Abstract
Mycoplasma genitalium is a cause of nongonococcal urethritis, particularly in patients not infected with Chlamydia trachomatis. A quantitative 5′ nuclease assay (TaqMan PCR) was developed and validated. The assay detected a fragment of the MgPa adhesin gene by use of a TaqMan MGB (minor groove binder) probe and included an internal processing control to detect PCR inhibition. Urethral swab specimens and first-void urine samples from M. genitalium-positive men were examined, and the M. genitalium DNA load was correlated to symptoms and signs. The assay consistently detected <5 genome copies without cross-reactions with other mycoplasmas. Urine and urethral swab specimens from men with urethritis had higher M. genitalium DNA loads than specimens from men without urethritis. However, a very broad overlap of DNA loads between patients with and without urethritis was observed. Urethral swab specimens from patients with urethral discharge had a significantly higher DNA load than specimens from patients without discharge. This correlation was not found in first-void urine specimens.
Mycoplasma genitalium was first isolated after prolonged incubation from 2 of 13 men with urethritis (26, 27). Despite repeated attempts by conventional culture techniques (21, 23), other urogenital isolates have been extremely rare and have been difficult to obtain (10, 17). Isolation of M. genitalium in cultures of throat and synovial fluid specimens has also been recorded, although these isolates were mixed with Mycoplasma pneumoniae (1, 25).
M. genitalium and M. pneumoniae share several structural properties, such as the flask shape and the terminal tip-like structure, and a significant antigenic relationship between the two mycoplasma species has hampered diagnostic serology (15, 16).
Because traditional procedures for the diagnosis of M. genitalium infection, like culture and serology, have failed, more extensive studies indicating that M. genitalium is a cause of sexually transmitted infections had to await the development of the PCR (11, 18).
The first two PCR-based studies of M. genitalium in patients with nongonococcal urethritis (NGU) were reported in 1993 (12). In those studies (16) M. genitalium was found significantly more often in men with NGU than in those without the condition. Furthermore, M. genitalium was found more often in men with Chlamydia trachomatis-negative NGU than in those with chlamydial NGU, indicating that the two microbes may act as separate causes of urethritis (6, 12). Several case-control studies of NGU patients and controls have confirmed this finding (for a review, see references 8 and 24). Subsequent studies have shown that M. genitalium is more closely associated with symptomatic urethritis than with asymptomatic urethritis (2; P. J. Horner and D. Taylor-Robinson, Letter, Lancet 343:790-791, 1994), particularly in those patients with an observable discharge (7). Recently, chronic NGU was also demonstrated to be associated with M. genitalium infection (5)
The detection of M. genitalium in higher numbers in urogenital specimens from patients with NGU than in those without the condition would further strengthen the evidence that M. genitalium is a cause of NGU (22). Recently, data supporting the presence of a higher number of M. genitalium DNA copies in first-void urine (FVU) from patients with nonchlamydial NGU (NCNGU) than from asymptomatic men was presented (28). Those researchers applied a quantitative TaqMan 5′ nuclease assay based on 16S rRNA gene sequences to M. genitalium-positive specimens. However, only two asymptomatic men were examined; therefore, we decided to study a larger series of patients. In order to do so, we developed a TaqMan quantitative PCR assay based on a conserved region of the MgPa gene and examined stored clinical material with the purpose of confirming the findings. The 16S rRNA gene of M. genitalium is very homologous to that of M. pneumoniae, and furthermore, the preponderance of secondary structures inherent to this gene precluded the design of specific and sensitive primers and probes for the TaqMan assay. We have previously demonstrated that the M. genitalium MgPa adhesin gene contains conserved regions interspersed with hypervariable sequences and that the hypervariable regions correlated with the MgPa repeated regions, which can be found in nine different positions in the genome (19). From previous experience, however, we knew that results obtained with the MgPa1 and MgPa3 primer set (11) correlated very well with those of a 16S rRNA gene-based assay (9); and since partial MgPa sequence information was available for four Danish strains (19), we selected primers and probes from conserved regions of the MgPa gene. The assay was validated regarding specificity against all human mycoplasmas and a range of phylogenetically related species. Sensitivity was validated by comparing the results of the PCR for detection of the 16S rRNA gene; by analyzing the standard curve in the presence of different clinical specimen types; and by adding known amounts of M. genitalium to clinical specimens to control for the matrix effect, i.e., the effect of irrelevant DNA and other components in clinical specimens.
(These data were presented in part at the 14th Meeting of The International Organization for Mycoplasmology, July 2002, Vienna, Austria.)
MATERIALS AND METHODS
Patients and specimens.
As part of an ongoing study of the clinical and epidemiological aspects of M. genitalium infection, male patients attending a sexually transmitted disease (STD) clinic in Huddinge, Stockholm, Sweden, between August 1997 and November 2001 were enrolled after providing informed consent. Patients without urethritis attended the clinic for a checkup for STDs, testing for human immunodeficiency virus infection, or treatment for condyloma or as part of contact tracing of patients with STDs. The local ethical committee approved the study. Urethral discharge specimens for smears were obtained with a plastic loop, and the smears were stained with methylene blue. Patients with a smear showing ≥5 polymorphonuclear leukocytes (PMNLs) per high-power (×1,000) microscopic field (hpf) were considered to have urethritis. Patients with 5 to 10 PMNLs/hpf were defined as having low-grade urethritis, whereas those with >10 PMNLs/hpf were defined as having high-grade urethritis.
After the discharge specimen for the smear was taken, a urethral swab sample was collected with a cotton-tipped swab for culture of Neisseria gonorrhoeae. Then, an ear-nose-throat cotton-tipped aluminum swab was inserted 3 to 4 cm into the urethra and subsequently placed in a tube with 1.8 ml of SP4 mycoplasma broth medium (10). This specimen was used for detection of M. genitalium and C. trachomatis by PCR. After the clinical examination, the patient was asked to collect 15 to 20 ml of FVU; 12 ml was used for the routine detection of C. trachomatis by PCR at the local diagnostic laboratory, and the remaining FVU was sent together with the urethral swab specimen by ordinary mail to the laboratory in Copenhagen. No information about the clinical status of the patients was given to the laboratory.
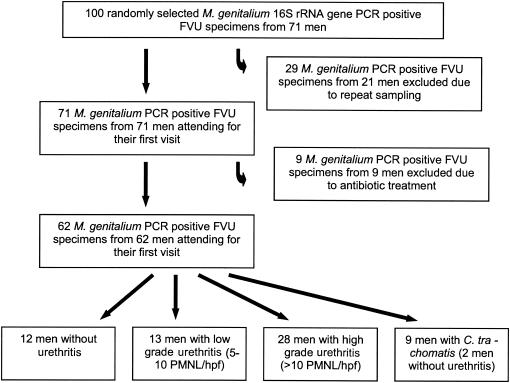
One hundred FVU specimens were randomly selected from among those found to be M. genitalium positive by a conventional PCR assay for detection of the 16S rRNA gene (9). After the code regarding the identity and clinical symptoms of the patients was broken, it became apparent that 21 men provided more than one FVU specimen, thus excluding 29 FVU specimens. In several cases, the patients were considered by the clinicians to have a new infection, but only the specimen from the first visit was included for analysis of the correlation between M. genitalium DNA load and clinical findings in the present study. Nine men had received antibiotics for a urogenital tract infection within the previous 6 months and were excluded. Thus, specimens from 62 men were included in the study. Nine of the 62 patients were also positive for C. trachomatis, and specimens from these patients were analyzed as a separate group. A flowchart illustrating the selection and elimination of patients and specimens is presented in Fig. 1.
FIG. 1.
Flowchart illustrating selection and elimination of FVU specimens from men attending an STD clinic. A random sample of 100 FVU specimens found to be positive by the conventional PCR for detection of the M. genitalium 16S rRNA gene were blindly tested by the TaqMan quantitative PCR. After the code regarding the identities of the patients and recent antibiotic treatment was broken, specimens were excluded from the subsequent analysis.
A matching urethral swab specimen was available for 93 of the 100 FVU specimens, and 13 of these were M. genitalium negative by conventional PCR. Urethral swab specimens from 56 of the 62 men finally included in the study were analyzed, and 8 of these were M. genitalium negative by conventional PCR.
Organisms and growth conditions.
M. genitalium strains G-37T, R 32G, Tw 10-6G, Tw 10-5G, Tw 48-5G, UTMB-10G, and M30 and M. genitalium strains isolated in our laboratory, designated M2282, M2298, M2300, M2321, M2341, M6090, and M6151, were grown in modified Friis's FF medium containing horse serum (10). M. genitalium strain M30 p7 (passage 7) was received from The Mollicutes Collection of Cultures and Antisera, Gainesville, Fla. (http://mycoplasmas.vm.iastate.edu/IOM/culturecollect.htm), as a freeze-dried culture. Attempts to regrow the strain were not successful, and consequently, the nonviable cells resuspended in modified Friis's FF medium were treated as viable broth cultures. The following species (strains) were grown in modified Hayflick's medium (16) and harvested by centrifugation in the late log phase: M. pneumoniae (FHT, Mac, M129-B8, M129-B170 and two clinical isolates), Mycoplasma hominis (PG21T, H34, H27, and three clinical isolates), Mycoplasma salivarium (PG20T), Mycoplasma buccale (CH 20247T), Mycoplasma orale (Patt and one clinical isolate), Mycoplasma fermentans (GT and S38), Mycoplasma faucium (DC 333T), Mycoplasma primatum (Navel), Mycoplasma pirum (Zeus), Mycoplasma lipophilum (Maby BT), Mycoplasma penetrans (GTU), Mycoplasma hyorhinis (GDL), Mycoplasma arginini (G230T), Mycoplasma gallisepticum (15302), Mycoplasma iowae (695), Mycoplasma immitans (4229), Mycoplasma testudinis (Hill), Mycoplasma alvi (Isley), and Acholeplasma laidlawii (AT). Ureaplasma urealyticum (serotypes I [F. Black 7] and VIII [F. Black 960T]) were grown in U10C medium (20). DNA from M. genitalium G-37T and M. pneumoniae strain Mac was extracted with chloroform as described previously (11). DNA was quantified spectrophotometrically and by visual comparison after gel electrophoresis and ethidium bromide staining. DNA from the other mycoplasma species tested was released by resuspending the pellet from 2 ml of broth culture obtained after centrifugation at 30,000 × g for 15 min at 4°C in 100 μl of lysis buffer (10 mM Tris HCl [pH 8.0], 1 mM EDTA, 0.5% Tween 20, 0.5% Nonidet P-40) containing 200 μg of proteinase K per ml. The samples were incubated at 55°C for 30 min, the proteinase was inactivated at 94°C for 15 min, and the tubes were briefly centrifuged to collect condensation droplets.
Sample preparation for PCR.
Sample preparation was performed before the specimens were frozen. A 100-μl aliquot of the swab specimen in SP4 mycoplasma broth medium was mixed with 300 μl of a 20% (wt/vol) Chelex 100 slurry (Bio-Rad, Richmond, Calif.) in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and the mixture was vigorously vortexed for 60 s and incubated at 95°C for 10 min. After centrifugation at 20,000 × g for 5 min, 5 μl of the supernatant, which corresponded to approximately 2 μl of the original swab specimen, was used for PCR. Urine specimens were concentrated by centrifugation of 1,800 μl at 20,000 × g for 15 min. The supernatant was discarded, with care taken not to dislodge the pellet yet to leave only a minimal amount of urine in the tube. The Chelex 100 slurry (300 μl) was added, and the mixture was treated as described above for the swab specimens. A total of 5 μl of the supernatant, which corresponded to approximately 36 μl of the FVU specimen, was used for PCR.
Selection of primers and probes.
Partial sequences of the M. genitalium MgPa adhesin gene from five different strains (M. genitalium G-37T, M2288, M2300, M2321, and M2341) (19) were aligned. The Primer Express program (version 2.0; Applied Biosystems, Foster City, Calif.) was used to find compatible primer and probe pairs from the M. genitalium G-37T sequence, and the selected sequences were compared with the alignment. Since the selected TaqMan probes were very long due to the low G+C contents of the selected regions, a new search was performed by using the TaqMan MGB (minor groove binder) primer and probe design option. TaqMan MGB probes are conjugated to the 3′ end with MGB groups. They form extremely stable duplexes with single-stranded DNA targets, allowing shorter probes to be designed (14), and they are supplied with nonfluorescent quenchers, which increases the possibilities for multiplexing. The selected primers amplified a 78-bp fragment with a 35% G+C content positioned between bases 1420 and 1497 in the MgPa operon sequence (GenBank accession no. M31431). The 18-bp MgPa-380 probe (Applied Biosystems) was labeled with 6-carboxyfluorescein (FAM) at the 5′ end (Table 1).
TABLE 1.
Primers and probes deduced from the M. genitalium MgPa gene sequencea used for TaqMan PCR and primers for construction of the IPC
| Primer or probe | Primer or probe name | Sequenceb | Nucleotide position |
|---|---|---|---|
| Primers for TaqMan assay | MgPa-355F | GAGAAATACCTTGATGGTCAGCAA | 1420 in MgPa operon sequence |
| MgPa-432R | GTTAATATCATATAAAGCTCTACCGTTGTTATC | 1497 in MgPa operon sequence | |
| Primers for construc- tion of IPC | MgPa-IPC-F | GAGAAATACCTTGATGGTCAGCAATTTCCGGGACGT ATCATGCT | 13915 in phage lambda sequence |
| MgPa-IPC-R | GTTAATATCATATAAAGCTCTACCGTTGTTATCACCG CTCAGGCATTTGCT | 14061 in phage lambda sequence | |
| MgPa-380 | FAM-ACTTTGCAATCAGAAGGT-MGB | 1445 in MgPa operon sequence | |
| Probe | Phage lambda IPC-R | TAMRA-TCCTTCGTGATATCGGACGTTGGCTG-BHQ2 | 14011 in phage lambda sequence |
The GenBank accession number for the M. genitalium MgPa sequence is M31431.
Sequences in boldface correspond to the phage lambda sequence (GenBank accession number J02459). MGB denotes a minor groove binder and a nonfluorescent quencher, and BHQ-2 denotes Black Hole Quencher 2 (nonfluorescent quencher).
Construction of an IPC for inhibition.
In order to detect Taq DNA polymerase inhibitors or suboptimal reaction conditions, an internal process control (IPC) was constructed as described previously (9). In brief, primers for amplification of parts of the phage lambda genome were synthesized with a tail that included the sequence of each of the MgPa primers added to the 5′ end of the corresponding phage lambda primer (Table 1). PCR products of 204 bp thus containing the binding sites of the MgPa primers were obtained by amplification of 1 ng of purified phage lambda DNA. The amplicons were gel purified, and a 10-fold titration of the IPC was added to separate master mixtures. The dilution of the IPC that had no influence on the threshold cycle (Ct) number for purified M. genitalium DNA was used in the assay. The phage lambda IPC-R probe (TAG Copenhagen, Copenhagen, Denmark) was 5′ labeled with 6-carboxytetramethylrhodamine (TAMRA) and quenched with Black Hole Quencher 2.
TaqMan assay.
Initial TaqMan assays were performed with the ABI TaqMan Universal PCR Master mix No AmpErase UNG (Applied Biosystems). However, Ct values were consistently lower and the relative increase in fluorescence was higher with the in-house master mixture described below. In particular, the IPC probe produced very poor amplification curves with the ABI master mixture regardless of the concentration. The assay with the in-house master mixture was performed in a 50-μl final reaction volume containing 1× PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl; Platinum; Invitrogen, Carlsbad, Calif.) with 5 mM MgCl2; 1 μM each primer MgPa-355F and MgPa-432R; 225 nM TAMRA-labeled phage lambda IPC-R Taqman probe; 75 nM FAM-labeled MgPa TaqMan MGB probe (Table 1); 62.5 μM each dATP, dGTP, and dCTP; 125 μM dUTP; 10% glycerol (Sigma-Aldrich Denmark A/S, Copenhagen, Denmark); 1 μl of 6-carboxy-′x'-rhodamine reference dye (Invitrogen); 5 μl of the appropriate dilution of IPC; and 2 U of Taq DNA polymerase (Platinum Taq; Invitrogen). An ABI 7900HT PRISM SDS real-time PCR instrument (Applied Biosystems) was used with a 96-well block and MicroAmp Optical 96-well reaction plates covered with ABI PRISM Optical Adhesive Covers (Applied Biosystems). The SDS instrument was programmed to a cycle of 50°C for 1 s (stage 1), 95°C for 10 min (stage 2), and 50 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. The primers amplified the 78-bp MgPa gene fragment and a 204-bp IPC fragment. Standard curves were produced by analyzing 10-fold dilutions of M. genitalium DNA containing 5 to 500,000 genome equivalents (geq). The M. genitalium DNA was diluted in TE buffer (pH 8.0) containing 1 μg of calf thymus DNA (D-8661; Sigma-Aldrich) per ml. All tests were performed in duplicate with 5 μl of template DNA. The Chelex 100-treated specimens had been stored at −20°C for up to 5 years (median storage time, 24 months; storage time range, 9 to 60 months).
Validation of the TaqMan assay.
The specificity of the TaqMan assay was evaluated by testing the members of the class Mollicutes, mentioned above, which covered all known human mycoplasma species and a range of other species covering phylogenetically related species and representatives of species from other branches. Furthermore, all currently available cultured strains of M. genitalium were included. The matrix effect was studied by adding 5 μl of each of 82 different Chelex 100-extracted M. genitalium-negative clinical specimens to a fixed amount of M. genitalium DNA and comparing the calculated DNA load to that found in wells receiving only water. The specimens stemmed from 38 male patients and 2 female patients, with each male patient providing an FVU specimen and a urethral swab specimen and each female patient providing an FVU specimen and urethral and cervical swab specimens. The capability of the assay to quantify M. genitalium DNA in the presence of clinical specimens was investigated by adding the M. genitalium DNA dilutions used to establish the standard curve to a fixed amount of 5 μl of Chelex 100-extracted clinical specimen. Six standard curves were prepared, with one curve each prepared for male FVU specimens, male urethral swab specimens, female FVU specimens, female urethral and cervical swab specimens, and water. The theoretical quantity assigned for a range of relevant Ct values was estimated for each of the standard curves.
In order to determine the limit of detection, purified DNA corresponding to 4 geq was tested in 12 replicates, and similarly, 24 replicates each of DNA corresponding to 3, 2, and 1 geq were tested. Logistic regression analysis was used to determine the amount of DNA that could be detected with a 95% likelihood.
Two hundred randomly selected urogenital specimens from patients found to be negative by conventional M. genitalium PCR (9) were tested by the TaqMan assay in order to document the clinical specificity and to compare the occurrence of inhibition or reaction failures.
In order to estimate the reproducibility of M. genitalium DNA load determination between sample preparations, frozen FVU and urethral swab specimens positive for M. genitalium by PCR were examined; 12 FVU specimens and 10 urethral swab specimens were divided before being subjected to sample preparation, and each of the two preparations were tested in six replicates by the TaqMan assay.
DNA sequencing.
In order to confirm the sequence conservation in the primer and probe binding sites, the DNA in 32 specimens from 30 randomly selected M. genitalium-positive patients were amplified with the MgPa1 and MgPa3 primer set (11). Lysates were also prepared from the broth cultures of 10 available M. genitalium strains (Table 2), and the DNA was amplified with the same primers. The amplicons were sequenced in both directions with the MgPa1 and MgPa3 primers with the ABI PRISM BigDye Primer Cycle Sequencing kit with dye terminators (Applied Biosystems). Sequences were read on an ABI PRISM 377 sequencer (Applied Biosystems).
TABLE 2.
M. genitalium strains sequenced from positions 1271 to 1501 of the MgPa operon sequence as part of the present study
| M. genitalium strain designation | Isolate source | Source | Refer- ence |
|---|---|---|---|
| R 32G | Human throat | J. G. Tully | 1 |
| Tw 10-6G | Human throat | J. G. Tully | 1 |
| Tw 10-5G | Human throat | J. G. Tully | 1 |
| Tw 48-5G | Human throat | J. G. Tully | 1 |
| UTMB-10G | Synovial fluid | J. G. Tully | 25 |
| M30 | Urethra | J. G. Tully | 27 |
| M2282 | Urethra | J. S. Jensen | 25 |
| M6090 | Urethra | Specimen provided by Bertille de Barbeyrac, Bordeaux, France; isolated in Copenhagen, Denmark. | |
| M6151 | Urethra | Specimen provided by Bertille de Barbeyrac, Bordeaux, France; isolated in Copenhagen, Denmark; isolated from the same patient from whom M6090 was isolated | |
| M30 p7, early passage | Urethra | The Mollicutes Collection of Cultures and Antisera, Gainesville, Fla. | 27 |
RESULTS
Validation of the TaqMan assay.
The assay with the M. genitalium probe and primer set selected was very robust and sensitive. It was found that the use of calf thymus DNA in the buffer for dilution of the M. genitalium DNA standards was important for the reproducibility, particularly for the lowest DNA concentrations. The standard curve generated by 10-fold dilutions of M. genitalium DNA was linear over a range of 5 log units, as illustrated by an r2 value of >0.99 for 10 consecutive setups. By using a model in which variances depend exponentially on log quantity and by also taking into account correlations between measurements originating from the same level of dilution of the same DNA standard, the 95% confidence intervals for true values of DNA loads could be estimated by use of fictitious duplicate Ct values. Examples of such calculations as well as the coefficients of variation (CVs) for a range of selected Ct values are given in Table 3.
TABLE 3.
Estimated M. genitalium DNA loads, 95% confidence intervals, and CVs for a range of selected fictitious duplicate Ct valuesa
| Mean Ctb | Estimated quantity (geq) | CV (%) | 95% CIc of estimated quantity |
|---|---|---|---|
| 20 | 537,442 | 11 | 431,245-669,789 |
| 22 | 139,856 | 9 | 116,811-167,448 |
| 24 | 36,394 | 8 | 31,249-42,387 |
| 26 | 9,470 | 7 | 8,173-10,973 |
| 28 | 2,464 | 9 | 2,072-2,931 |
| 30 | 641 | 12 | 506-811 |
| 32 | 166 | 17 | 118-235 |
| 34 | 43 | 26 | 26-74 |
| 36 | 11 | 43 | 5-26 |
| 38 | 3 | 71 | 0.7-12 |
The quantities were determined by use of a model in which variances depend exponentially on log quantity and also modeling of the correlations between repetitions for the same DNA standard.
The mean of two observations.
CI, confidence interval.
The calculated limit of detection that would be positive for 95% of the wells analyzed was 4.65 geq; i.e., the assay was consistently capable of detecting less than 5 geq of M. genitalium. By use of the sample preparation method described here, this would correspond to 3,500 geq/ml for urethral swab specimens or 6,300 geq per swab and 140 geq/ml of FVU.
The TaqMan assay was less prone to inhibition than our conventional PCR for detection of the 16S rRNA gene, as experienced during parallel testing by the two assays. Among 200 M. genitalium-negative specimens, 18 (9%) were initially inhibited or failed to amplify the IPC in the PCR for detection of the 16S rRNA gene, whereas none were inhibited or failed to amplify the IPC in the TaqMan assay (P < 0.0001, McNemar's test). Even with a crude DNA preparation simply prepared by boiling the clinical specimen in a suspension of Chelex 100 resin, 5 μl of the lysate could be used for the TaqMan assay without interfering with the quantification. This was demonstrated by plating a master mixture containing 100 geq of M. genitalium into 82 wells of the PCR plate and adding 5 μl of a Chelex 100-treated clinical specimen found to be negative by conventional PCR. The two wells receiving sterile water had a calculated DNA load of 75 geq, whereas the calculated DNA loads of those wells receiving a clinical specimen were in the range of 58 to 169 (mean, 101), demonstrating the lack of inhibition and good reproducibility regardless of the presence of human DNA. The lower calculated loads in the wells receiving sterile water were seen repeatedly and are probably an effect of the human DNA present in the clinical specimens, which may act as a carrier.
The presence of different types of clinical specimens had only minor influences on the standard curve. The modeling of the standard curves described above showed that the slopes of the standard curves generated in the presence of male urethral swab specimens or cervical swab specimens were statistically significantly different from the slopes of the curves generated without clinical material. However, the practical robustness of the method was illustrated when the six individual standard curves with and without clinical specimens were used to calculate the DNA loads and CVs from a range of fictitious duplicate Ct values (Table 4). By using a mean Ct of 30, DNA loads that varied between 598 and 722 (mean, 643) were returned.
TABLE 4.
Estimated M. genitalium DNA loads and CVs for a range of selected fictitious duplicate Ct valuesa
| Ct | Water
|
Male FVU
|
Male urethral swab
|
Female FVU
|
Female urethral swab
|
Cervical swab
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimated quantity (geq) | CV (%) | Estimated quantity (geq) | CV (%) | Estimated quantity (geq) | CV (%) | Estimated quantity (geq) | CV (%) | Estimated quantity (geq) | CV (%) | Estimated quantity (geq) | CV (%) | |
| 20 | 537,442 | 11 | 551,208 | 7 | 712,258 | 24 | 578,001 | 10 | 566,598 | 14 | 575,016 | 9 |
| 26 | 9,470 | 7 | 9,287 | 5 | 10,455 | 15 | 9,354 | 6 | 9,858 | 9 | 10,450 | 5 |
| 30 | 641 | 12 | 610 | 7 | 626 | 13 | 598 | 7 | 662 | 10 | 722 | 7 |
| 34 | 43 | 26 | 40 | 12 | 38 | 16 | 38 | 10 | 44 | 13 | 50 | 12 |
| 38 | 3 | 71 | 3 | 20 | 2 | 22 | 2 | 17 | 3 | 19 | 3 | 25 |
Quantities were determined by use of a model in which variances depend exponentially on log quantity and also modeling of the correlations between repetitions for the same DNA standard. Modeling was based on standard curves generated with various urogenital specimen types.
The variation contributed by the type of sample preparation was studied for 12 FVU specimens and 10 urethral swab specimens, which were divided before sample preparation. Each of the two preparations was tested in six replicates, and the means and CVs for the two preparations were compared. As illustrated in Table 5, less than a twofold difference in the M. genitalium DNA load was found for all specimens containing a mean of >5 geq, and linear regression revealed a correlation coefficient of 0.9996 (r2 = 0.9992).
TABLE 5.
Mean M. genitalium DNA loads in FVU and urogenital swab specimens subjected to sample preparation in parallel and analyzed by the TaqMan assay in six replicates
| Specimen no. | Mean M. genitalium DNA load (geq)
|
CV (%) between sample preparations | |
|---|---|---|---|
| Sample preparation 1 | Sample preparation 2 | ||
| FVU 307 | 2.3 | 2.1 | 6 |
| FVU 309 | 1.2 | 2.8 | 57 |
| FVU 327 | 15 | 22 | 27 |
| FVU 351 | 108 | 131 | 14 |
| FVU 434 | 13 | 19 | 27 |
| FVU 475 | 5,514 | 4,864 | 9 |
| FVU 490 | 153 | 238 | 31 |
| FVU 533 | 629 | 544 | 10 |
| FVU 555 | 1.7 | 3.1 | 41 |
| FVU 634 | 134 | 195 | 26 |
| FVU 639 | 17 | 13 | 19 |
| FVU 666 | 11 | 14 | 17 |
| Swab 80 | 2.8 | 0.6 | 92 |
| Swab 86 | 0.5 | 0.9 | 40 |
| Swab 277 | 7.3 | 11 | 29 |
| Swab 310 | 3.6 | 1 | 80 |
| Swab 352 | 4.9 | 7.1 | 26 |
| Swab 428 | 2.9 | 3.9 | 21 |
| Swab 640 | 7.3 | 13 | 40 |
| Swab 1601 | 16 | 13 | 15 |
| Swab 4181 | 22 | 18 | 14 |
| Swab 4207 | 97 | 136 | 24 |
The wide range of mycoplasma species mentioned in Materials and Methods gave negative reactions by the TaqMan assay, and the 200 urogenital specimens negative by conventional PCR were also negative by the M. genitalium TaqMan assay.
The primer and probe sequences were selected from the available sequences from isolate G-37T (variants US and DK) as well as Danish isolates M2288, M2300, M2321, and M2341 (19). These sequences were supplemented with five sequences of strains from the respiratory tract and synovial fluid as well as the sequence of a late passage of strain M30, which is a urethral isolate (Table 2).
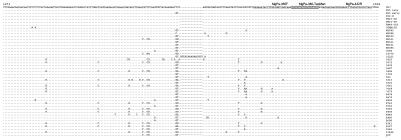
The primer and probe sequences selected from all of the cultured strains initially sequenced were 100% identical, as can be seen in the alignment shown in Fig. 2. However, it was decided that additional sequence information should be collected in order to provide additional information about the assay, and therefore, the relevant part of the MgPa genes of three recent isolates obtained in our laboratory as well as an early passage (passage 7) of urethral strain M. genitalium M30 were sequenced together with 30 randomly selected amplicons produced with the MgPa1 and MgPa3 primer set from samples from 28 patients. To our surprise, 10 amplicons produced from samples from 9 of the 28 patients studied carried one or two mutations in the forward primer. Five divergent types were found (Fig. 2). With the aim of studying the influences of these mutations on the detection limit and linearity of the assay, clinical specimens with each of the divergent sequences were tested undiluted and at fivefold dilutions (to 1:3,125) in TE buffer containing 1 μg of calf thymus DNA per ml. The same amount of the diluted specimen (5 μl) was subjected to amplification by the PCR for detection of the 16S rRNA gene and the TaqMan assay. For all of the mutation types, the detection limit was the same or fivefold lower by the TaqMan assay than by the conventional PCR. Since four of the five undiluted clinical specimens contained less than 250 geq, the linearity could not be determined with certainty since the CVs for low DNA loads were too high to allow the construction of a standard curve.
FIG. 2.
Alignment of MgPa sequences from positions 1271 to 1501 of the MgPa operon sequence (GenBank accession number M31431) from cultured M. genitalium strains and PCR products from 30 specimens from 28 patients. The sequences of the primers used in the TaqMan assay are underlined, and the sequence of the TaqMan MGB probe is shaded.
Sequence variation in the MgPa gene.
The remarkable heterogeneity found by restriction enzyme digestion of the amplicon generated with the MgPa1-MgPa3 primer set has been reported previously (11). These findings were confirmed in the present study. The variability in this area is surprising since this part of the MgPa gene is present only in a single copy in the M. genitalium G-37T genome sequence. The 15-bp insert found in specimen 16221 was noteworthy, and this region of the gene seems to be rather variable. Whether the variability reflects differences in strain pathogenicity cannot be established in the present study.
The significant homology between the sequences of strain G-37T and the respiratory and synovial fluid strains as well as the late passage of strain M30 is striking and may indicate a clonal origin, which was also demonstrated in a previous study by typing by amplified-fragment length polymorphism analysis (13). The early passage of strain M30, on the other hand, had two base substitutions that led to the loss of the EcoRI restriction enzyme site present in the late-passage strain and the type strain.
Patients and specimens.
Of the 100 FVU specimens with a positive result by conventional PCR, all were positive by the TaqMan assay. Of the 87 urethral swab specimens, 11 were M. genitalium negative by conventional PCR, but 2 of these were positive by the TaqMan assay. One of the 76 urethral swab specimens positive by conventional PCR was negative by the TaqMan assay.
The median age of the 62 M. genitalium-positive men who provided the FVU specimens that were included in the study was 27 years (age range, 18 to 52 years); 14 men did not have urethritis. Two of these men were also positive for C. trachomatis. Fourteen men had low-grade urethritis, defined as 5 to 10 PMNLs/hpf in the urethral discharge. One man was concomitantly positive for C. trachomatis. High-grade urethritis was found in 34 men, of whom 6 were also positive for C. trachomatis. There was no difference in the age distributions between the groups.
M. genitalium DNA load in FVU specimens.
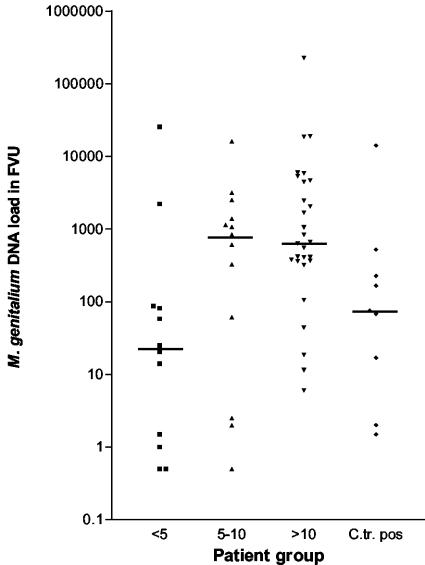
C. trachomatis-negative patients without urethritis (n = 12) had a median M. genitalium DNA load of 22.7 geq (range, 0.5 to 25,709 geq), whereas the 13 men with low-grade NCNGU had a median load of 837 geq (range, 0.5 to 16,093 geq) and the 28 men with high-grade NCNGU (>10 PMNLs/hpf in the urethral secretion) had a median load of 647 geq (range, 6 to 225,657 geq). Only two of the nine C. trachomatis-positive patients did not have urethritis; consequently, C. trachomatis-positive patients were considered as a separate group and had a median M. genitalium DNA load of 75 geq (range, 1.5 to 14,092 geq) (Fig. 3). When the M. genitalium DNA loads in the C. trachomatis-negative groups were compared, they were found to be different (P = 0.025, Kruskall-Wallis test). As is illustrated in Fig. 3, the variation within each group was considerable.
FIG. 3.
M. genitalium DNA load in FVU specimens from patients without urethritis (<5 PMNLs/hpf), low-grade NCNGU (5 to 10 PMNLs/hpf), and high-grade NCNGU (>10 PMNLs/hpf) and patients with a concomitant C. trachomatis infection (C.tr. pos). The median DNA load for each group is marked by a horizontal line.
M. genitalium-positive patients with NCNGU (n = 41) and complaints of dysuria (n = 28) had a median DNA load of 1,105 geq (range, 0.5 to 225,657 geq), whereas those without dysuria (n = 13) had a median DNA load of 411 geq (range, 2 to 2,514 geq). The difference did not reach statistical significance, however (P = 0.07, Mann-Whitney test). A self-reported observation of a discharge (n = 26) was not associated with a higher M. genitalium DNA load in men with NCNGU, and the same was the case when a discharge was observed at the physical examination (n = 20). A discharge was observed by the patient or was detected at examination for 29 patients, and these patients had a median M. genitalium DNA load of 1,057 geq, whereas the 12 patients without a discharge had a median DNA load of 328 geq; the difference did not reach statistical significance (P = 0.11, Mann-Whitney test).
No correlation between age and M. genitalium DNA load was found (Spearman's rank correlation).
M. genitalium DNA load in urethral swab specimens.
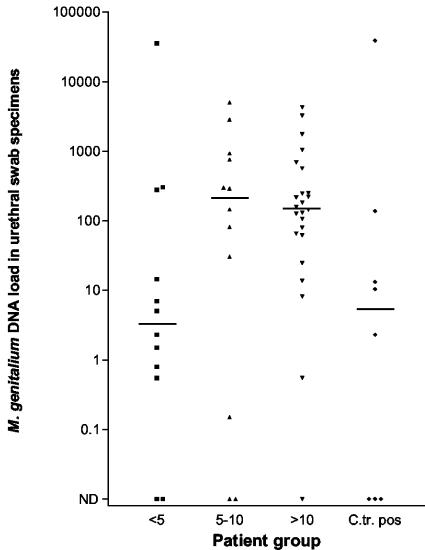
Fifty-six men provided a urethral swab specimen together with the FVU specimen. The urethral swab specimen, however, was negative by the conventional PCR for detection of the 16S rRNA gene for eight patients, and the same specimens from seven of these patients were negative by the TaqMan assay. The specimen from one patient was negative by the TaqMan assay and positive by the conventional PCR; only 3 geq of M. genitalium was detected in the FVU specimen from this patient. He had attended the clinic for a test for human immunodeficiency virus infection and had no symptoms and low-grade urethritis. Eight patients were positive for C. trachomatis and were analyzed as a separate group. The M. genitalium DNA loads in patients without urethritis (n = 12), low-grade NCNGU (n = 12), and high-grade NCNGU (n = 24) were not statistically different (Fig. 4). When the M. genitalium DNA loads in patients with or without urethritis were compared without distinguishing between low- and high-grade NCNGU, patients with urethritis had significantly higher DNA loads than those without urethritis (P = 0.03).
FIG. 4.
M. genitalium DNA loads in urethral swab specimens from patients without urethritis (<5 PMNLs/hpf), low-grade NCNGU (5 to 10 PMNLs/hpf), and high-grade NCNGU (>10 PMNLs/hpf) and patients with a concomitant C. trachomatis infection (C.tr. pos). Specimens without detectable M. genitalium DNA are marked as not detectable (ND). The median DNA load for each group marked by a horizontal line.
The M. genitalium DNA load was significantly higher in urine than in the urethral swab specimens (P = 0.0038, Wilcoxon signed rank test). This also held true if the eight TaqMan PCR-negative urethral swab specimens were excluded from the comparison. However, by taking into account the fact that the volume of FVU specimen analyzed was larger than the volume of urethral swab transport medium, this difference was no longer statistically significant. A linear correlation between the DNA loads in the two specimen types was found (P < 0.0001) by using nonparametric linear regression, but the 95% confidence interval for the slope was between 0.4 and 3.5, indicating the rather low correlation at the patient level.
A discharge was observed by the patient or was detected at examination for 27 C. trachomatis-negative patients, regardless of the presence or absence of urethritis; and these patients had a significantly higher median M. genitalium DNA load of 248 geq, whereas the 21 patients without a discharge had a median load of 5 geq (P = 0.0001, Mann-Whitney test). In patients with NCNGU, a strong correlation between the median M. genitalium DNA load and the presence of a discharge reported by the patient or detected by examination was also found. The median M. genitalium DNA load for patients with a discharge and NCNGU (n = 25) was 248 geq, whereas those 11 without a discharge had a median load of 13 geq (P = 0.002, Mann-Whitney test).
The urethral swab specimens of the C. trachomatis-negative patients complaining of dysuria (n = 29) did not have significantly higher M. genitalium DNA loads than the swab specimens of those 19 who did not experience this symptom.
Other findings.
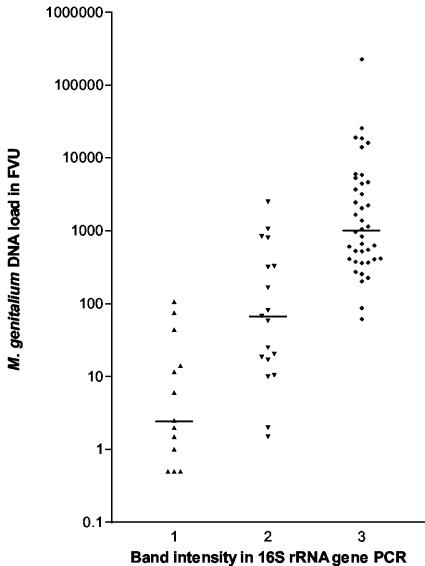
Comparison of the intensity of the 16S rRNA gene amplicons from clinical specimens on an ethidium bromide-stained agarose gel with the numbers of M. genitalium geq showed a strong correlation between the intensity of the band and the DNA load (Fig. 5).
FIG. 5.
M. genitalium DNA loads in 71 FVU specimens from men who attended the STD clinic for the first time during the study and who were grouped by the intensity of the 16S rRNA gene amplicon detected in an ethidium bromide-stained agarose gel. Column 1, a faint band; column 2, a clear band but the IPC is still visible; column 3, a strong band with no IPC visible. Data were recorded during the screening of the specimens. The median DNA load for each group is marked by a horizontal line.
Of the 74 urethral swab specimens positive by conventional PCR, 21 (28%) contained <10 geq; and of the 100 FVU specimens, 14 (14%) contained <10 geq. These findings emphasize the need for highly sensitive assays for clinical studies.
DISCUSSION
The quantitative 5′ nuclease real-time assay developed for the present study was sensitive, specific, and robust. Despite the unexpected finding of five different sequence variants in the forward primer, the sensitivity of the assay was not impaired, as demonstrated by the fact that all 100 FVU specimens found to be positive by the conventional PCR for detection of the 16S rRNA gene were positive by the TaqMan assay and 2 of the 10 urethral swab specimens found to be negative by the conventional PCR were positive by the new assay. Only one urethral swab specimen positive by conventional PCR was negative by the quantitative assay. The result of repeat testing by PCR for detection of the 16S rRNA gene was negative, indicating that the DNA may have deteriorated during storage. The specificity for other mycoplasma species was perfect, but more importantly, the specificity for urogenital specimens found to be M. genitalium negative by conventional PCR was also excellent.
In order to prove that M. genitalium is a cause of urogenital tract infections, modifications of the original Henle-Koch postulates must be used. Taylor-Robinson devised such modifications (22), and one of the modified criteria was the need to detect the infecting microbe at higher numbers in infected patients than in asymptomatic carriers. M. genitalium should then be detected in higher numbers in urogenital specimens from patients with NGU than in those without NGU. Recently, Yoshida et al. (28) found evidence of higher M. genitalium DNA loads in FVU specimens from patients with NCNGU than in those from asymptomatic men. However, only two asymptomatic men were examined, and the specimen from one of them was actually negative by the TaqMan assay, which makes one question the sensitivity of the assay. Furthermore, the specificity of the assay may not be optimal, since the primer sequences were located in parts of the 16S rRNA gene that are shared with M. pneumoniae and no information about the specificity for M. genitalium-negative clinical specimens was given. On the other hand, the results for FVU specimens found in the present study are in good agreement with those reported in the study by Yoshida et al. (28), since we found a significant difference in the DNA loads in FVU specimens from patients without urethritis and those from patients with NCNGU. Surprisingly, although a correlation between the DNA loads in FVU specimens and urethral swab specimens was demonstrated, the correlation between DNA load and the presence of urethritis could not be confirmed with the series of urethral swab specimens used when the distinction between low- and high-grade urethritis was maintained. When all samples from patients with NCNGU defined as ≥5 PMNLs/hpf were examined, however, patients with urethritis had significantly higher DNA loads than those without urethritis. The strong association between a high M. genitalium DNA load and the presence of a discharge, which was demonstrated for urethral swab specimens only, may have obscured the relationship when the urethritis group was divided between those with low- and high-grade urethritis. It may not be surprising that the DNA load was higher in urethral swab specimens from patients with a discharge since the amount of secretions collected on the swab would be higher in this situation. On the other hand, one would have expected the same correlation to be found for the FVU specimens, in which the secretions are collected by the washing effect of the urine, but the difference did not reach statistical significance for FVU specimens, although a clear trend was observed. One confounder could be the fact that for all patients the swab specimen was taken before the FVU specimen was collected, and thus, the main part of the secretion may have been collected on the swab due to “milking” of the urethra during the examination. Against this theory would be the finding of a generally higher M. genitalium DNA load in the FVU specimens than in the urethral swab specimens. Also, for 11 of the 87 complete sets of specimens, the urethral swab specimens were negative by conventional PCR, whereas the FVU specimens were positive. By selection of the patients for the study according to a positive result for FVU specimens by conventional PCR, one would expect that not all urethral swab specimens would be positive, but the results are in good agreement with our experience that approximately 95% of the FVU specimens in sample sets are positive by conventional PCR, whereas 87% of the urethral swab specimens are positive by using the sample preparation method described here (J. S. Jensen, B. Dohn, E. Björnelius, and P. Lidbrink, Abstr., Proc. 13th Int. Congr. Int. Org. Mycoplasmol., p. 248, 2000).
A trend toward a higher M. genitalium DNA load in the FVU specimens from patients with dysuria was observed, but it did not reach statistical significance (P = 0.07). It would probably be rewarding to examine the responses to treatment for patients who have been treated with antibiotics but who fail to clear the organism. These patients often experience a transient clinical improvement which may be associated with a decreased M. genitalium DNA load, but their symptoms often recur after treatment (4).
Twenty-eight percent of the urethral swab specimens and 14% of the FVU specimens contained less than 10 geq of M. genitalium DNA, and likewise, 20 and 13% of the two types of specimens, respectively, had less than 5 geq, which emphasizes the need for highly sensitive assays for clinical purposes and for improvements in specimen preparation techniques.
The sequencing of both cultured M. genitalium strains and positive clinical specimens provided new insight into the high level of variation found in the MgPa gene, even in parts believed to be conserved. The possibility that mutations have been introduced during the first stages of PCR amplification, particularly in those sequences determined directly from a clinical specimen, cannot be excluded. However, except for the 15-bp insertion, it appears that most of the polymorphisms are found in more than one strain, making this explanation less likely. The observation that the sequences of an early-passage (passage 7) strain and the late-passage strain of one of the original urogenital isolates, strain M30, appear to be different is in need of further investigation. It has previously been reported (19) that the sequence of an early-passage strain of M. genitalium G-37T received directly from David Taylor-Robinson (G-37 variant DK) was different from that of the strain used for determination of the genome sequence of M. genitalium (G-37 variant US). This polymorphism, however, was apparently confined to parts of the MgPa gene present as multiple repeats (the B region) and could be explained by a single recombination event. Since the polymorphism in strain M30 is outside the repeat regions, other explanations must be applied. Since the MgPa1-MgPa3 primer set used for determination of the sequences has a high sensitivity when it is applied directly to clinical specimens, this part of the MgPa gene shows promise for use in molecular epidemiological studies of sexual networks (i.e., networks of sexual partners) and recurrent infections after antibiotic treatment.
The quantitative TaqMan assay described here may prove to be very useful for several types of studies. In clinical studies, the assay may provide important information on the effect of treatment, which may otherwise be difficult to obtain due to the lack of good culture methods for M. genitalium. Since conventional PCR is unable to distinguish between viable and dead mycoplasmas, information about the relative increase or decrease in organism load may be helpful (3). In the laboratory, the assay may help improve sample preparation methods and provide new information about the effects of storage and freeze-thaw cycles. This may have important implications in the interpretation of the results of studies performed with archival specimens.
Acknowledgments
Determination of the sequences of the amplicons obtained with primer set MgPa1-MgPa3 from specimens 15776, 16221, and 11844 was kindly performed by Estrid Høgdall, Statens Serum Institut.
Jesper Madsen, Department of Biostatistics, Statens Serum Institut, provided helpful advice regarding statistical methods and performed the modeling for determination of the CVs and confidence intervals for the standard curves.
This study was approved by the local ethical committee, and all participants provided informed consent.
No author had any conflict of interest, either financial or personal, that may have biased his or her actions.
REFERENCES
- 1.Baseman, J. B., S. F. Dallo, J. G. Tully, and D. L. Rose. 1988. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J. Clin. Microbiol. 26:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björnelius, E., P. Lidbrink, and J. S. Jensen. 2000. Mycoplasma genitalium in non-gonococcal urethritis—a study in Swedish male STD patients. Int. J. STD AIDS 11:292-296. [DOI] [PubMed] [Google Scholar]
- 3.Deguchi, T., T. Yoshida, S. Yokoi, M. Ito, M. Tamaki, H. Ishiko, and S. Maeda. 2002. Longitudinal quantitative detection by real-time PCR of Mycoplasma genitalium in first-pass urine of men with recurrent nongonococcal urethritis. J. Clin. Microbiol. 40:3854-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falk, L., H. Fredlund, and J. S. Jensen. 2003. Tetracycline treatment does not eradicate Mycoplasma genitalium. Sex. Transm. Infect. 79:318-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horner, P., B. Thomas, C. Gilroy, M. Egger, M. McClure, and D. Taylor-Robinson. 2003. Antibodies to Chlamydia trachomatis heat-shock protein 60 kDa and detection of Mycoplasma genitalium and Ureaplasma urealyticum are associated independently with chronic nongonococcal urethritis. Sex. Transm. Dis. 30:129-133. [DOI] [PubMed] [Google Scholar]
- 6.Horner, P. J., C. B. Gilroy, B. J. Thomas, R. O. Naidoo, and D. Taylor-Robinson. 1993. Association of Mycoplasma genitalium with acute non-gonococcal urethritis. Lancet 342:582-585. [DOI] [PubMed] [Google Scholar]
- 7.Horner, P. J., B. Thomas, C. B. Gilroy, M. Egger, and D. Taylor-Robinson. 2002. Do all men attending departments of genitourinary medicine need to be screened for non-gonococcal urethritis? Int. J. STD AIDS 13:667-673. [DOI] [PubMed] [Google Scholar]
- 8.Jensen, J. S. 2004. Mycoplasma genitalium—the aetiologic agent of urethritis and other sexually transmitted diseases. J. Eur. Acad. Dermatol. Venereol. 18:1-11. [DOI] [PubMed]
- 9.Jensen, J. S., M. B. Borre, and B. Dohn. 2003. Detection of Mycoplasma genitalium by PCR amplification of the 16S rRNA gene. J. Clin. Microbiol. 41:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen, J. S., H. T. Hansen, and K. Lind. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen, J. S., S. A. Uldum, J. Søndergård-Andersen, J. Vuust, and K. Lind. 1991. Polymerase chain reaction for detection of Mycoplasma genitalium in clinical samples. J. Clin. Microbiol. 29:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, J. S., R. Ørsum, B. Dohn, S. Uldum, A. M. Worm, and K. Lind. 1993. Mycoplasma genitalium: a cause of male urethritis? Genitourin. Med. 69:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokotovic, B., N. F. Friis, J. S. Jensen, and P. Ahrens. 1999. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J. Clin. Microbiol. 37:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kutyavin, I. V., I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov, A. A. Gall, R. Dempcy, M. W. Reed, R. B. Meyer, and J. Hedgpeth. 2000. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lind, K. 1982. Serological cross-reactions between “Mycoplasma genitalium” and M. pneumoniae. Lancet ii:1158-1159. [DOI] [PubMed] [Google Scholar]
- 16.Lind, K., B. Ø. Lindhardt, H. J. Schütten, J. Blom, and C. Christiansen. 1984. Serological cross-reactions between Mycoplasma genitalium and Mycoplasma pneumoniae. J. Clin. Microbiol. 20:1036-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, D., W. Xu, G. Liang, S. Wang, Z. Wang, Z. Bi, and W. Zhu. 1999. Isolation and identification of Mycoplasma genitalium from high risk populations of sexually transmitted diseases in China. Chin. Med. J. 112:489-492. [PubMed] [Google Scholar]
- 18.Palmer, H. M., C. B. Gilroy, P. M. Furr, and D. Taylor-Robinson. 1991. Development and evaluation of the polymerase chain reaction to detect Mycoplasma genitalium. FEMS Microbiol. Lett. 61:199-203. [DOI] [PubMed] [Google Scholar]
- 19.Peterson, S. N., C. C. Bailey, J. S. Jensen, M. B. Borre, E. S. King, K. F. Bott, and C. A. Hutchison. 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. USA 92:11829-11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Razin, S., and J. G. Tully. 1983. Mycoplasma characterization, p. 140. In Methods in mycoplasmology, vol. 1. Academic Press, Inc., New York, N.Y.
- 21.Samra, Z., M. Borin, Y. Bukowsky, Y. Lipshitz, and D. Sompolinsky. 1988. Non-occurrence of Mycoplasma genitalium in clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 7:49-51. [DOI] [PubMed] [Google Scholar]
- 22.Taylor-Robinson, D. 1983. The role of mycoplasmas in non-gonococcal urethritis: a review. Yale J. Biol. Med. 56:537-543. [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor-Robinson, D., P. M. Furr, and N. F. Hanna. 1985. Microbiological and serological study of non-gonococcal urethritis with special reference to Mycoplasma genitalium. Genitourin. Med. 61:319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor-Robinson, D., and P. J. Horner. 2001. The role of Mycoplasma genitalium in non-gonococcal urethritis. Sex. Transm. Infect. 77:229-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tully, J. G., D. L. Rose, J. B. Baseman, S. F. Dallo, A. L. Lazzell, and C. P. Davis. 1995. Mycoplasma pneumoniae and Mycoplasma genitalium mixture in synovial fluid isolate. J. Clin. Microbiol. 33:1851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288-1291. [DOI] [PubMed] [Google Scholar]
- 27.Tully, J. G., D. Taylor-Robinson, D. L. Rose, R. M. Cole, and J. M. Bove. 1983. Mycoplasma genitalium, a new species from the human urogenital tract. Int. J. Syst. Bacteriol. 33:387-396. [Google Scholar]
- 28.Yoshida, T., T. Deguchi, M. Ito, S. Maeda, M. Tamaki, and H. Ishiko. 2002. Quantitative detection of Mycoplasma genitalium from first-pass urine of men with urethritis and asymptomatic men by real-time PCR. J. Clin. Microbiol. 40:1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]