Abstract
A total of 15 of 101 (14.8%) nasal methicillin-resistant Staphylococcus aureus (MRSA) isolates exhibited mupirocin resistance (Mupr) compared with 1 of 154 (0.6%) methicillin-susceptible Staphylococcus aureus isolates. A total of 14 (93%) isolates exhibiting high-level Mupr belonged to a single clone. Horizontal plasmid transfer and transmission of Mupr strains contribute to a high incidence of Mupr MRSA at our institution.
Mupirocin (Mup) is a topical antibacterial agent that interferes with protein synthesis by competitively inhibiting bacterial isoleucyl-tRNA synthetase (11, 19). A 2% Mup calcium ointment (Bactroban Nasal; GlaxoSmithKline) applied topically to the anterior nares eradicates carriage of Staphylococcus aureus and prevents infection in certain settings (2, 7, 8, 13). An important concern, however, is the development of Mup resistance (Mupr) (14, 17), of which there are two types. Low-level Mupr (MIC, 8 to 256 mg/liter) is usually associated with a mutation in the gene for target enzyme, while high-level Mupr (Hi-Mupr) (MIC, >256 mg/liter) is mediated by a plasmid containing the ileS2 gene that encodes an additional isoleucyl-tRNA synthetase enzyme (3). Such transmissible resistance raises concern about the spread of Mupr as Mup usage becomes more widespread (9, 10, 17). The objectives of this study were to determine (i) the prevalence of Mupr in methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) isolates from nasal samples; (ii) the location, if present, of the ileS2 gene in Mupr isolates; and (iii) the organism genotype, as defined by pulsed-field gel electrophoresis (PFGE).
Nasal samples submitted to the Clinical Microbiology Laboratory of the Hospital Universitario Doce de Octubre for isolation of S. aureus between October 2001 and October 2002 were included in the study. Specimens were obtained from two groups. Group I comprised adults who underwent elective heart surgery, before which a sample from the anterior nares was taken to determine whether the patient was a carrier of S. aureus. Group II included all new cases of MRSA infection diagnosed at the hospital that were associated with concurrent nasal carriage. Since 1996, we have instituted the use of topical Mup ointment to reduce the prevalence of nasal MRSA. Samples were inoculated onto phenol-red mannitol salt agar plates that were incubated at 37°C for 48 h. Isolation and identification of S. aureus were based upon standard microbiological procedures. All isolates were screened for resistance to Mup on Mueller-Hinton agar with a 5-μg disk (Oxoid). A zone of inhibition ≤ 13 mm in diameter was considered to reflect Mupr (5). Mupr organisms underwent MIC analysis by the E-test strip method (AB Biodisk). Susceptibility testing with other antibiotics was performed by disk diffusion (12, 16). All isolates were confirmed as MRSA by PCR detection of the mecA gene (6). PCR was also performed on all Mupr isolates to detect the plasmid-associated ileS2 gene (1).
Mupr isolates were typed by PFGE following DNA extraction and digestion with SmaI (4). Restriction fragments were separated in a CHEF DRIII PFGE system (Bio-Rad Laboratories). Migration of DNA fragments was normalized using an appropriate size marker. Computer-assisted analysis of PFGE patterns was carried out using GelCompar software (Applied Maths). PFGE types (designated by letters) differed by <7 fragments, while subtypes (designated by Arabic numerals) had indistinguishable patterns (15). Plasmid DNA was extracted by a rapid minipreparation procedure (QIAprep spin plasmid kit from Qiagen) and digested with HindIII. Southern blot analysis of PFGE-separated SmaI digests of genomic DNA and HindIII plasmid fragments was performed with an ECL system (Amersham) using a 456-bp PCR-amplified ileS2 gene fragment as a probe.
Among patients in group I, S. aureus was isolated from 159 of 689 (23%) nasal swabs. Of these isolates, 154 (96.9%) were MSSA and 5 (3.1%) were MRSA. In contrast, 96 of 137 (70.1%) patients in group II yielded MRSA culture-positive nasal swabs. A total of 15 of 101 (14.8%) MRSA isolates were Mupr compared with 1 of 154 (0.6%) isolates of MSSA. Of the Mupr isolates, 14 of 16 (87.5%) exhibited Hi-Mupr and gave a positive result by ileS2 PCR (Table 1).
TABLE 1.
Mupirocin-resistant isolates of S. aureus from the nostrils
| Isolate | Mupirocin resistance pattern (MIC [μg/ml])a | ileS2 PCR result | Resistance patternb | mecA PCR result | PFGE pattern | ileS2 locus polymorph |
|---|---|---|---|---|---|---|
| 1 | R (>1,024) | + | Met/Cip/Ery/Gen | + | A1 | I |
| 2 | R (>1,024) | + | Met/Cip/Ery/Clin | + | A4 | II |
| 3 | R (>1,024) | + | Met/Cip/Ery/Gen/Fus | + | A2 | I |
| 4 | I (8) | − | Met/Cip/Ery/Clin/Gen | + | A3 | |
| 5 | R (>1,024) | + | Met/Cip/Ery/Gen | + | A1 | II |
| 6 | R (>1,024) | + | Met | + | A5 | II |
| 7 | I (16) | − | Met/Cip/Ery/Clin/Gen | + | B | |
| 8 | R (>1,024) | + | Met/Cip/Ery/Clin/Gen | + | A2 | II |
| 9 | R (>1,024) | + | Met/Cip/Ery/Gen | + | A2 | I |
| 10 | R (>1,024) | + | Met/Cip/Ery/Clin/Gen | + | A1 | II |
| 11 | R (>1,024) | + | Met/Cip/Ery | + | A2 | I |
| 12 | R (>1,024) | + | Met/Cip/Ery | + | A1 | II |
| 13 | R (>1,024) | + | Susceptible | − | C | II |
| 14 | R (>1,024) | + | Met/Cip | + | A1 | II |
| 15 | R (>1,024) | + | Met/Cip/Ery | + | A1 | II |
| 16 | R (>1,024) | + | Met/Cip | + | A1 | II |
R, resistant; I, intermediate.
Cip, ciprofloxacin; Clin, clindamycin; Fus, fusidic acid; Ery, erythromycin; Gen, gentamicin; Met, methicillin.
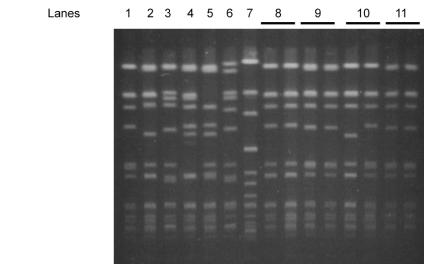
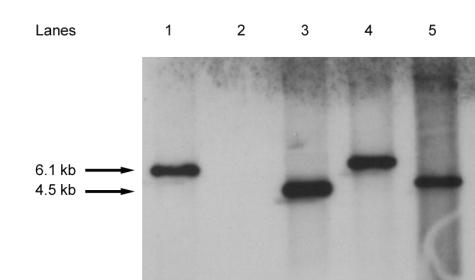
Among the 16 Mupr isolates, PFGE identified one major clone (PFGE type A) containing 14 of 15 (93%) isolates of MRSA and 13 of 14 (93%) Hi-Mupr isolates. The remaining two isolates belonged to two other PFGE types (Fig. 1). All nasal carriers of type A Mupr MRSA also provided other specimens from which MRSA of the same PFGE subtype was isolated. Patients with PFGE type A were located in different hospital wards, with the exception of those who yielded isolates 12 and 15 (both subtype A1), who were resident in the Trauma Ward. Southern analysis of HindIII-digested plasmid DNA confirmed the plasmid location of the ileS2 gene in all 14 Hi-Mupr isolates. Different sizes of hybridizing HindIII plasmid fragments distinguished two ileS2 gene polymorphs. Regions homologous to the ileS2 probe were found on HindIII fragments of 6.1 kb (polymorph I; 10 isolates) or 4.5 kb (polymorph II; 4 isolates) (Fig. 2). Most Hi-Mupr MRSA isolates of PFGE type A (9 of 13, 69%) carried the ileS2 probe region on the 4.5-kb fragment, as did the only Hi-Mupr MSSA isolate (PFGE type C) (Fig. 1 and 2). No positive results were observed in Southern blot analysis of PFGE-separated SmaI-digested genomic DNA. This is because the ileS2-containing plasmids (even prior to digestion with SmaI) are smaller than can be detected at the lower limit of resolution and migrate out of the gel during the course of electrophoresis.
FIG. 1.
PFGE DNA patterns of Mupr S. aureus isolates from nasal samples. Lanes 1 to 7 represent genotypes A1 (isolate 1), A2 (isolate 3), A3 (isolate 4), A4 (isolate 2), A5 (isolate 6), B (isolate 7), and C (isolate 13), respectively. Lanes 8 to 11 represent the banding patterns of paired Mups and subsequently recovered Mupr S. aureus isolates (isolates 5, 12, 14, and 15) from nasal samples of four patients.
FIG. 2.
Southern blot of HindIII-digested plasmid DNA from selected Mupr S. aureus isolates hybridized with an ileS2 probe. Lane 1, MRSA isolate 1 (Hi-Mupr); lane 2, MRSA isolate 4 (low-level Mupr); lane 3, MRSA isolate 5 (Hi-Mupr); lane 4, MRSA isolate 8 (Hi-Mupr); lane 5, MSSA isolate 13 (Hi-Mupr).
We identified four MRSA-infected patients with nasal colonization by Mup-susceptible (Mups) organisms; Mupr bacteria were subsequently isolated (isolates 5, 12, 14, and 15) from these patients 7 to 30 days after intranasal application of Mup ointment (Table 1). The Mups and Mupr isolates from three of the four patients were of the same PFGE subtype, while in the case of the fourth patient there was a difference of a single band (Fig. 1). In these patients, Mup treatment probably exerted selective pressure for organisms which had preexisting high-level resistance and which subsequently recolonized their nasal passages (18).
We detected a much higher percentage of Mupr among isolates of MRSA (14.8%) than among isolates of MSSA (0.6%). Two epidemiological phenomena probably contribute to Hi-Mupr in S. aureus. First, Southern blots of plasmid DNA located the ileS2 resistance gene on two different plasmid fragments, indicating that at least two plasmids or plasmid variants harbor this gene. One of these variants was implicated in horizontal gene transfer and spread of Hi-Mupr between MRSA and MSSA. This was demonstrated by the identification of a 4.5-kb ileS2-hybridizing plasmid fragment in two isolates (one of MRSA and the other of MSSA) with distinctly different PFGE genotypes. Second, identification of the same PFGE subtypes and ileS2 hybridization and antibiotic resistance patterns among Hi-Mupr isolates (Table 1) suggests that patient-patient transmission also occurs. Mup treatment should therefore be used cautiously to avoid the emergence of Hi-Mupr MRSA and the spread of resistance in hospitals in which MRSA is frequently isolated.
Acknowledgments
We thank Tobin Hellyer for suggestions and comments and Mar Aguilera and Antonia Martín for excellent technical assistance.
REFERENCES
- 1.Anthony, R. M., A. M. Connor, E. G. M. Power, and G. L. French. 1999. Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 18:30-34. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, J. M. 1996. Preventing staphylococcal infections by eradicating nasal carriage of Staphylococcus aureus: proceeding with caution. Infect. Control Hosp. Epidemiol. 17:775-779. [DOI] [PubMed] [Google Scholar]
- 3.Cookson, B. D. 1998. The emergence of mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J. Antimicrob. Chemother. 41:11-18. [DOI] [PubMed] [Google Scholar]
- 4.Dominguez, M. A., H. De Lencastre, J. Liñares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs, P. C., R. N. Jones, and A. L. Barry. 1990. Interpretive criteria for disk diffusion susceptibility testing of mupirocin, a topical antibiotic. J. Clin. Microbiol. 28:608-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geha, D. J., J. R. Uhl, C. A. Gustaferro, and D. H. Persing. 1994. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 32:1768-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herwaldt, L. A. 1998. Reduction of Staphylococcus aureus nasal carriage and infection in dialysis patients. J. Hosp. Infect. 40(Suppl. B):S13-S23. [DOI] [PubMed] [Google Scholar]
- 8.Kalmeijer, M. D., H. Coertjens, P. M. van Nieuwland-Bollen, D. Bogaers-Hofman, G. A. J. de Baere, A. Stuurman, A. van Belkum, and J. A. J. W. Kluytmans. 2002. Surgical site infections in orthopedic surgery: the effect of mupirocin nasal ointment in a double-blind, randomized, placebo-controlled study. Clin. Infect. Dis. 35:353-358. [DOI] [PubMed] [Google Scholar]
- 9.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leski, T. A., M. Gniadkowski, A. Skoczynska, E. Stefaniuk, K. Trzcinski, and W. Hryniewicz. 1999. Outbreak of mupirocin-resistant staphylococci in a hospital in Warsaw, Poland, due to plasmid transmission and clonal spread of several strains. J. Clin. Microbiol. 37:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morton, T. M., J. L. Johnston, J. Patterson, and G. L. Archer. 1995. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob. Agents Chemother. 39:1272-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Perl, T. M., J. J. Cullen, R. P. Wenzel, M. B. Zimmerman, M. A. Pfaller, D. Sheppard, J. Twombley, P. P. French, L. A. Herwaldt, and the Mupirocin and the Risk of Staphylococcus aureus Study Team. 2002. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N. Engl. J. Med. 346:1871-1877. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz, F. J., E. Lindenlauf, B. Hofmann, A. D. Fluit, J. Verhoef, H. P. Heinz, and M. E. Jones. 1998. The prevalence of low- and high-level mupirocin resistance in staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 42:489-495. [DOI] [PubMed] [Google Scholar]
- 15.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toma, E., and D. Barriault. 1995. Antimicrobial activity of fusidic acid and disk diffusion susceptibility testing criteria for gram-positive cocci. J. Clin. Microbiol. 33:1712-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasquez, J. E., E. S. Walker, B. W. Franzus, B. K. Overbay, D. R. Reagan, and F. A. Sarubbi. 2000. The epidemiology of mupirocin resistance among methicillin-resistant Staphylococcus aureus at a Veterans' Affairs Hospital. Infect. Control Hosp. Epidemiol. 21:459-464. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe, H., H. Masaki, N. Asoh, K. Watanabe, K. Oishi, S. Kobayashi, A. Sato, R. Sugita, and T. Nagatake. 2001. Low concentrations of mupirocin in the pharynx following intranasal application may contribute to mupirocin resistance in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:3775-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao, J. D. C., and R. C. Moellering, Jr. 1999. Antibacterial agents, p. 1474-1504. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.