα7 and Dβ1-Dβ3) and a similar number have been identified in other insect species. The focus of the present study is the Dα5, Dα6 and Dα7 subunits, which are distinguished by their sequence similarity to one another and also by their close similarity to the vertebrate α7 nAChR subunit.
Results
A full-length cDNA clone encoding the Drosophila nAChR Dα5 subunit has been isolated and the properties of Dα5-, Dα6- and Dα7-containing nAChRs examined in a variety of cell expression systems. We have demonstrated the functional expression, as homomeric nAChRs, of the Dα5 and Dα7 subunits in Xenopus oocytes by their co-expression with the molecular chaperone RIC-3. Also, using a similar approach, we have demonstrated the functional expression of a heteromeric ‘triplet’ nAChR (Dα5 + Dα6 + Dα7) with substantially higher apparent affinity for acetylcholine than is seen with other subunit combinations. In addition, specific cell-surface binding of [125I]-α-bungarotoxin was detected in both Drosophila and mammalian cell lines when Dα5 was co-expressed with Dα6 and RIC-3. In contrast, co-expression of additional subunits (including Dα7) with Dα5 and Dα6 prevented specific binding of [125I]-α-bungarotoxin in cell lines, suggesting that co-assembly with other nAChR subunits can block maturation of correctly folded nAChRs in some cellular environments.
Conclusion
Data are presented demonstrating the ability of the Drosophila Dα5 and Dα7 subunits to generate functional homomeric and also heteromeric nAChRs.
Background
Nicotinic acetylcholine receptors (nAChRs) are excitatory neurotransmitter receptors that are found in both vertebrate and invertebrate species. In insects, nAChRs are expressed throughout the nervous system and are the site of action for economically important insecticides such as spinosyns and neonicotinoids [1,2]. Detailed information is available concerning the structure of nAChRs, as a consequence of studies conducted with receptors purified from the electric organ of the marine ray Torpedo[3] and from X-ray crystallographic studies conducted with nAChR fragments [4] and also with the closely related acetylcholine binding protein [5]. Nicotinic receptors are assembled from five subunits arranged around a central cation-selective pore [6,7]. Conventional agonists, such as acetylcholine, activate the receptor by binding at an extracellular site located at the interface between two subunits [8], although recent evidence indicates that nAChRs can also be activated by ligands binding to an allosteric transmembrane site [9].
Ten nAChR subunits (Dα1-Dα7 and Dβ1-Dβ3) have been identified in the model insect Drosophila melanogaster and a similar number of subunits have been identified in other insect species [1,2]. Despite considerable efforts, there has been only limited success in expressing insect nAChRs in artificial expressions systems [10,11] and, where functional expression has been achieved, ion channel currents have tended to be small or have been generated in response to relatively high agonist concentrations [12-14]. Experimental approaches that have had some success in overcoming problems associated with expression of insect nAChRs include the expression of subunit chimeras containing domains from other neurotransmitter receptors [15], co-expression of insect nAChRs with vertebrate subunits [16,17] or a combination of these approaches [18]. Co-expression with vertebrate nAChR subunits is an approach that has been used in the characterization of nAChR subunits cloned from insect pest species such as the aphid Myzus persicae[19,20] and the brown planthopper Nilaparvata lugens[21,22]. However, for most insect species for which nAChRs have been cloned, there have been no reports of successful heterologous expression. This includes nAChRs cloned from the honeybee Apis mellifera[23-25], diamondback moth Plutella xylostella[26,27], house fly Musca domestica[28-30], locust Locusta migratoria[31], mosquito Anopheles gambiae[25], red flour beetle Tribolium castaneum[25,32], silkworm Bombyx mori[25,33] and tobacco hornworm Manduca sexta[34].
RIC-3 is a nAChR-associated molecular chaperone that was originally characterised in the nematode Caenorhabditis elegans[35] but has also been identified in several other species, including mammals and insects [36]. It is a transmembrane protein that is able to enhance maturation (folding and assembly) of several nAChR subtypes [36]. For example, co-expression of RIC-3 with the vertebrate nAChR α7 subunit enhances levels of functional expression in Xenopus oocytes [35] and is able to facilitate the functional expression of α7 nAChRs in mammalian cell lines that are otherwise non-permissive for expression of α7 [37,38]. In some cell types it has been found that the α7 subunit can be expressed (subunit protein can be detected) but, in the absence of RIC-3, is unable to fold into a conformation that can be detected by radioligand binding or form functional nAChRs [37,38]. In addition, some success has been achieved in overcoming difficulties associated with expression of insect nAChRs by the co-expression with RIC-3 [39,40].
The Dα5, Dα6 and Dα7 subunits of Drosophila show close sequence similarity to one another (53-63% amino acid identity [41]) and also have close similarity to the vertebrate nAChR α7 subunit (42-46% amino acid identity [42]). Of the three Drosophila subunits, Dα5 and Dα7 have the closest sequence similarity to one another and Dα6 has the highest sequence similarity to the vertebrate α7 [43]. In the present study, we report the molecular cloning of the Dα5 subunit, the only Drosophila nAChR subunit for which a full-length cDNA clone was not previously available in our laboratory. Heterologous expression studies with Dα5, Dα6 and Dα7 are described in three host cell types: Drosophila S2 cells, human tsA201 cells and Xenopus oocytes. Functional expression of several subunit combinations has been achieved in Xenopus oocytes and has enabled the pharmacological properties of recombinant nAChRs to be examined. Evidence is provided that demonstrates the ability of subunits to form both homomeric and heteromeric nAChRs. Of particular note is evidence that Dα5 can generate functional homomeric channels and that Dα7 can form both homomeric and heteromeric channels. We are not aware of any previous studies demonstrating the ability of Dα5 and Dα7 subunits to generate such recombinant nAChRs, either with subunits cloned from Drosophila or with analogous nAChR subunits from other insect species.
Results
Molecular cloning of Dα5
A full-length cDNA encoding the Drosophila nAChR Dα5 subunit was isolated from a preparation of Drosophila embryo mRNA. The Dα5 cDNA encodes an open reading frame of 807 amino acids corresponding to the previously described Dα5 isoform B [41]. In agreement with previous studies [41], the Dα5 cDNA isolated in this study contains an open reading frame encoding an unusually large N-terminal domain, extending some 300 amino acids upstream of the start methionine in most nAChR subunits.
Heterologous expression of Dα5 in Drosophila and human cell lines
The full-length coding sequence of the Dα5 cDNA was sub-cloned into the Drosophila expression vector pRmHa3 (to facilitate expression in Drosophila S2 cells) and into pRK5 (to facilitate expression in human tsA201 cells). In cells transfected with pRmHa3-Dα5 or pRK5-Dα5 alone, no evidence of specific high-affinity binding of nicotinic radioligands (125I]-α-bungarotoxin, 3 H]-epibatidine or 3 H]-methyllycaconitine) could be detected. The Dα5 subunit was also co-expressed with an extensive series of Drosophila nAChR subunit combinations. Expression studies with more than 100 different subunit combinations (containing between 2 and 10 different Drosophila nAChR subunit subtypes) have been examined in our laboratory. However, no specific binding was detected with any these combinations (in the absence of any co-expressed chaperone proteins, see later). To illustrate the extent of these studies, details of all Drosophila nAChR subunit combinations containing the Dα5 subunit are listed in Table 1. It is possible that the lack of radioligand binding is a consequence of the expressed subunit proteins failing to undergoing appropriate maturation (folding and assembly) due to a requirement for specific chaperone proteins, as has been reported for other nAChR subunits [37,38], or due to a requirement for additional nAChR subunits.
Table 1.
Radioligand binding to Drosophila nAChR subunit combinations
| Subunit combination | [125I]-α-BTX Binding | [3 H]-epibatidine binding | ||
|---|---|---|---|---|
| |
- RIC3 |
+ RIC3 |
- RIC3 |
+ RIC3 |
| Dα5 |
– |
– |
– |
– |
| Dα5/Dα1 |
– |
– |
– |
– |
| Dα5/Dα2 |
– |
– |
– |
– |
| Dα5/Dα3 |
– |
– |
– |
– |
| Dα5/Dα4 |
– |
– |
– |
– |
| Dα5/Dα6 |
– |
+ |
– |
– |
| Dα5/Dα7 |
– |
– |
– |
– |
| Dα5/Dβ1 |
– |
– |
– |
– |
| Dα5/Dβ2 |
– |
– |
– |
– |
| Dα5/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2 |
– |
– |
– |
– |
| Dα5/Dα1/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα2/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα2/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα3/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα3/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα3/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα4/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα4/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα4/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα6/Dα7 |
– |
– |
– |
– |
| Dα5/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dβ1/Dβ3 |
– |
– |
– |
– |
| Dα5/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα3/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα1/Dα3/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα3/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα4/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα1/Dα4/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα4/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα2/Dα3/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα2/Dα3/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα2/Dα3/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα2/Dα4/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα2/Dα4/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα2/Dα4/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα3/Dα4/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα3/Dα4/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα3/Dα4/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα6/Dα7/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα6/Dα7/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα6/Dα7/Dβ3 |
– |
– |
– |
– |
| Dα5/Dβ1/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα3/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα4/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα2/Dα3/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα2/Dα4/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα3/Dα4/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα3/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα4/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dβ1/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα2/Dα3/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα2/Dα4/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα2/Dβ1/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα3/Dα4/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα3/Dβ1/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dα4/Dβ1 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dα4/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dα4/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα6/Dα7/Dβ1/Dβ2/Dβ3 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dα4/Dα6/Dα7/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dα4/Dα6/Dα7/Dβ1/Dβ2 |
– |
– |
– |
– |
| Dα5/Dα1/Dα2/Dα3/Dα4/Dα6/Dα7/Dβ1/Dβ2/Dβ3 | – | – | – | – |
To illustrate the extent of radioligand binding studies undertaken, the Table lists all subunit combinations containing Dα5 that were examined in transfected Drosophila S2 cells. Binding studies were performed with [125I]-α-bungarotoxin (10 nM) and [3 H]-epibatidine (30 nM). Combinations of Drosophila nAChR subunit cDNAs were transfected in the absence or presence of RIC-3 cDNA. Data indicating presence or absence of specific binding are derived from at least 3 independent experiments.
Previous studies have shown that, when co-expressed with a vertebrate β2 subunit, some Drosophila nAChR α-subunits can generate functional recombinant nAChRs and form high-affinity binding sites for nicotinic radioligands (see for example [16,44]). However, when Dα5 was co-expressed with vertebrate β2 in Drosophila S2 cells or in human tsA201 cells, no specific radioligand binding could be detected. These findings with Dα5 are similar to those conducted previously with the closely related Drosophila Dα6 and Dα7 subunits [15]. However, in control experiments conducted in parallel, high levels of specific radioligand binding were detected after co-expression of Drosophila Dα2 and Dα3 subunits with the rat β2 (Rβ2) subunit. This is in agreement with previous studies conducted with the Dα2 + Rβ2 and Dα3 + Rβ2 subunit combinations [17,45].
Dα5/5HT3A subunit chimera
As has been described previously for the Dα6 and Dα7 subunits [15], a chimera was constructed containing the N-terminal ligand-binding domain of the Dα5 subunit fused to the transmembrane and C-terminal regions of the mouse 5-HT3A subunit (5HT3A). Despite the inability of the intact Dα5 subunit to be detected by 125I]-α-bungarotoxin binding when expressed in Drosophila S2, expression of the Dα5/5HT3A chimera resulted in high levels of cell-surface 125I]-α-bungarotoxin binding (Figure 1). These data from recombinant subunit chimeras is consistent with evidence derived from native Drosophila nAChRs that Dα5 forms part of an α-bungarotoxin binding nAChR [46]. Expression studies with the intact and chimeric Dα5 subunit indicate that, in common with the Dα6 and Dα7 subunits, inefficient folding and assembly can be attributed to domains present in the C-terminal subunit domain. Similar conclusions have also been made concerning the closely related vertebrate α7 subunit [47].
Figure 1.
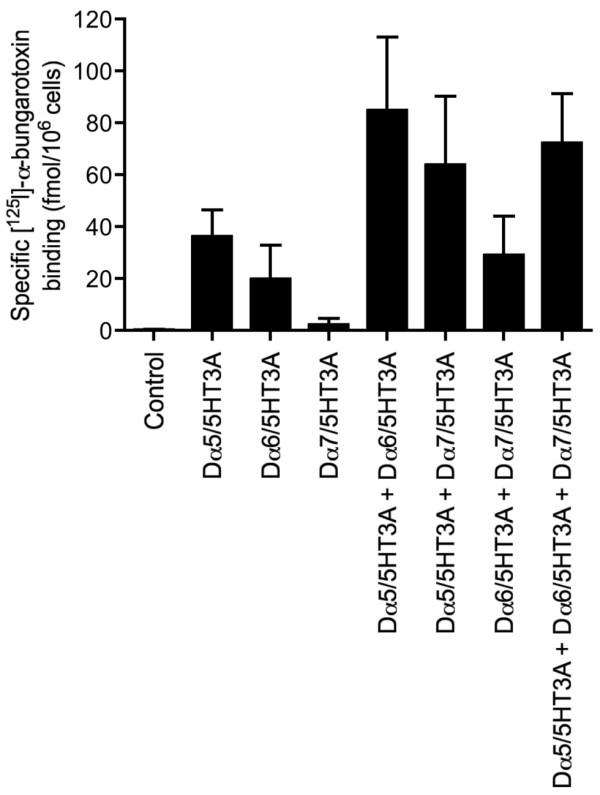
Radioligand binding to nAChR subunit chimeras expressed in Drosophila S2 cells. Cell surface [125I]-α-bungarotoxin binding to transiently transfected Drosophila S2 cells with subunit chimeras (Dα5/5HT3A, Dα6/5HT3A and Dα7/5HT3A). Experiments were performed in triplicate and are means ± SEM of 4–14 independent experiments.
The influence of co-expressing combinations of subunit chimeras was also examined. In comparisons to the level of 125I]-α-bungarotoxin binding detected with Dα5/5HT3A alone, higher levels of specific cell-surface binding were detected when the Dα5 chimera was co-expressed with other subunit chimeras (Dα6/5HT3A and Dα7/5HT3A; Figure 1). However, the levels of specific binding detected were not significantly higher than would have been expected from a possible additive effect of co-expressing these chimeras. Consequently, this data cannot be used as evidence to support the possibility of heteromeric co-assembly, as was the case previously for studies conducted with the Dα6 and Dα7 subunit chimeras [15].
Heterologous expression with RIC-3
Previous studies have demonstrated that the molecular chaperone protein RIC-3 can enhance maturation of several nAChRs [36]. This finding has prompted us to examine the influence of co-expressing Dα5 with RIC-3 in cultured cell lines, as we have done previously for other Drosophila nAChR subunits [39]. Various combinations (see Table 1 and Figure 2 for details) of Dα5, Dα6 and Dα7 were expressed with either CeRIC-3 or DmRIC-3 in both Drosophila S2 cells and human tsA201 cells. No specific cell-surface 125I]-α-bungarotoxin binding was detected when any of these subunits were expressed individually with RIC-3. However, specific binding was detected when Dα5 was co-expressed with Dα6 (Figure 2), albeit at a lower level than seen with the subunit chimeras (Figure 1). Interestingly, no specific binding was detected when these two subunits were also co-expressed with Dα7 (or other subunits; see Table 1), suggesting that Dα7 may co-assemble with Dα5 or Dα6 and, in doing so, impair receptor assembly or maturation. Similar results were obtained in both cell types examined (Figure 2), although specific binding was detected in mammalian cells only when they were cultured at a temperature lower than 37°C. As has been reported previously [15,17], lowering the temperature of transfected mammalian cells from 37°C to 25°C for 24 hours (the temperature at which Drosophila S2 cells are maintained) facilitates receptor assembly and cell-surface expression. As has been discussed previously with respect to insect nAChR subunits [15,17], the detection of specific radioligand binding only in mammalian cells cultured at 25°C is likely to be a consequence of more efficient subunit folding and assembly at lower temperatures.
Figure 2.
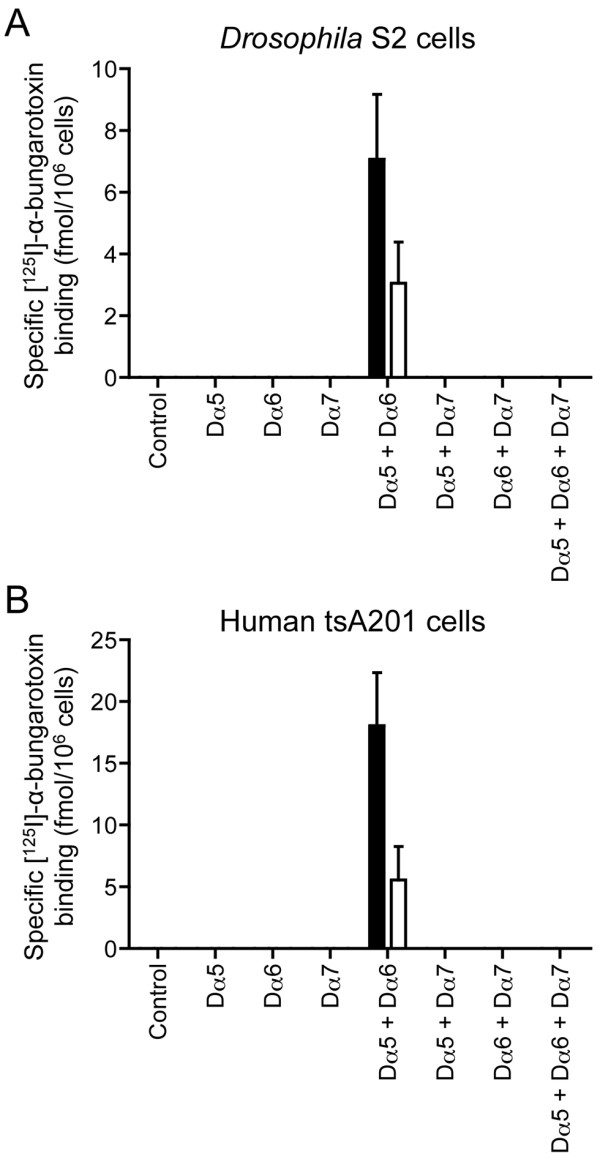
Radioligand binding to Drosophila nAChR subunit combinations in cultured cell lines. Cell surface [125I]-α-bungarotoxin binding to cell lines transiently transfected with combinations of Dα5, Dα6 and Dα7 subunits. In all cases, subunit combinations were co-transfected with either CeRIC-3 (filled bars) or DmRIC-3 (open bars). No specific binding was detected for any subunit combination in the absence of RIC-3 (not shown). Data are presented for Drosophila S2 cells (A) and for human tsA201 cells cultured at 25 °C (B). Controls represent mock-transfected cells. Experiments were performed in triplicate and are means ± SEM of 5–8 independent experiments.
Expression in Xenopus oocytes
Xenopus oocytes were injected with cRNA encoding various combinations of the Drosophila nAChR subunits Dα5, Dα6 and Dα7. No evidence of functional expression was detected for any subunit combination in the absence of co-expressed RIC-3. However, when co-expressed with CeRIC-3, functional responses to acetylcholine were detected in oocytes injected with either the Dα5 or the Dα7 subunit, indicating the presence of functional homomeric Dα5 and Dα7 nAChRs (Figure 3). However, even when co-expressed with RIC-3, functional expression was somewhat inconsistent, being observed in some but not all batches of oocytes tested (responses greater than 5 nA were observed in only about a third of the oocyte batches tested). Dose–response curves indicate that acetylcholine has a similar EC50 for these two homomeric receptor subtypes (8.8 ± 2.5 μM 6.7 ± 1.7 μM, respectively). In contrast, no functional expression was detected when Dα6 was co-expressed with CeRIC-3.
Figure 3.
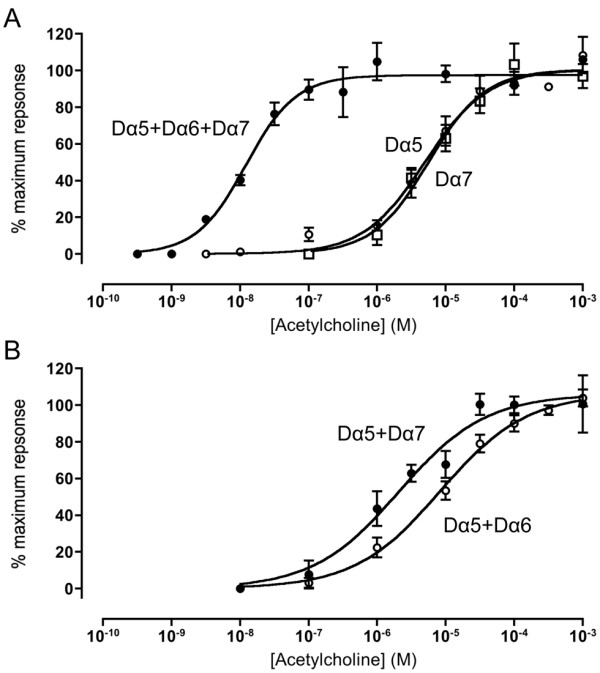
Functional expression of Drosophila nAChR subunit combinations in Xenopus oocytes. A) Dose–response curves for acetylcholine are shown for homomeric Dα5 nAChRs (open circles) homomeric Dα7 nAChRs (open squares) and for triplet Dα5 + Dα6 + Dα7 nAChRs (closed circles). B) Dose–response curves for acetylcholine are shown for heteromeric Dα5 + Dα6 nAChRs (open circles) and Dα5 + Dα7 nAChRs (closed circles) In all cases, nAChR subunits were co-expressed with CeRIC-3. Data are means ± SEM of 3–8 independent experiments.
Expression of pairwise subunit combinations (with CeRIC-3) gave dose–response curves that were not significantly different (P > 0.05) to that of homomeric Dα5 or Dα7 nAChRs (Table 2 and Figure 3). Consequently, it was not possible to conclude whether pairwise heteromeric receptors were expressed. One pairwise combination (Dα6 + Dα7) failed to generate consistent responses, an indication that co-assembly of Dα6 with Dα7 blocks formation of functional nAChRs in oocytes. However, when all three subunits (Dα5, Dα6 and Dα7) were co-expressed with CeRIC-3, dose–response data indicated a single population of receptors with a significantly higher (ANOVA, P < 0.05; Student’s t-test P < 0.01) apparent affinity for acetylcholine (13.5 ± 1.7 nM; Figure 3) than that of either of the two homomeric nAChRs (Dα5 or Dα7) or any of the putative pairwise subunit combinations (Table 2). For all subunit combinations examined (see Table 2), functional responses to acetylcholine were completely blocked by a 10 min pre-incubation with 100nM α-bungarotoxin. This block was completely reversible but occurred on a slow timescale, recovery taking, typically, about 15 minutes (Figure 4). No significant differences were observed in pharmacological properties when nAChRs were co-expressed with DmRIC-3 [39], rather than CeRIC-3 (data not shown).
Table 2.
Functional properties of recombinant nAChRs expressed in Xenopus oocytes
| Subunits | EC 50(μM or nM)* | Hill slope | n ** | I max[ I mean] † (nA) |
|---|---|---|---|---|
| Dα5 |
8.8 ± 2.5 μM |
1.1 ± 0.3 |
6 |
200 [141 ± 25] |
| Dα6 |
– |
– |
‡ |
– |
| Dα7 |
6.7 ± 1.7 μM |
1.0 ± 0.3 |
4 |
86 [45 ± 13] |
| Dα5 + Dα6 |
8.6 ± 2.4 μM |
1.0 ± 0.1 |
5 |
47 [20 ± 8] |
| Dα5 + Dα7 |
1.6 ± 0.3 μM |
1.0 ± 0.1 |
3 |
53 [39 ± 8] |
| Dα6 + Dα7 |
– |
– |
‡ |
– |
| Dα5 + Dα6 + Dα7 | 13.5 ± 1.7 nM* | 1.2 ± 0.3 | 6 | 150 [107 ± 12] |
Note, in all cases, nAChR subunits were co-expressed with CeRIC-3.
* Note, EC50 value for Dα5 + Dα6 + Dα7 is expressed as nM, rather than μM.
**EC50 and Hill slopes are means ± SEM of separate fits to dose–response curves derived from independent experiments (n = 3–6).
† Relatively small currents were detected with all subunit combinations, as indicated by the size of the maximum current that was detected with each subunit combination (Imax) and the mean maximum current (Imean) from between 6–20 different oocytes.
‡ Subunit combinations that failed to generate functional responses are indicated by a dash. This is based on studies conducted with at least 5 batches of oocytes that generated functional nAChRs with other subunit combinations and at least 10 oocytes from each batch (n > 50).
Figure 4.
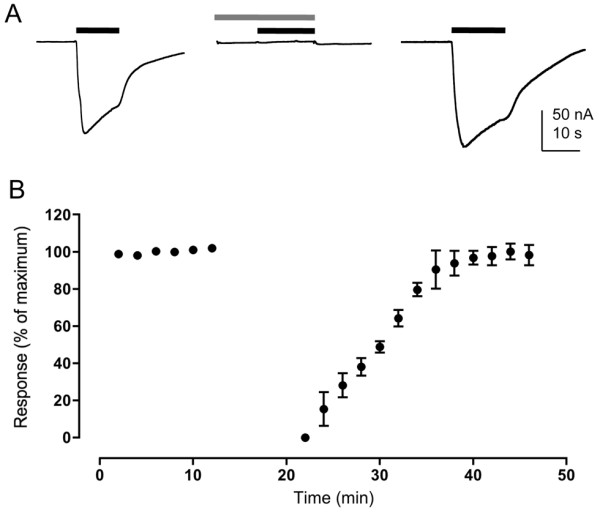
Antagonism of Dα5 nAChRs by α-bungarotoxin. A) Representative responses to acetylcholine (100 μM; black bar) are shown (left), together with block after a 10 min pre-incubation with α-bungarotoxin (100 nM; grey bar) (middle). Recovery from α-bungarotoxin block after 10 minutes is also illustrated (right). Data shown are for homomeric Dα5 nAChRs (co-expressed with CeRIC-3) but the results (complete block with full recovery) were observed for all subunit combinations that generated functional nAChRs (see Table 2). B) Data indicating the time course for recovery after a 10 min incubation with α-bungarotoxin (100 nM; illustrated by the grey bar). Data points are normalised to the maximum response prior to block by α-bungarotoxin and are means ± SEM of 3 independent experiments.
Discussion
The Dα5, Dα6 and Dα7 subunits differ from other Drosophila nAChR subunits in their close sequence similarity to the vertebrate α7 nAChR subunit [41,48], a subunit that is notable for its ability to form both homomeric and heteromeric nAChRs [49-52]. In addition to being one of the best characterised homomeric nAChRs, the vertebrate α7 subunit can co-assemble into heteromeric nAChRs by co-assembly with the α8 subunit (in avian speices) [50]. Although an α8 subunit is not present in mammals, recent evidence indicates that the mammalian α7 subunit can also form functional heteromeric nAChRs by co-assembly with β2 [51,52].
Relatively limited information is available about the physiological roles of the Dα5, Dα6 and Dα7 subunits in Drosophila, or about the role of analogous subunits in other insect species. There is, however, evidence from studies of native nAChRs in Drosophila that Dα5 forms part of a nAChR that is sensitive to α-bungarotoxin [46], Dα6 forms part of the spinosad-sensitive nAChR [53] and that Dα7 is required for the visually-mediated cholinergic escape response [54].
As has been discussed elsewhere [10,11], difficulties have been encountered in the efficient functional expression of insect nAChRs. Here we report the cloning of a full-length cDNA of the Drosophila Dα5 subunit corresponding to a previously described isoform B [41]. Other isoforms of Dα5 described previously (isoforms A and C) [41] are a consequence of alternative splicing and have fewer exons than isoform B. Isoform A lacks exon 7, which codes for part of the second transmembrane domain, whilst isoform C lacks exon 5, which codes for the region containing the extracellular Cys-loop. The cloning of the Dα5 subunit was first reported in 2002 [41] but no expression studies were described at that time. More recently, it has been reported that Dα5 does not generate functional homomeric nAChRs when expressed in Xenopus oocytes, even when co-expressed with RIC-3 [40]. Functional expression was, however, reported in the same study when Dα5 was co-expressed with Dα6 and RIC-3 [40]. In the present study, we have detected functional responses when Dα5 is co-expressed with Dα6 but, in contrast to the previous study [40], we have also obtained evidence for the functional expression of homomeric Dα5 nAChRs. Similarly, we have demonstrated that Dα7 can form both homomeric and heteromeric nAChRs. As far as we are aware, there have been no previous reports of the successful functional expression of Dα7, as either a homomeric or a heteromeric nAChR. Given the difficulties encountered in obtaining reproducible functional expression of insect recombinant nAChRs, it is not surprising that there may be some apparent differences in subunit combinations found to generate functional receptors in this and previous studies, particularly since the focus of the most detailed previous study was the identification of a spinosyn-sensitive nAChRs [40].
Our studies conducted in cell lines provided evidence that the pairwise combination Dα5 + Dα6 generates a high affinity radioligand binding site, a finding that agrees with previous studies demonstrating functional expression of Dα5 + Dα6 nAChRs in oocytes [40]. Interestingly, we have found no evidence of specific binding when Dα7 was co-expressed with Dα5 and Dα6 in the same cell lines. This lack of specific binding would seem to suggest that, in the two cell lines examined, co-assembly of Dα7 with the Dα5 and Dα6 subunit interferes with the formation of correctly assembled complexes. We observed a somewhat similar situation in oocytes, where expression of Dα7 alone generates functional nAChRs but it fails to do so when co-expressed with Dα6. This may reflect a tendency for Dα6 and Dα7 to assemble into non-functional complexes. The one situation where this tendency is not dominant is when all three subunits (Dα5 + Dα6 + Dα7) are co-expressed with RIC-3 in oocytes, where they are able to form a functional ‘triplet’ nAChR with high apparent affinity for acetylcholine.
The present findings suggest that the environment provided by the host cell exerts a substantial effect on the assembly of these nAChR subtypes, a phenomena that has been reported previously for other nAChRs [47,55,56]. Previous studies by another research group [40] support the conclusion that co-assembly of Dα5 + Dα6 nAChRs is somewhat inefficient. Not only was functional expression of the Dα5 + Dα6 subunit combination found to be inconsistent in the previous study, but it also appeared to be dependent on the ratio of cRNAs injected [40]. Perhaps this inconsistent functional expression reflects a tendency for some subunit combinations to assemble into non-functional complexes and that this may be more prevalent in certain subunit stoichiometries. It is possible that, in the native cellular environment, factors determining efficiency of subunit assembly and maturation may differ, perhaps as a consequence of a different array of endogenous chaperone proteins. This conclusion is supported by previous studies that have indicated that influence of RIC-3 on maturation of nAChRs is influenced by the host cell [39] and may help to explain the differences that we have observed in the ability of some subunit combinations to assemble into nAChRs in different expression systems.
The data obtained from expression studies in Drosophila and human cell lines is broadly similar. However, successful expression in human cells required incubation at a temperature lower than they would normally be maintained at (25°C, rather than 37°C) [note: Drosophila S2 cells are routinely maintained at 25°C]. Previous studies have demonstrated that the folding and assembly of the nAChRs from insects [17] and from some other non-insect species, such as the cold-water ray Torpedo[57], can be influenced by temperature. This temperature dependence appears to be a consequence of inefficient protein folding and/or subunit assembly at higher temperatures. Previously, due to difficulties in expression of Dα6 and Dα7 nAChR subunits, we examined the ability of subunit chimeras to assemble into complexes capable of binding 125I]-α-bungarotoxin [15]. From such studies, it was possible to conclude that the Dα6 and Dα7 subunits were capable of heterometic co-assembly. In the present study the data from subunit chimeras is less clear cut. Although higher levels of 125I]-α-bungarotoxin were seen consistently when the Dα5 chimera was co-expressed with either the Dα6 and Dα7 chimeras, it was not clear in all cases whether this was greater than an additive effect. Nevertheless these findings are consistent with the conclusion that Dα5 is able to co-assemble into heteromeric complexes. For all subunit combinations examined, responses to acetylcholine were completely blocked by α-bungarotoxin, a finding that is consistent with previous studies conducted with native nAChRs purified from Drosophila which demonstrated that Dα5 is part of an α-bungarotoxin binding nAChR [46].
As mentioned above, a previous study has reported the functional expression of heteromeric Dα5 + Dα6 nAChRs (co-expressed with RIC-3) in Xenopus oocytes and also the inability of either Dα5 or Dα6 to form functional homomeric nAChRs [40]. Significantly, the authors of this earlier study describe substantial difficulties in achieving reliable functional expression. In the present study, despite demonstrating the functional expression of several combinations of the Dα5, Dα6 and Dα7 subunits, we have also encountered a much lower success rate than is typically achieved with other nAChRs. In both transfected cell lines and in Xenopus oocytes, we occasionally failed to detect evidence of radioligand binding or functional expression, despite success with other nAChRs that were expressed as positive controls (for example the mammalian α7 nAChR). The difficulties that we and others have encountered may be associated with a tendency for these subunits to co-assemble into non-functional complexes. It is possible that this may reflect a requirement for additional chaperone proteins. Indeed, a study conducted with a C. elegans nAChR has demonstrated a requirement for three different chaperone proteins for efficient functional heterologous expression [58].
Conclusions
In summary, whereas it has been reported previously that Dα5 and Dα6 can form a functional heteromeric nAChR (albeit inefficiently) when expressed in Xenopus oocytes [40], this is the first evidence that either Dα5 or Dα7 can form functional homomeric nAChRs. It is also the first demonstration that Dα7 can form a functional heteromeric nAChR. Of particular interest is the evidence that the three subunits examined in this study can co-assemble to form a functional triplet (Dα5 + Dα6 + Dα7) nAChR with a high apparent affinity for acetylcholine.
Methods
Plasmids and cRNA synthesis
Subcloning of Drosophila nAChR subunit cDNAs Dα1(ALS), Dα2(SAD), Dα3, Dα4, Dα6, Dα7, Dβ1(ARD), Dβ2(SBD) and Dβ3 [alternative subunit nomenclature in parenthesis] into expression vectors pRmHa3 and pRK5 has been described previously [15,17,59,60]. Construction and subcloning of Dα6/5HT3A and Dα7/5HT3A subunit chimeras has also been described previously [15]. For expression studies in Xenopus oocytes, subunit cDNAs were subcloned into pGEMHE [61] downstream of the SP6 promoter. Plasmid constructs (pGEMHE) containing nAChR subunit cDNAs were linearized with NheI and purified with QIAQuik PCR purification kit (Qiagen). In vitro synthesis of cRNA was performed using mMessage mMachine SP6 transcription kit (Ambion). C. elegans RIC-3 (CeRIC-3) cDNA [35] was provided by Millet Treinin (Hebrew University, Israel). The D. melanogaster RIC-3 (DmRIC-3) cDNA used in this study corresponds to the previously described splice variant DmRIC-37a,9[39].
Molecular cloning of Dα5
Oligonucleotide primers were synthesized which correspond to the predicted 5′ and 3′ untranslated regions of transcript CG32975 identified by the GadFly Drosophila genome annotation project (flybase.org). A first-strand cDNA synthesis kit (G.E. Healthcare) was used to isolate cDNA from Drosophila melanogaster embryo Poly A + RNA (Clontech). A 2425 bp fragment was amplified using KOD hot start polymerase (Novagen) and was subcloned into plasmid pCRII (Invitrogen). The cDNA construct was sequenced and found to correspond to the previously identified isoform B [41] [note: isoforms A and C show alternative splicing and have fewer exons than isoform B]. The cDNA fragment was subcloned into the EcoRI site of pRmHa3 and pGEMHE to create pRmHa3-Dα5 and pGEMHE-Dα5.
Construction of Dα5/5HT3A chimera
To construct a Dα5/5HT3A chimera, a similar approach was used to that described previously for the Drosophila Dα6 and Dα7 subunits [15] and for mammalian nAChR subunits [47,62]. A BclI site was introduced into pRmHa3-Dα5 by means of the QuikChange site-directed mutagenesis method (Stratagene) at a position equivalent to V201 in the previously described mammalian α7/5HT3A chimera [62]. The C-terminal region of Dα5 was removed by digestion with BclI and SmaI and the corresponding region of the mouse 5HT3A subunit [63] ligated to create the construct pRmHa3-Dα5/5HT3A. The chimeric cDNA was subcloned into plasmid pRK5 by excising the construct from pRmHa3 with restriction enzymes EcoRI and XbaI to create pRK5-Dα5/5HT3A.
Heterologous expression in cultured cell lines
Schneider’s Drosophila S2 cells [64] were obtained from Dr Thomas Bunch, University of Arizona, and grown in Shields and Sang M3 medium (Sigma) containing 12.5% heat inactivated foetal calf serum (First Link), 100U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) at 25°C. Exponentially growing S2 cells were transfected by a modified calcium phosphate method as described previously [65]. Cells were transiently transfected with plasmid pRmHa3 were induced by the addition of CuSO4 (0.6 mM) for 24 h. Human kidney tsA201 cells [66] were obtained from Dr William Green, University of Chicago, and cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% foetal calf serum (First Link) and 100U/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were maintained in a humidified incubator containing 5% CO2 at 37°C. Cells were transfected using Effectene (Qiagen) according to the manufacturer’s instructions and incubated overnight at 37°C. To facilitate efficient folding and assembly of Drosophila nAChR subunits, cells were incubated at 25°C for a further 24 hours, before being assayed for radioligand binding.
Radioligand binding
3 H]-epibatidine (56.31 Ci/mmol) and 125I]-α-bungarotoxin (2200 Ci/mmol) were purchased from Perkin Elmer. 3 H]-methyllycaconitine (100 Ci/mmol) was purchased from American Radiolabeled Chemicals. Radioligand binding to transiently transfected S2 or tsA201 cells (both whole cell or membrane preparations) using tritiated ligands has been described previously [45]. Samples were assayed by filtration onto Whatman GF/B filters pre-soaked in 0.5% polyethylenimine (PEI) followed by rapid washing using a Brandel cell harvester. Samples assayed using the ligand 125I]-α-bungarotoxin were incubated in buffer containing 0.5% BSA and harvested onto Whatman GF/A filters pre-soaked in 0.5% PEI, as described previously [15]. Preliminary experiments were carried out to ensure that incubation times were long enough to enable radioligand binding to reach equilibrium. Amounts of total cellular protein were determined by a Bio-Rad DC protein assay using bovine serum albumin standards.
Oocyte electrophysiology
Adult female Xenopus laevis frogs were obtained from the European Xenopus Resource Centre (University of Portsmouth). Oocytes were isolated and defolliculated as described previously [67] following procedures that have been approved by both UCL’s Biological Services Management Group and the UK Home Office (under licences PIL70/23585 and PPL70/06819). For heterologous expression, cRNA (6–12 ng) was injected into the oocyte cytoplasm in a volume of 32.2 nl, using a Nanoject II microinjector (Drummond Scientific). Experiments were performed, typically, 2–4 days after injection of oocytes. Two electrode voltage-clamp recordings (with the oocyte membrane potential held at -60 mV) were performed essentially as described previously [67], using a Warner Instruments OC-725 C amplifier (Harvard Apparatus), PowerLab 8SP and Chart 5 software (AD Instruments). Agonists were applied to oocytes using a BPS-8 solenoid valve solution exchange system (ALA Scientific), controlled by Chart software. Data were analyzed using GraphPad Prism software. For multiple comparisons, statistical significance was determined by ANOVA with Tukey’s post-hoc test. Additional pair-wise comparisons were performed by Student’s t-test.
Abbreviations
nAChR, nicotinic acetylcholine receptor.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SJL performed the experimental work (molecular biological, pharmacological and electrophysiological), interpreted the data and helped to write the manuscript. TC performed some of the molecular biological work. JG was involved in planning experiments and assisted in interpretation of the data. NSM designed the study, assisted in interpreting the data and helped to write the manuscript. All authors read and approved the final manuscript.
Contributor Information
Stuart J Lansdell, Email: s.lansdell@ucl.ac.uk.
Toby Collins, Email: tobythrower@aol.com.
Jim Goodchild, Email: jim.goodchild@syngenta.com.
Neil S Millar, Email: n.millar@ucl.ac.uk.
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) via an Industrial Partnership Award grant (BB/G009392/1) that was co-funded by Syngenta. TC was supported by a BBSRC doctoral training account PhD studentship.
References
- Millar NS, Denholm I. Nicotinic acetylcholine receptors: targets for commercially important insecticides. Invert Neurosci. 2007;7:53–66. doi: 10.1007/s10158-006-0040-0. [DOI] [PubMed] [Google Scholar]
- Jones AK, Brown AM, Sattelle DB. Insect nicotinic acetylcholine receptor gene families: from genetic model organisms to vector, pest and beneficial species. Invert Neurosci. 2007;7:67–73. doi: 10.1007/s10158-006-0039-6. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Dellisanti CD, Yao Y, Stroud JC, Wang Z-Z, Chen L. Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94Å resolution. Nature Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Taly A, Corringer P-J, Guedin D, Lestage P, Changeux J-P. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nature Rev Drug Discovery. 2009;8:733–750. doi: 10.1038/nrd2927. [DOI] [PubMed] [Google Scholar]
- Albuquerque EX, Pereira FR, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias HR. Localization of agonist and competitive antagonist binding sites on nicotinic acetylcholine receptors. Neurochem Int. 2000;36:595–645. doi: 10.1016/s0197-0186(99)00154-0. [DOI] [PubMed] [Google Scholar]
- Gill JK, Savolainen M, Young GT, Zwart R, Sher E, Millar NS. Agonist activation of α7 nicotinic acetylcholine receptors via an allosteric transmembrane site. Proc Natl Acad Sci USA. 2011;108:5867–5872. doi: 10.1073/pnas.1017975108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar NS. Heterologous expression of mammalian and insect neuronal nicotinic acetylcholine receptors in cultured cell lines. Biochem Soc Trans. 1999;27:944–950. doi: 10.1042/bst0270944. [DOI] [PubMed] [Google Scholar]
- Millar NS. A review of experimental techniques used for the heterologous expression of nicotinic acetylcholine receptors. Biochem Pharmacol. 2009;78:766–776. doi: 10.1016/j.bcp.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Marshall J, Buckingham SD, Shingai R, Lunt GG, Goosey MW, Darlison MG, Sattelle DB, Barnard EA. Sequence and functional expression of a single α subunit of an insect nicotinic acetylcholine receptor. EMBO J. 1990;9:4391–4398. doi: 10.1002/j.1460-2075.1990.tb07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawruk E, Schloss P, Betz H, Schmitt B. Heterogeneity of Drosophila nicotinic acetylcholine receptors: SAD, a novel developmentally regulated α-subunit. EMBO J. 1990;9:2671–2677. doi: 10.1002/j.1460-2075.1990.tb07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgard F, Fraser SP, Katkowska MJ, Djamgoz MBA, Dunbar SJ, Windass JD. Cloning and functional characterisation of two novel nicotinic acetylcholine receptor α subunits from the insect pest Myzus persicae. J Neurochem. 1998;71:903–912. doi: 10.1046/j.1471-4159.1998.71030903.x. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Millar NS. Molecular characterisation of Dα6 and Dα7 nicotinic acetylcholine receptor subunits from Drosophila: formation of a high-affinity α-bungarotoxin binding site revealed by expression of subunit chimeras. J Neurochem. 2004;90:479–489. doi: 10.1111/j.1471-4159.2004.02499.x. [DOI] [PubMed] [Google Scholar]
- Bertrand D, Ballivet M, Gomez M, Bertrand S, Phannavong B, Gundelfinger ED. Physiological properties of neuronal nicotinic receptors reconstituted from the vertebrate β2 subunit and Drosophila α subunits. Eur J Neurosci. 1994;6:869–875. doi: 10.1111/j.1460-9568.1994.tb00997.x. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Schmitt B, Betz H, Sattelle DB, Millar NS. Temperature-sensitive expression of Drosophila neuronal nicotinic acetylcholine receptors. J Neurochem. 1997;68:1812–1819. doi: 10.1046/j.1471-4159.1997.68051812.x. [DOI] [PubMed] [Google Scholar]
- Bass C, Lansdell SJ, Millar NS, Schroeder I, Turberg A, Field LM, Williamson MS. Molecular characterisation of nicotinic acetylcholine receptor subunits from the cat flea, Ctenocephalides felis (Siphonaptera: Pulicidae) Insect Biochem Mol Biol. 2006;36:86–96. doi: 10.1016/j.ibmb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Huang Y, Williamson MS, Devonshire AL, Windass JD, Lansdell SJ, Millar NS. Molecular characterization and imidacloprid selectivity of nicotinic acetylcholine receptor subunits from the peach-potato aphid Myzus persicae. J Neurochem. 1999;73:380–389. doi: 10.1046/j.1471-4159.1999.0730380.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Williamson MS, Devonshire AL, Windass JD, Lansdell SJ, Millar NS. Cloning, heterologous expression and co-assembly of Mpβ1, a nicotinic acetylcholine receptor subunit from the aphid Myzus persicae. Neurosci Lett. 2000;284:116–120. doi: 10.1016/s0304-3940(00)00969-1. [DOI] [PubMed] [Google Scholar]
- Liu Z, Williamson MS, Lansdell SJ, Denholm I, Han Z, Millar NS. A nicotinic acetylcholine receptor mutation conferring target-site resistance to imidacloprid in Nilaparvata lugens (brown planthopper) Proc Natl Acad Sci USA. 2005;102:8420–8425. doi: 10.1073/pnas.0502901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Williamson MS, Lansdell SJ, Han Z, Denholm I, Millar NS. A nicotinic acetylcholine receptor mutation (Y151S) causes reduced agonist potency to a range of neonicotinoid insecticides. J Neurochem. 2006;99:1273–1281. doi: 10.1111/j.1471-4159.2006.04167.x. [DOI] [PubMed] [Google Scholar]
- Thany SH, Lenaers G, Crozatier M, Armengaud C, Gauthier M. Identification and localization of the nicotinic acetylcholine receptor alpha3 mRNA in the brain of the honeybee, Apis mellifera. Insect Mol Biol. 2003;12:255–262. doi: 10.1046/j.1365-2583.2003.00409.x. [DOI] [PubMed] [Google Scholar]
- Thany SH, Crozatier M, Raymond-Delpech V, Gauthier M, Lenaers G. Apisα2, Apisα7-1 and Apisα7-2: three new neuronal nicotinic acetylcholine receptor α-subunits in the honeybee brain. Gene. 2005;344:125–132. doi: 10.1016/j.gene.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Jin Y, Tian N, Cao J, Liang J, Yang Z, Lv J. RNA editing and alternative splicing of the insect nAChR subunit alpha6 transcript: evolutionary conservation, divergence and regulation. BMC Evolution Biol. 2007;7:98. doi: 10.1186/1471-2148-1187-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SW, Chen M, Dawson A, Zhao J-Z, Vogel H, Shelton AM, Heckel DG, Jiggins CD. Mis-spliced transcripts of nicotinic acetylcholine receptor α6 are associated with field evolved spinosad resistance in Plutella xylostells (L.) PLoS Genetics. 2010;6:e1000802. doi: 10.1371/journal.pgen.1000802. 1000810.1000137/journal.pgen.1000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich FD, Chen M, Shelton AM, Scott JG. Transcripts of the nicotinic acetylcholine receptor subunit gene Pxylα6 with premature stop codons are associated with spinosad resistance in diamondback moth, Plutella xylostella. Invert Neurosci. 2010;10:25–33. doi: 10.1007/s10158-010-0102-1. [DOI] [PubMed] [Google Scholar]
- Gao J-R, Deacutis JM, Scott JG. The nicotinic acetylcholine receptor subunit Mdα5 and Mdβ3 on autosome 1 of Musca domestica are not involved in spinosad resistance. Insect Mol Biol. 2007;16:691–701. doi: 10.1111/j.1365-2583.2007.00770.x. [DOI] [PubMed] [Google Scholar]
- Gao J-R, Deacutis JM, Scott JG. Characterization of the nicotinic acetylcholine receptor gene Mdα2 from the house fly Musca domestica. Arch Insect Biochem Physiol. 2007;64:30–42. doi: 10.1002/arch.20158. [DOI] [PubMed] [Google Scholar]
- Gao J-R, Deacutis JM, Scott JG. The nicotinic acetylcholine receptor subunit Mdα6 from Musca domestica is diversified via post-transcriptional modification. Insect Mol Biol. 2007;16:325–334. doi: 10.1111/j.1365-2583.2007.00730.x. [DOI] [PubMed] [Google Scholar]
- Hermsen B, Stetzer E, Thees R, Heiermann R, Schrattenholz A, Ebbinghaus U, Kretschmer A, Methfessel C, Reinhardt S, Maelicke A. Neuronal nicotinic receptors in the Locust Locusta migratoria: cloning and expression. J Biol Chem. 1998;273:18394–18404. doi: 10.1074/jbc.273.29.18394. [DOI] [PubMed] [Google Scholar]
- Rinkevich FD, Scott JG. Transcriptional diversity and allelic variation in nicotinic acetylcholine receptor subunits of the red flour beetle, Tribolium castaneum. Insect Mol Biol. 2009;18:233–242. doi: 10.1111/j.1365-2583.2009.00873.x. [DOI] [PubMed] [Google Scholar]
- Shao Y-M, Dong K, Zhang C-X. The nicotinic acetylcholine receptor gene family of the silkworm. Bombyx mori. BMC Genomics. 2007;8:324. doi: 10.1186/1471-2164-8-324. 310.1186/1471-2164-1188-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastham HM, Lind RJ, Eastlake JL, Clarke BS, Towner P, Reynolds SE, Wolstenholme AJ, Wonnacott S. Characterization of a nicotinic acetylcholine receptor from the insect Manduca sexta. Eur J Neurosci. 1998;10:879–889. doi: 10.1046/j.1460-9568.1998.00095.x. [DOI] [PubMed] [Google Scholar]
- Halevi S, McKay J, Palfreyman M, Yassin L, Eshel M, Jorgensen E, Treinin M. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 2002;21:1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar NS. RIC-3: a nicotinic acetylcholine receptor chaperone. Br J Pharmacol. 2008;153:S177–S183. doi: 10.1038/sj.bjp.0707661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Burton B, Urrutia A, Shcherbatko A, Chavez-Noriega LE, Cohen CJ, Aiyar J. Ric-3 promotes functional expression of the nicotinic acetylcholine receptor α7 subunit in mammalian cells. J Biol Chem. 2005;280:1257–1263. doi: 10.1074/jbc.M410039200. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Gee VJ, Harkness PC, Doward AI, Baker ER, Gibb AJ, Millar NS. RIC-3 enhances functional expression of multiple nicotinic acetylcholine receptor subtypes in mammalian cells. Mol Pharmacol. 2005;68:1431–1438. doi: 10.1124/mol.105.017459. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Collins T, Yabe A, Gee VJ, Gibb AJ, Millar NS. Host-cell specific effects of the nicotinic acetylcholine receptor chaperone RIC-3 revealed by a comparison of human and Drosophila RIC-3 homologues. J Neurochem. 2008;105:1573–1581. doi: 10.1111/j.1471-4159.2008.05235.x. [DOI] [PubMed] [Google Scholar]
- Watson GB, Chouinard SW, Cook KR, Geng C, Gifford JM, Gustafson GD, Hasler JM, Larrinua IM, Letherer TJ, Mitchell JC. et al. A spinosyn-sensitive Drosophila melanogaster nicotinic acetylcholine receptor identified through chemically induced target site resistance, resistance gene identification, and heterologous expression. Insect Biochem Mol Biol. 2010;40:376–384. doi: 10.1016/j.ibmb.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Grauso M, Reenan RA, Culetto E, Sattelle DB. Novel putative nicotinic acetylcholine receptor subunit genes, Dα5, Dα6 and Dα7, in Drosophila melanogaster identify a new and highly conserved target of adenosine deaminase acting on RNA-mediated A-to-I pre-mRNA editing. Genetics. 2002;160:1519–1533. doi: 10.1093/genetics/160.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Sattelle DB. Diversity of insect nicotinic acetylcholine receptor subunits. Adv Exp Med Biol. 2009;683:25–43. doi: 10.1007/978-1-4419-6445-8_3. [DOI] [PubMed] [Google Scholar]
- Sattelle DB, Jones AK, Sattelle BM, Matsuda K, Reenan R, Biggin PC. Edit, cut and paste in the nicotinic acetylcholine receptor gene family of Drosophila melanogaster. Bioessays. 2005;27:366–376. doi: 10.1002/bies.20207. [DOI] [PubMed] [Google Scholar]
- Schulz R, Sawruk E, Mülhardt C, Bertrand S, Baumann A, Phannavong B, Betz H, Bertrand D, Gundelfinger ED, Schmitt B. Dα3, a new functional a subunit of nicotinic acetylcholine receptors from Drosophila. J Neurochem. 1998;71:853–862. doi: 10.1046/j.1471-4159.1998.71020853.x. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Millar NS. The influence of nicotinic receptor subunit composition upon agonist, α-bungarotoxin and insecticide (imidacloprid) binding affinity. Neuropharmacol. 2000;39:671–679. doi: 10.1016/s0028-3908(99)00170-7. [DOI] [PubMed] [Google Scholar]
- Wu P, Ma D, Pierzchala M, Wu J, Yang L-C, Mai X, Chang X, Schmidt-Glenewinkel T. The Drosophila acetylcholine receptor subunit Dα5 is part of an α-bungarotoxin binding acetylcholine receptor. J Biol Chem. 2005;280:20987–20994. doi: 10.1074/jbc.M409639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper ST, Millar NS. Host cell-specific folding of the neuronal nicotinic receptor α8 subunit. J Neurochem. 1998;70:2585–2593. doi: 10.1046/j.1471-4159.1998.70062585.x. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Couturier S, Bertrand D, Matter JM, Hernandez MC, Bertrand S, Millar N, Valera S, Barkas T, Ballivet M. A neuronal nicotinic acetylcholine receptor subunit (α7) is developmentally regulated and forms a homo-oligomeric channel blocked by α-BTX. Neuron. 1990;5:847–856. doi: 10.1016/0896-6273(90)90344-f. [DOI] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Maggi R, Longhi R, Hanke W, Klinke N, Clementi F. α7 and α8 nicotinic receptor subtypes immunoprecipitated from chick retina have different immunological, pharmacological and functional properties. Eur J Neurosci. 1997;9:1201–1211. doi: 10.1111/j.1460-9568.1997.tb01475.x. [DOI] [PubMed] [Google Scholar]
- Khiroug SS, Harkness PC, Lamb PW, Sudweeks SN, Khiroug L, Millar NS, Yakel JL. Rat nicotinic receptor α7 and β2 subunits co-assemble to form functional heteromeric nicotinic receptor channels. J Physiol. 2002;540:425–434. doi: 10.1113/jphysiol.2001.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Huang Y, Xue F, Simard A, DeChon J, Li G, Zhang J, Lucero L, Wang M, Sierks M. et al. A novel nicotinic acetylcholine receptor subtype in basal forebrain cholinergic neurons with high sensitivity to amyloid peptides. J Neurosci. 2009;29:918–929. doi: 10.1523/JNEUROSCI.3952-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry T, McKenzie JA, Batterham P. A Dα6 knockout strain of Drosophila melanogaster confers a high level of resistance to spinosad. Insect Biochem Mol Biol. 2007;37:184–188. doi: 10.1016/j.ibmb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Fayyazuddin A, Zaheer MA, Hiesinger PR, Bellen HJ. The nicotinic acetylcholine receptor Dα7 is required for an escape behavior in Drosophila. PLoS Biol. 2006;4(3):e63. doi: 10.1371/journal.pbio.0040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper ST, Millar NS. Host cell-specific folding and assembly of the neuronal nicotinic acetylcholine receptor α7 subunit. J Neurochem. 1997;68:2140–2151. doi: 10.1046/j.1471-4159.1997.68052140.x. [DOI] [PubMed] [Google Scholar]
- Lewis TM, Harkness PC, Sivilotti LG, Colquhoun D, Millar NS. The ion channel properties of a rat recombinant neuronal nicotinic receptor are dependent on the host cell type. J Physiol. 1997;505:299–306. doi: 10.1111/j.1469-7793.1997.299bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Claudio T. Temperature-sensitive expression of all-Torpedo and Torpedo-rat hybrid AChR in mammalian muscle cells. J Cell Biol. 1990;110(5):1705–1717. doi: 10.1083/jcb.110.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulin T, Gielen M, Richmond JE, Williams DC, Paoletti P, Bessereau J-L. Eight genes are required for functional reconstitution of the Caenorhabditis elegans levamisole-sensitive acetylcholine receptor. Proc Natl Acad Sci USA. 2008;105:18590–18595. doi: 10.1073/pnas.0806933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdell SJ, Millar NS. Cloning and heterologous expression of Dα4, a Drosophila neuronal nicotinic acetylcholine receptor subunit: identification of an alternative exon influencing the efficiency of subunit assembly. Neuropharmacol. 2000;39:2604–2614. doi: 10.1016/s0028-3908(00)00111-8. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Millar NS. Dβ3, an atypical nicotinic acetylcholine receptor subunit from Drosophila: molecular cloning, heterologous expression and coassembly. J Neurochem. 2002;80:1009–1018. doi: 10.1046/j.0022-3042.2002.00789.x. [DOI] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Eiselé J-L, Bertrand S, Galzi J-L, Devillers-Thiéry A, Changeux J-P, Bertrand D. Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature. 1993;366:479–483. doi: 10.1038/366479a0. [DOI] [PubMed] [Google Scholar]
- Maricq AV, Peterson AS, Brake AJ, Myers RM, Julius D. Primary structure and functional expression of the 5HT3 receptor, a serotonin-gated ion channel. Science. 1991;254:432–437. doi: 10.1126/science.1718042. [DOI] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morph. 1972;27:353–365. [PubMed] [Google Scholar]
- Millar NS, Buckingham SD, Sattelle DB. Stable expression of a functional homo-oligomeric Drosophila GABA receptor in a Drosophila cell line. Proc R Soc Lond B. 1994;258:307–314. doi: 10.1098/rspb.1994.0178. [DOI] [PubMed] [Google Scholar]
- Margolskee RF, McHendry-Rinde B, Horn R. Panning transfected cells for electrophysiological studies. Biotechniques. 1993;15:906–911. [PubMed] [Google Scholar]
- Young GT, Broad LM, Zwart R, Astles PC, Bodkin M, Sher E, Millar NS. Species selectivity of a nicotinic acetylcholine receptor agonist is conferred by two adjacent extracellular β4 amino acids that are implicated in the coupling of binding to channel gating. Mol Pharmacol. 2007;71:389–397. doi: 10.1124/mol.106.030809. [DOI] [PubMed] [Google Scholar]


