Abstract
Cell-to-cell signaling involving N-acyl-homoserine lactone compounds termed autoinducers (AIs) is instrumental to virulence factor production and biofilm development by Pseudomonas aeruginosa. In order to determine the importance of cell-to-cell signaling during the colonization of mechanically ventilated patients, we collected 442 P. aeruginosa pulmonary isolates from 13 patients. Phenotypic characterization showed that 81% of these isolates produced the AI-dependent virulence factors elastase, protease, and rhamnolipids. We identified nine genotypically distinct P. aeruginosa strains. Six of these strains produced AIs [N-butanoyl-homoserine lactone or N-(3-oxo-dodecanoyl)-homoserine lactone] and extracellular virulence factors (elastase, total exoprotease, rhamnolipid, hydrogen cyanide, or pyocyanin) in vitro. Three of the nine strains were defective in the production of both AIs and extracellular virulence factors. Two of these strains had mutational defects in both the lasR and rhlR genes, which encode the N-acyl-homoserine lactone-dependent transcriptional regulators LasR and RhlR, respectively. The third of these AI-deficient strains was only mutated in the lasR gene. Our observations suggest that most, but not all, strains colonizing intubated patients are able to produce virulence factors and that mutations affecting the cell-to-cell signaling circuit are preferentially located in the transcriptional regulator genes.
The gram-negative bacterium Pseudomonas aeruginosa is a major nosocomial pathogen responsible for severe pulmonary infections in mechanically ventilated patients, a complication associated with high mortality and excessive hospital costs (7, 27). Among ventilator-associated pneumonia (VAP) caused by P. aeruginosa, 95% of cases are preceded by colonization of the respiratory tract, a well-recognized risk factor for pulmonary infection (20). The risk of colonization of intubated patients by P. aeruginosa is directly linked to the presence of the endotracheal intubation device and increases with the duration of intubation (29).
P. aeruginosa regulates the production of several extracellular virulence factors by at least two cell-to-cell signaling systems, called lasRI and rhlRI (60). This circuit is organized in a hierarchical cascade in which the lasRI system is required for full expression of the rhlRI system (28). Accumulation of the two autoinducers (AIs) N-(3-oxododecanoyl)-homoserine lactone (3-oxo-C12-HSL) (39) and N-butanoyl-homoserine lactone (C4-HSL) (40), the products of the LasI and RhlI enzymes, respectively, allows the bacteria to sense their own cell density (17) in order to coordinate the production of virulence factors (9, 38, 60). As a result, the secretion of extracellular virulence factors such as elastase increases strongly once a threshold bacterial cell density has been reached (38). This phenomenon has been suggested to play an important role during acute P. aeruginosa infections, overwhelming host defenses and explaining the frequently dramatic clinical evolution (60). Cell-to-cell signaling has also been suggested to be required for the establishment of a P. aeruginosa biofilm (8, 15). This growth pattern is required for P. aeruginosa to colonize inert surfaces and possibly also the lung of cystic fibrosis (CF) patients (6, 53). Several studies with various animal models and las/rhl mutants have clearly demonstrated the importance of a functional cell-to-cell signaling circuit for the full virulence of P. aeruginosa (48). The in vivo production of AIs has been demonstrated in a chronic pneumonia mouse model (64). However, only few data exist concerning the role of cell-to-cell signaling during P. aeruginosa infections in humans. Clinical isolates deficient in the production of extracellular virulence factors regulated by cell-to-cell signaling, such as elastase and exotoxin A, have been described previously. Most isolates were specifically affected in the expression of the lasB (elastase) and toxA (exotoxin A) genes (22, 43, 47), whereas cell-to-cell signaling-deficient clinical isolates have been described only sporadically (19, 22, 54). One lasR mutant still able to produce cell-to-cell signals has been isolated from a wound (22), and one rhlR mutant has been isolated from a patient with a urinary tract infection (54). The production of AIs by clinical isolates has been demonstrated in vitro for P. aeruginosa strains isolated from CF patients (19) and for ex vivo in sputum of CF patients colonized by P. aeruginosa (53). We have detected AIs in situ in CF lung tissue (14). These results have been recently confirmed by the detection of AIs in the sputum of CF patients (11, 33). In contrast, no data are available concerning the production of AIs by P. aeruginosa strains colonizing mechanically ventilated patients. We have recently isolated AIs from biofilms covering endotracheal intubation devices that were retrieved from patients colonized by P. aeruginosa (S. Favre-Bonté et al., unpublished data), demonstrating that AIs are effectively produced in this clinical setting.
To extend our understanding of the role of cell-to-cell signaling during pulmonary colonization of intubated patients, we collected 442 P. aeruginosa isolates colonizing the respiratory tract of 13 intubated patients during the first 3 days of documented colonization. Invasive isolates responsible for acute pulmonary infections in these patients were also collected. The entire collection was screened semiquantitatively for the production of three cell-to-cell signaling-dependent virulence factors. In this collection, we identified nine genotypically different strains and quantified their production of various extracellular virulence factors and cell-to-cell signaling molecules, along with their adherence and biofilm formation capacities. We describe the characterization of three unusual clinical strains that are unable to produce cell-to-cell signals as a result of mutations in the lasR and/or rhlR transcriptional regulator genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
We collected daily pulmonary aspirates from 13 intubated patients that were hospitalized in the intensive care units of the University Hospital Geneva and were colonized by P. aeruginosa. The aspirates were plated on selective agar plates (cetrimide [0.03%]) in order to identify Pseudomonas isolates (14). From each aspirate, 10 to 17 P. aeruginosa isolates were collected individually and stored at −70°C. The genetically marked strains and plasmids used in the present study are listed in Table 1. Both Escherichia coli and P. aeruginosa were routinely grown on nutrient agar plates, in nutrient yeast broth (55) or in Luria-Bertani (LB) broth (49) at 37°C. For pyocyanin extraction, P. aeruginosa strains were grown with aeration in glycerol-alanine medium (16). For quantitative hydrogen cyanide (HCN) determination, P. aeruginosa strains were grown under oxygen limitation in tightly closed 125 ml-bottles containing 40 ml of glycine minimal medium (4). For adhesion and biofilm formation assays, P. aeruginosa strains were grown in M63 medium supplemented with 0.05% Casamino Acids and 0.2% glucose (36) or in AP medium (100 mM monosodium glutamate, 100 mM sodium gluconate, 7.5 mM NaH2PO4, 16.3 mM K2HPO4, 10 mM MgSO4 · H2O [pH 7.0]) supplemented with 0.3 M NaCl (59). Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml, gentamicin at 10 μg/ml, and kanamycin at 50 μg/ml for E. coli and carbenicillin at 250 μg/ml and gentamicin at 10 μg/ml for P. aeruginosa. To counterselect E. coli S17-1 donor cells in matings with P. aeruginosa, chloramphenicol was used at 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | 55 |
| 4.1A1 | Clinical respiratory isolate, genotype L | This study |
| 3D10 | Clinical respiratory isolate, genotype C | This study |
| 7B5 | Clinical respiratory isolate, genotype B | This study |
| 19G12 | Clinical respiratory isolate, genotype H | This study |
| 36D5 | Clinical respiratory isolate, genotype A | This study |
| 60B5 | Clinical respiratory isolate, genotype D | This study |
| 22D10 | Clinical respiratory isolate, genotype F, lasR | This study |
| 1.1A1 | Clinical respiratory isolate, genotype K, lasR, rhlR | This study |
| 18F7 | Clinical respiratory isolate, genotype I, lasR, rhlR | This study |
| 64C4 | Clinical blood isolate, genotype F | This study |
| 64C2 | Clinical blood isolate, genotype I | This study |
| 43A2 | Clinical respiratory isolate, genotype A | This study |
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR Δ(lacZYA-argF) U169 λ(φ80dlacZΔM15) | 49 |
| S17-1 | thi pro hsdR+recA/RP4-2 Tc::Mu, Km::Tn7 (RP4 to 2 = RP4ΔTn1) | 52 |
| SM10/λpir | thi-1 thr-1 leuB26 tonA21 lacY1 supE44 recA integrated RP4-2 Tcr::Mu Kmr/λpir | 35 |
| Plasmids | ||
| pLP105 | pSW200 carrying lasRI on a 2.4-kb BamHI-XhoI fragment; Apr | 51 |
| pME3280a | Mini-Tn7 gene delivery vector based on pUX-BF5 with a HindIII-SmaI-KpnI-NcoI-SphI-StuI-SpeI multiple cloning site; Gmr Apr | 65 |
| pME3280b | Mini-Tn7 gene delivery vector based on pUX-BF5 with a HindIII-PstI-MluI-SpeI multiple cloning site; Gmr Apr | 65 |
| pME3827 | pME6001 carrying lasR on a 1.1-kb PvuI-KpnI fragment; Gmr | 42 |
| pME3840 | pME6001 carrying rhlRI on a 2-kb PstI fragment; Gmr | 42 |
| pME3870 | pME3280a carrying rhlR on a 1.25-kb HindIII-SpeI fragment; Apr Gmr | This study |
| pME3871 | pME3280b carrying rhlR1 on a 2-kb PstI fragment; Apr Gmr | This study |
| pME3872 | pME3280a carrying lasR on a 1.1-kb [PvuI]-KpnI fragment; Apr Gmr | This study |
| pUK21 | Cloning vector, ori from pMB1; Kmr | 63 |
| pUX-BF5 | “Carrier” plasmid containing the attTn7::mini-Tn7-Km system; Kmr | 1 |
| pUX-BF13 | “Helper” plasmid containing Tn7 transposition functions; R6K replicon; Apr | 1 |
Tcr, tetracycline resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Apr, ampicillin resistance.
Genotyping.
Individual isolates were genotyped by the random amplified polymorphic DNA (RAPD) method as previously described (32). The RAPD reactions were performed on cell pellets of overnight cultures by using the RAPD Ready-to-Go kit (Pharmacia) and primer 208 (30). One isolate per RAPD type was subsequently typed by the pulsed-field gel electrophoresis method as previously described (57). DNA was digested with the restriction enzyme SpeI.
Phage typing and mutator assay.
A set of 10 typing phages obtained from T. L. Pitt (Central Public Health Laboratory, London, United Kingdom) was used at routine test dilution as described by Bergan (2). The mutator phenotype was assessed essentially as described by Oliver et al. (37).
DNA manipulations and sequencing.
Small-scale preparations of plasmid DNA were carried out by the CTAB (cetyltrimethylammonium bromide) method (10), and large-scale preparations were performed by using Nucleobond AX100 columns (Macherey Nagel). Chromosomal DNA was extracted with phenol-chloroform (1:1) by a method described previously (18). Restriction digests, ligations, electrophoresis, and other manipulations of nucleic acid fragments were performed by standard techniques (49). Restriction fragments were purified from agarose gels with GeneClean II (Bio 101). Transformation of E. coli and P. aeruginosa was carried out by electroporation (13). For Southern blotting, chromosomal DNA was digested overnight with the appropriate enzymes, electrophoresed on a 0.8% agarose gel, and transferred to a Hybond-N nylon membrane (Amersham Biosciences). DNA probe labeling, hybridization, and detection were performed by using the DIG DNA labeling and detection kit (Roche) according to the manufacturer's procedures. To detect the rhlR/rhlI genes, chromosomal DNA was digested with PstI and BglII, and a 2-kb PstI probe containing rhlR and rhlI from strain PAO1 was obtained from plasmid pME3840 (42). To detect the lasR/lasI genes, chromosomal DNA was digested with PstI, SacII, or NcoI, respectively, and probed with a 1.7-kb SalI fragment containing the lasR/lasI region from plasmid pLP105 (51). Primers las1 (5′-CGCCGAACTGGAAAAGTGGC-3′) and las2 (5′-TGAGAGGCAAGATCAGAGG-3′) were used to amplify by PCR the entire 1.24-kb chromosomal lasR region including the lasR leader region. Nucleotide sequences were determined on both strands with a dye terminator kit (Perkin-Elmer) and an ABI Prism 373 Sequencer by using las1 and las2 primers.
Complementation by single-copy insertion.
Complementation was carried out by using a Tn7-based system for a single-copy insertion into chromosomes of gram-negative bacteria (24, 65). Genes of interest were cloned into plasmids pME3280a or pME3280b containing a mini Tn7-Gm transposon (Table 1). The 2-kb PstI fragment from pME3840 containing rhlRI was cloned into pME3280b to generate pME3871. To obtain pME3870, the rhlR gene was excised from pME3840 on a 1.25-kb PstI-BglII fragment and inserted via a pUK21-derived linker into pME3280a between HindIII and SpeI. Plasmid pME3872 carrying lasR was generated by inserting the 1.1-kb PvuI-KpnI fragment from pME3827 into pME3280a between StuI and KpnI. The PvuI end was made blunt by treatment with T4 DNA polymerase. Chromosomal insertion of the mini-Tn7 constructs carrying lasR, rhlR, and rhlRI, respectively, were obtained via three-parental matings as follows. Recipient P. aeruginosa strains, grown overnight at 43°C, were mixed with E. coli SM10/λpir carrying the pUX-BF13 transposase helper plasmid (1) and E. coli S17-1 carrying pME3871, pME3870, or pME3872.
AI estimation by thin-layer chromatography.
Clinical P. aeruginosa isolates were cultivated with shaking in 20 ml of nutrient yeast broth (in a 50-ml Erlenmeyer flask) at 37°C to obtain an optical density at 600 nm (OD600) of ∼2 or more. Cells were removed by centrifugation, and supernatants were adjusted to pH 5 prior to extraction with an equal volume of dichloromethane. Water was removed from the organic phase by using dry magnesium sulfate, and dichloromethane was evaporated by rotary evaporation. The total extract was concentrated 200-fold by dissolving it in 100 μl of 50% (vol/vol) acetonitrile in 5 mM KH2PO4-Na2HPO4 buffer (pH 7.0). An aliquot was loaded onto a C18 reversed-phase thin-layer chromatography plate. The chromatograms were developed with a methanol-water eluant (60:40 [vol/vol]) and revealed by the indicator organisms, Chromobacterium violaceum mutant CV026 (31) for C4-HSL and Agrobacterium tumefaciens (traG-lacZ) (5) for 3-oxo-C12-HSL. The content of AIs was estimated by comparison with C4-HSL and 3-oxo-C12-HSL standards. The absence of AI production by strains 22D10, 1.1A1, and 18F7 was confirmed by independent specific bioassays. Briefly, bacterial strains were grown in LB medium until they reached stationary phase. Aliquots of filtered culture supernatants were subjected to two extractions with 1 volume of acidified ethyl acetate (0.01% acetic acid). The extracted AIs were quantified by using E. coli MG4λI14(pPCS1) for 3-oxo-C12-HSL and P. aeruginosa PAO-JP2(pECP61.5) for C4-HSL as reporter strains (62). β-Galactosidase activity was determined as previously described (34).
Production of pyocyanin, HCN, elastase, and rhamnolipids and motility assay.
For pyocyanin quantification, clinical isolates were grown for 24 h in glycerol-alanine medium (20 ml in 50-ml Erlenmeyer flasks) at 37°C with shaking. Pyocyanin was extracted with chloroform from cell-free supernatants and assayed spectrophotometrically at 520 nm (12). For HCN determinations, the cultures were grown in glycine minimal medium at 37°C with gentle shaking for 16 h. HCN was quantified as described elsewhere (21). Total extracellular protease (61) and rhamnolipid (26) production were determined semiquantitatively by agar plate assays. Elastase production was measured quantitatively in culture supernatants of bacteria grown for 8 h in LB medium by the Elastin Congo red assay (41). For swimming motility 2 μl overnight suspensions were stabbed into swimming plates (LB medium containing 0.3% agar) incubated overnight at 20°C. Swarming motility was measured as described previously (25).
Adhesion capacity and biofilm formation.
Adhesion capacity was measured as previously described (36) with the following modifications. Strains were grown overnight without agitation in LB medium, resuspended in M63 medium supplemented with 0.05% Casamino Acids and 0.2% glucose to an OD600 of 1, and incubated in microtiter plates at 37°C for 1 h. Quantification of the adhering bacteria to microtiter dishes was determined by crystal violet staining (36). Biofilm formation occurred in static conditions during 3 days as previously described (15, 58). Briefly, P. aeruginosa strains were grown in LB medium with agitation at 37°C for 6 h. Sterile polyvinyl chloride (PVC) pieces of 1 cm2 were incubated for 1 h without agitation to allow bacteria to adhere, transferred to AP medium supplemented with 0.3 M NaCl (59), and incubated for 3 days without agitation at 37°C. Biofilm was quantified by crystal violet staining (36).
RESULTS
Collection of clinical isolates.
We collected a total of 442 P. aeruginosa isolates from thirteen intubated patients. This collection consisted of 34 isolates collected in pulmonary aspirates from each of the 13 intubated patients during the first 3 days of established colonization (Table 2). Six of the thirteen patients were severe trauma patients, two had experienced heart failure, one had non-Pseudomonal sepsis, one had brain hemorrhage, one had chronic obstructive pulmonary disease with bronchiectasis, one had bacterial non-Pseudomonas pneumonia, and one had taken a suicidal drug overdose. The duration of intubation before colonization by P. aeruginosa was detected varied between 1 and 14 days (Table 2). In order to determine the importance of cell-to-cell signaling during respiratory tract colonization of these patients, we determined semiquantitatively the production of the AI-dependent virulence factors elastase, total exoprotease, and rhamnolipids of the entire collection. A total of 81% of these isolates were able to produce elastase and protease, and 80% produced rhamnolipids (data not shown).
TABLE 2.
Patient characteristics and isolate collection
| Patient | Underlying diseasea | Duration (days) of intubation before documented colonization | No. of isolates analyzed (genotype) |
|---|---|---|---|
| 1 | Drug overdose | 10 | 34 (K) |
| 2 | Polytrauma | 9 | 34 (L) |
| 3 | COPD and bronchiectasis | 1 | 10 (C) |
| 24 (F) | |||
| 4 | Polytrauma | 10 | 11 (B) |
| 23 (C) | |||
| 5 | Nonpseudomonal sepsis | 2 | 8 (C) |
| 26 (I) | |||
| 6 | Nonpseudomonal pneumonia | 10 | 34 (H) |
| 7 | Brain hemorrhage | 14 | 34 (A) |
| 8 | Polytrauma | 9 | 6 (B) |
| 28 (D) | |||
| 9 | Myocardial infarct | 2 | 34 (B) |
| 10 | Polytrauma | 7 | 34 (A) |
| 11 | Polytrauma | 3 | 34 (B) |
| 12 | Polytrauma | 5 | 22 (B) |
| 12 (C) | |||
| 13 | Congestive heart failure | 2 | 34 (B) |
COPD, chronic obstructive pulmonary disease.
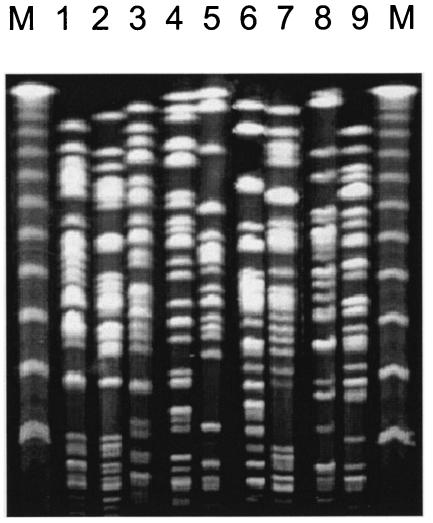
Among the 442 isolates, 9 genetically independent strains were identified by RAPD genotyping (data not shown), and the identifications were confirmed by pulsed-field gel electrophoresis (Fig. 1). Five patients were cocolonized with two genetically different strains at the same time: patient 3 by strains C and F, patient 4 by strains B and C, patient 5 by strains C and I, patient 8 by strains B and D, and patient 12 by strains B and C (Table 2). Patients 3, 5, and 7 presented an acute-pseudomonal pulmonary infection. For patient 7, the invasive isolate (43A2 [genotype A]) was isolated from a bronchoalveolar lavage at >104 CFU/ml; in association with a compatible clinical picture, this is diagnostic of a pulmonary infection (44). For patients 3 and 5, the invasive isolates (66C4 [genotype F] and 64C2 [genotype I], respectively) were recovered from the blood. All isolates belonging to the same genotype had the same AI-dependent virulence phenotype except for some isolates belonging to genotype C that showed fluctuations in rhamnolipid production (data not shown). Therefore, one representative isolate of each of the nine genotypes was selected and further characterized: patient 1, isolate 1.1A1 with genotype K; patient 2, isolate 4.1A1 with genotype L; patient 3, isolate 22D10 with genotype F; patient 4, isolate 3D10 with genotype C; patient 5, isolate 7B5 with genotype B; patient 6, isolate 18F7 with genotype I; patient 7, isolate 19G12 with genotype H; patient 8, isolate 36D5 with genotype A; and patient 8, isolate 60B5 with genotype D (Table 3).
FIG. 1.
PFGE profiles (SpeI digestion) of the 9 genotypes of P. aeruginosa. Lanes: 1, isolate 36D5, genotype A; 2, isolate 7B5, genotype B; 3, isolate 3D10, genotype C; 4, isolate 60B5, genotype D; 5, isolate 22D10, genotype F; 6, isolate 19G12, genotype H; 7, isolate 18F7, genotype I; 8, isolate 1.1A1, genotype K; 9, isolate 4.1A1, genotype L. M, molecular weight markers (lambda ladder).
TABLE 3.
Characterization and complementation of P. aeruginosa strains isolated from intubated patients
| Strain (genotype) | Productiona of:
|
Productionb of:
|
Elastaseb (OD495) | Productione of:
|
Adhesionb (%) | Biofilmb (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| C4-HSL (μM) | 3-Oxo-C12-HSL (nM) | Pyocyanin (μg/ml) | HCN (μM) | Rhamnolipids | Proteases | ||||
| PAO1 | 5 | 300 | 10 | 29.2 | 2.7 | +++ | +++ | 100 | 100 |
| 4.1A1 (L) | 5 | 250 | 1 | 52.8 | 2.7 | +++ | +++ | 97 | 88 |
| 3D10 (C) | 4.5 | 340 | 8.9 | 23.4 | 2.7 | + | ++ | 104 | 91 |
| 7B5 (B) | 3 | 125 | 12 | 15.3 | 2.7 | + | +++ | 147 | 150 |
| 19G12 (H) | 1 | 260 | 3.5 | 27.6 | 2.3 | + | +++ | 31 | 10 |
| 36D5 (A) | 3 | 340 | 5.1 | 43.6 | 1.2 | ++ | +++ | 130 | 76 |
| 60B5 (D) | 4 | 375 | 8 | 34.3 | 2.6 | + | +++ | 139 | 33 |
| 22D10 (F) | <0.5 | <50 | 2.5 | <5 | 0.03 | BD | + | 62 | 70 |
| lasR+ | 2 | 150 | 25 | 9.7 | 2.6 | +++ | +++ | NDd | ND |
| rhlR+ | <0.5 | <50 | 2.3 | <5 | 1.1 | BD | + | ND | ND |
| 1.1A1 (K) | <0.5 | <50 | <1 | <5 | BDc | BD | BD | 116 | 47 |
| lasR+ | 1 | 375 | <1 | 9.2 | 2.7 | BD | +++ | ND | ND |
| rhlR+ | <0.5 | <50 | 27.8 | <5 | 0.8 | + | + | ND | ND |
| 18F7 (I) | <0.5 | <50 | <1 | <5 | BD | BD | BD | 88 | 76 |
| lasR+ | 4 | 300 | <1 | 9.4 | 1.2 | BD | ++ | ND | ND |
| rhlR+ | <0.5 | <50 | 27.4 | <5 | 0.6 | + | + | ND | ND |
Values are means of three independent experiments with a standard deviation of 10%.
Values are means of three independent experiments with a standard deviation of <10%.
BD, below detection.
ND, not done.
+++, high level; ++, intermediate level; +, low level.
Phenotypic characterization of colonizing P. aeruginosa strains. (i) Phage typing and hypermutable phenotype.
The strains 7B5, 3D10, 60B5, 22D10, 18F7, 1.1A1, and 4.1A1 each gave distinct phage typing patterns, confirming molecular typing, whereas strains 36D5 and 19G12 were insensitive to the phages used (data not shown). None of the nine strains showed a hypermutable phenotype in that none demonstrated an increased level of spontaneous rifampin-resistant mutants compared to wild-type PAO1.
(ii) Adhesion and biofilm formation capacity.
Compared to strain PAO1, the majority of the strains had an equivalent or higher capacity to adhere to PVC (Table 3). Only strains 22D10, 19G12, and 18F7 had a reduced capacity to adhere in our assay. Notably, strain 19G12 had a drastically reduced adherence capacity compared to strain PAO1. As expected for a strain with reduced adherence capacity, strain 19G12 formed almost no biofilm. All strains were able to form biofilm in a static biofilm assay, however, with a somewhat reduced capacity compared to strain PAO1, with the exception of strain 7B5 (Table 3).
(iii) Production of cell-to-cell signaling-dependent virulence factors.
To characterize further the nine genotypically different strains, we quantified their in vitro production of several extracellular virulence factors that are dependent upon an active cell-to-cell signaling circuitry, namely, pyocyanin, hydrogen cyanide (HCN), and elastase (60), as described in Materials and Methods. The production of pyocyanin was below the detection threshold of our assay (1 μg/ml) for strains 18F7 and 1.1A1 and low for strain 4.1A1 compared to the genetically well characterized wild-type strain PAO1 (Table 3). Similarly, HCN could not be detected in culture supernatants of strains 18F7 and 1.1A1 (detection limit, 5 μM). In contrast to pyocyanin, HCN was elevated in the supernatant of strain 4.1A1 and could not be detected in supernatants of strain 22D10 (Table 3). All strains, except 22D10, 18F7, and 1.1A1, produced significant amounts of elastase (Table 3). These results show that six of nine P. aeruginosa strains colonizing the 13 intubated patients were able to produce extracellular virulence factors controlled by the cell-to-cell signaling circuitry. The lack of protease, elastase, rhamnolipid, and HCN production by the three remaining strains (22D10, 1.1A1, and 18F7) suggested that these three strains might have a defective cell-to-cell signaling circuit.
(iv) Production of cell-to-cell signals.
At an OD600 of 2.0 (late exponential phase), six of the nine strains produced amounts of 3-oxo-C12-HSL comparable to those of strain PAO1 (Table 3). Strains 22D10, 1.1A1, and 18F7 produced less than 50 nM 3-oxo-C12-HSL, which was the detection threshold of the thin-layer chromatography assay. The production of C4-HSL was determined at an OD600 of 2.0 (Table 3) and also after overnight incubation (OD600 = 4 to 5) (data not shown). At an OD600 of 2.0, all strains except four (19G12, 22D10, 1.1A1, and 18F7) produced amounts of C4-HSL comparable to those of strain PAO1. In late-stationary-phase cultures (OD600 = 4 to 5), strain 19G12 produced normal amounts of C4-HSL, whereas the production of this AI remained below the detection limit (0.5 μM) in the supernatants of the other three strains (data not shown). Even the more sensitive bioassays (detection limits of 1 pM for 3-oxo-C12-HSL and 1 nM for C4-HSL) (62) were unable to detect AIs in supernatants of strains 22D10, 1.1A1, and 18F7 (data not shown).
The production of the two AIs by six of nine strains, representing 81% of the 442 isolates, suggested that a majority of the P. aeruginosa strains colonizing the 13 intubated patients may have active lasRI and rhlRI cell-to-cell signaling systems. In contrast, strains 22D10, 1.1A1, and 18F7 seemed to have a defective cell-to-cell signaling circuit.
Analysis of the rhlR/rhlI and lasR/lasI genes in cell-to-cell signaling-deficient strains.
We checked the integrity of the cell-to-cell signaling genes rhlR/rhlI and lasR/lasI in all nine clinical strains by Southern blotting. All nine strains and the reference strain PAO1 gave identical hybridization signals of 0.73- and 1.25-kb after digestion with PstI-BglII, using a rhlR/rhlI probe (data not shown), suggesting that all strains possessed rhlR and rhlI, without an obvious deletion or insertion in these genes. Six AI-producing strains, as well as the two AI-negative strains 1.1A1 and 18F7, gave the same hybridization pattern (signals of 0.41, 0.59, and 1.0 kb) with a lasR/lasI probe, as did strain PAO1 when chromosomal DNA was digested with PstI (data not shown). However, the AI-deficient strain 22D10 and the AI-proficient strain 19G12 showed a different pattern. In strain 22D10, the 0.41-kb PstI band corresponding to the central lasR region was missing, suggesting either a DNA polymorphism in the PstI sites or a deletion. For strain 19G12, the absence of the 0.59-kb PstI band corresponding to the 3′ end of lasI, combined with the appearance of a new high-molecular-weight band, suggested either a polymorphism, a deletion, or an insertion at the end of the lasI gene.
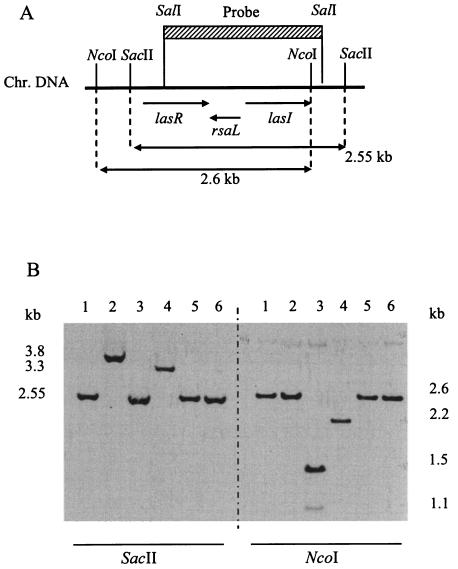
In order to include the lasR promoter region (not detected by PstI digestion), we performed two other chromosomal DNA digests, using either NcoI or SacII (Fig. 2A). For strain 22D10, digestion with SacII led to a shift of the expected 2.55-kb band to ∼3.3 kb, whereas digestion with NcoI led to a shift from the expected 2.6-kb band to ∼2.2 kb (Fig. 2B). This band pattern suggested the presence of a deletion in the lasR gene of strain 22D10. This was confirmed by PCR amplification of the lasR gene region of strain 22D10, which yielded a fragment that was smaller by ∼460 bp than the functional 1.24-kb lasR gene fragment of the reference strain PAO1 (data not shown). For strains 1.1A1 and 18F7, restriction with one enzyme gave the expected fragment lengths (2.6 kb for NcoI in strain 1.1A1 and 2.55 kb for SacII in strain 18F7), whereas restriction with the second enzyme (SacII for strain 1.1A1 and NcoI for strain 18F7) generated new fragments, suggesting a polymorphism for both strains (Fig. 2B). Two bands (1.1 and 1.5 kb) were generated by the NcoI digest of strain 18F7, suggesting the creation of a new NcoI site. For strain 19G12, no indication of a deletion or insertion was found (Fig. 2B).
FIG. 2.
Southern hybridization of chromosomal DNA from a subset of P. aeruginosa isolates to a lasR-lasI′ probe. (A) Location of the probe relative to an extended lasR-lasI region; (B) Southern blot of chromosomal DNA digested with SacII (left part of panel) or NcoI (right part of panel). Lanes: 1, PAO1; 2, 1.1A1; 3, 18F7; 4, 22D10; 5, 19G12; 6, 3D10.
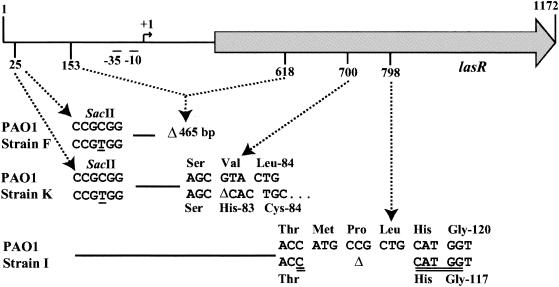
In order to corroborate these results, we sequenced the lasR gene of strains 22D10, 1.1A1 and 18F7 (Fig. 3). A deletion of 465 bp, including the lasR promoter in strain 22D10, a 9-bp in-frame deletion of strain 18F7, generating a new NcoI site, as suggested by the band pattern in Fig. 2B, and a 1-bp frameshift, as well as several point mutations for strain 1.1A1, including mutations leading to the loss of the SacII site upstream of lasR, explain the band pattern in Fig. 2B. These results show that the three clinical strains 22D10, 1.1A1, and 18F7 bear mutations in the lasR gene. This could explain their virulence factor-negative phenotype, as well as their deficiency in AI production. In order to determine whether lasR and/or rhlR mutations might also be present in cell-to-cell signaling-proficient strains, we sequenced several isolates belonging to genotype L. No mutations were found in three different isolates (data not shown).
FIG. 3.
Location of mutations in the lasR gene of P. aeruginosa isolates 22D10 (genotype F), 1.1A1 (genotype K), and 18F7 (genotype I) relative to the lasR gene of strain PAO1. Shaded arrow, lasR gene; −35, −10, +1, elements of lasR promoter; Δ, deletion; underlined bases, polymorphism in the SacII site at position 25; double-underlined bases, new NcoI site. Coordinates refer to the nucleotide sequence of lasR: the “1” in this figure corresponds to nucleotide 1557718 (56).
Complementation of strains 22D10, 1.1A1, and 18F7.
To confirm the role of the lasR mutations in strains 22D10, 1.1A1 and 18F7, we introduced the functional lasR gene of strain PAO1 by chromosomal insertion into these three strains, as described in Materials and Methods. The insertion of a single lasR+ copy into strain 22D10 restored the production of both AIs, as well as the production of pyocyanin, elastase, and HCN (Table 3). The chromosomal insertion of lasR+ into strains 1.1A1 and 18F7 restored the production of AIs, elastase, and HCN. However, the production of pyocyanin was not restored (Table 3). Interestingly, the production of pyocyanin could be restored by the insertion of the rhlR+ gene alone; however, this did not restore the production of AIs and of HCN by these two strains (Table 3). The rhlR genes of strains 1.1A1 and 18F7 were therefore isolated by PCR amplification and sequenced. The rhlR gene of strain 18F7 had a stop codon (TAG) instead of a glutamine codon (CAG) at position 25, whereas the rhlR gene of strain 1.1A1 differed from that of strain PAO1 at several positions (CCC to TCC [P65S], GAT to GAG [D81Q], and GTG to GCG [V94A]). These data indicate that the RhlR protein is truncated in strain 18F7 and probably also nonfunctional in strain 1.1A1 and that, in both strains, rhlR+ could restore the defect in pyocyanin production, as expected based on the findings of a previous study (3).
These results confirm that the lasR deletion of strain 22D10 accounts for the observed phenotype. Concerning strains 1.1A1 and 18F7, the combination of lasR and rhlR mutations probably explains their AI and exoproduct deficiencies. As the genetic differences of strains 22D10, 1.1A1, and 18F7 were further confirmed by phage typing (see above), these strains are unrelated. Thus, the emergence of cell-to-cell signaling-negative variants is not limited to a specific genotype.
Cell-to-cell signaling in three invasive P. aeruginosa strains.
Three of the 13 patients—patients 3, 5, and 7—presented a P. aeruginosa pneumonia as a complication of their respiratory colonization. For patients 3 and 5, a P. aeruginosa bacteremia was documented. The three isolates responsible for an invasive complication (strains 64C4, 64C2, and 43A2, respectively) were genotypically different. In all cases the invasive strain belonged to a genotype that had been previously recovered as a colonizing strain (64C4, genotype F; 64C2, genotype I; and 43A2, genotype A). The virulence phenotypes and AI production, as well as the adherence and biofilm formation capacities of the invasive isolates, were identical to those of their colonizing counterparts (data not shown). The AI-dependent virulence factor production of isolate 43A2, responsible for pneumonia in patient 7, was comparable to PAO1 levels. In contrast, the bacteremic isolates 64C4 and 64C2, which were recovered from patients 3 and 5, respectively, were significantly impaired in their capacity to produce AI-dependent virulence factors due to mutations in the lasR gene, as well as in rhlR for strain 64C2. Strikingly, patients 3 and 5 were both cocolonized by strains of the genotype C, producing extracellular virulence factors and signal molecules at PAO1 levels (Table 2).
DISCUSSION
P. aeruginosa is responsible for severe nosocomial pneumonia in mechanically ventilated patients (46). Since cell-to-cell signaling-deficient mutants are significantly less virulent in pneumonia animal models, it has been postulated that cell-to-cell signaling might play a major role in the pathogenesis of P. aeruginosa lung infections (48, 60). However, only few data are available so far on the function of cell-to-cell signaling in pulmonary isolates of P. aeruginosa.
In the present study, we measured the production of three AI-dependent virulence factors as indicators of signaling in 442 P. aeruginosa isolates collected during the first 3 days of documented colonization of 13 intubated patients. A total of 81% of these isolates produced significant amounts of AI-dependent virulence factors, suggesting that these isolates probably were signaling proficient. This observation is in agreement with the expectation that cell-to-cell signaling would be important during colonization of intubated patients by P. aeruginosa. However, since we did not determine the in vivo activity of cell-to-cell signaling during colonization, the importance of AI-mediated signaling in this process remains speculative. Moreover, since our patients were not routinely screened for P. aeruginosa colonization, the duration of colonization (about 3 days) monitored in the present study might fluctuate between 1 and 5 days.
Among the 442 isolates we identified nine genotypically different strains. We showed that six of these nine genotypically different P. aeruginosa strains had a functional cell-to-cell signaling circuit. These six strains were able to produce in vitro the two AIs 3-oxo-C12-HSL and C4-HSL, as well as AI-dependent extracellular virulence factors, including total exoprotease, elastase, HCN, pyocyanin, and rhamnolipids, at levels equivalent to those of the reference strain PAO1. All strains except one were able to adhere to inert surfaces (i.e., PVC) and to produce biofilms. However, in general biofilm formation was less pronounced than in strain PAO1. These results suggest that P. aeruginosa strains colonizing intubated patients, in contrast to strains colonizing CF patients (53), do not have an increased capacity for biofilm production. This is surprising considering the importance of adhesion to, as well as biofilm formation on the endotracheal intubation device during upper respiratory tract colonization of intubated patients. It is also interesting that for the three AI-deficient strains (22D10, 1.1A1, and 18F7) the abilities to adhere to PVC and to form biofilms were comparable to those of AI-proficient strains. The only strain exhibiting a relatively low adhesion and biofilm formation capacity (strain 19G12) was essentially AI proficient.
In the three strains deficient in cell-to-cell signaling (22D10, 1.1A1, and 18F7), the lasR gene encoding the LasR transcriptional regulator was inactivated by various mutations. Two of these strains also had mutations in the rhlR gene, probably explaining the defect in both AIs and extracellular virulence factor production. Since the AI production of these strains was complemented by the chromosomal insertion of the wild-type lasR and rhlR genes, additional mutations outside of the lasI and rhlI genes, encoding the AI synthases LasI and RhlI, are unlikely. Apparently, the lasR and rhlR genes are more frequently subject to mutations affecting the activity of the cell-to-cell signaling circuit than are the AI synthase genes lasI and rhlI. To our knowledge, this is the first description and characterization of clinical isolates unable to produce cell-to-cell signals as a result of both lasR and rhlR mutations. Indeed, only mutants (i) affected in the expression of lasB and toxA (22, 43, 47), (ii) mutated in lasR (22) but still able to produce cell-to-cell signals, or (iii) mutated in rhlR (54) but not characterized have been described so far. The three AI-deficient genotypes F, K, and I together represented 19% of the 442 isolates.
Surprisingly, two of three invasive strains (64C4 and 64C2) isolated from blood belonged to these lasR or lasR/rhlR mutants (genotypes F and I, respectively). This was unexpected since, in another study, 90% of 270 bacteremic P. aeruginosa isolates produced elastase and rhamnolipids, suggesting that their cell-to-cell signaling circuit was active (E. Boffi El Amari and C. van Delden, unpublished results). It has been recently suggested that cytotoxicity mediated by type III secretion could be more important than cell-to-cell signaling in the pathogenesis of P. aeruginosa pneumonia (23, 45, 50). However, the three invasive strains of the present study were not as cytotoxic as the cytotoxic reference strain PA103. In addition, none of these three strains was resistant to serum bactericidal activity (data not shown), and none was a hypermutator, which is in agreement with a previous report suggesting that hypermutable strains are mainly found among respiratory strains isolated from CF patients but are uncommon in other clinical situations (37). Very intriguing is the observation that the two patients infected with the AI-deficient strains 64C4 and 64C2 were initially cocolonized with strains of the cell-to-cell signaling-positive genotype C. This might simply be a coincidence, considering the small sample size. On the other hand, further investigations should examine whether, in some circumstances, cell-to-cell signaling-negative strains could profit from the extracellular enzymes produced by cell-to-cell signaling positive partners. One appealing hypothesis could be a relatively lower fitness of cell-to-cell signaling proficient isolates compared to cell-to-cell signaling deficient ones. The cell-to-cell signaling machinery might represent an important burden for proficient isolates under certain conditions in vivo. Further research on the role of the cell-to-cell signaling circuit during colonization of mechanically ventilated patients will hopefully give us clues as to how P. aeruginosa pneumonia in intubated patients can be better prevented or treated.
Acknowledgments
This study was supported by the program “Génie Biomédical,” the Swiss National Foundation for Scientific research (project 31-56608.99 to D.H. and projects 3231-051940.97 and 3200-052189.97 to C.V.D.), and the Herbette Foundation (to V.K.).
We thank R. Comte for outstanding technical assistance, T. Köhler for helpful discussions, and B. Ricou, J. A. Romand, and J. Pugin for help with the collection of clinical samples.
REFERENCES
- 1.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 2.Bergan, T. 1978. Phage typing of Pseudomonas aeruginosa. Methods Microbiol. 10:169-199. [Google Scholar]
- 3.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castric, P. A. 1975. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21:613-618. [DOI] [PubMed] [Google Scholar]
- 5.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Craven, D. E., and K. A. Steger. 1995. Epidemiology of nosocomial pneumonia: new perspectives on an old disease. Chest 108:1S-16S. [DOI] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to cell signals in the development of bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Sal, G., G. Manfioletti, and C. Schneider. 1988. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 16:9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson, D. L., R. Endersby, A. Kirkham, K. Stuber, D. D. Vollman, H. R. Rabin, I. Mitchell, and D. G. Storey. 2002. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 70:1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farinha, M. A., and A. M. Kropinski. 1990. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol. Lett. 70:221-225. [DOI] [PubMed] [Google Scholar]
- 14.Favre-Bonté, S., J. C. Pache, J. Robert, D. Blanc, J. C. Pechere, and C. Van Delden. 2002. Detection of Pseudomonas aeruginosa cell-to-cell signals in lung tissue of cystic fibrosis patients. Microb. Pathog. 32:143-147. [DOI] [PubMed] [Google Scholar]
- 15.Favre-Bonté, S., T. Köhler, and C. Van Delden. 2003. Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 52:598-604. [DOI] [PubMed] [Google Scholar]
- 16.Frank, L. H., and R. D. DeMoss. 1959. On the biosynthesis of pyocyanin. J. Bacteriol. 77:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 18.Gamper, M., B. Ganter, M. R. Polito, and D. Haas. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226:943-957. [DOI] [PubMed] [Google Scholar]
- 19.Geisenberger, O., M. Givskov, K. Riedel, N. Hoiby, B. Tummler, and L. Eberl. 2000. Production of N-acyl-l-homoserine lactones by P. aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiol. Lett. 184:273-278. [DOI] [PubMed] [Google Scholar]
- 20.George, D. L., P. S. Falk, R. G. Wunderink, K. V. J. Leeper, G. U. Meduri, E. L. Steere, C. E. Corbett, and C. G. Mayhall. 1998. Epidemiology of ventilator-acquired pneumonia based on protected bronchoscopic sampling. Am. J. Respir. Crit. Care Med. 158:1839-1847. [DOI] [PubMed] [Google Scholar]
- 21.Gewitz, H. S., E. K. Pistorius, H. Voss, and B. Vennesland. 1976. Cyanide formation in preparations from Chlorella vulgaris Beijerinck: effect of sonication and amygdalin addition. Planta 131:145-148. [DOI] [PubMed] [Google Scholar]
- 22.Hamood, A. N., J. Griswold, and J. Colmer. 1996. Characterization of elastase-deficient clinical isolates of Pseudomonas aeruginosa. Infect. Immun. 64:3154-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauser, A. R., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 24.Hφjberg, O., U. Schnider, H. V. Winteler, J. Sφrensen, and D. Haas. 1999. Oxygen-sensing reporter strain of Pseudomonas fluorescens for monitoring the distribution of low-oxygen habitats in soil. Appl. Environ. Microbiol. 65:4085-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Köhler, T., L. K. Curty, F. Barja, C. Van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köhler, T., C. Van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kollef, M. H. 1999. The prevention of ventilator-associated pneumonia. N. Engl. J. Med. 340:627-634. [DOI] [PubMed] [Google Scholar]
- 28.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 29.Levine, S. A., and M. S. Niederman. 1991. The impact of tracheal intubation on host defenses and risks for nosocomial pneumonia. Clin. Chest Med. 12:523-543. [PubMed] [Google Scholar]
- 30.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 32.Mereghetti, L., N. Marquet-van der Mee, J. Loulergue, J. C. Rolland, and A. Audurier. 1998. Pseudomonas aeruginosa from cystic fibrosis patients: study using whole cell RAPD and antibiotic susceptibility. Pathol. Biol. 46:319-324. [PubMed] [Google Scholar]
- 33.Middleton, B., H. C. Rodgers, M. Camara, A. J. Knox, P. Williams, and A. Hardman. 2002. Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol. Lett. 207:1-7. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Miller, V. L., and J. J. Mekalonos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 37.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 38.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 39.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Role of Pseudomonas aeruginosa las and rhl quorum-sensing systems in the control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pritchard, A. E., and M. L. Vasil. 1990. Possible insertion sequences in a mosaic genome organization upstream of the exotoxin A gene in Pseudomonas aeruginosa. J. Bacteriol. 172:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugin, J., R. Auckenthaler, N. Mili, J. P. Janssens, P. D. Lew, and P. M. Suter. 1991. Diagnosis of ventilator-associated pneumonia by bacteriologic analysis of bronchoscopic and nonbronchoscopic “blind” bronchoalveolar lavage fluid. Am. Rev. Respir. Dis. 143:1121-1129. [DOI] [PubMed] [Google Scholar]
- 45.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 46.Ruimy, R., E. Genauzeau, C. Barnabe, A. Beaulieu, M. Tibayrenc, and A. Andremont. 2001. Genetic diversity of Pseudomonas aeruginosa strains isolated from ventilated patients with nosocomial pneumonia, cancer patients with bacteremia, and environmental water. Infect. Immun. 69:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 1999. Pseudomonas aeruginosa strains obtained from patients with tracheal, urinary tract and wound infection: variations in virulence factors and virulence genes. J. Hosp. Infect. 43:211-218. [DOI] [PubMed] [Google Scholar]
- 48.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2:1721-1731. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor N. Y.
- 50.Sawa, T., M. Ohara, K. Kurahashi, S. S. Twining, D. W. Frank, D. B. Doroques, T. Long, M. A. Gropper, and J. P. Wiener-Kronish. 1998. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect. Immun. 66:3242-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seed, P. C., L. Passador, and B. H. Iglewski. 1995. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering:transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 53.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 54.Sokurenko, E. V., V. Tchesnokova, A. T. Yeung, C. A. Oleykowski, E. Trintchina, K. T. Hughes, R. A. Rashid, J. M. Brint, S. L. Moseley, and S. Lory. 2001. Detection of simple mutations and polymorphisms in large genomic regions. Nucleic Acids Res. 29:E111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanisich, V. A., and B. W. Holloway. 1972. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. 19:91-108. [DOI] [PubMed] [Google Scholar]
- 56.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 57.Struelens, M. J., V. Schwam, A. Deplano, and D. Baran. 1993. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J. Clin. Microbiol. 31:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeoka, K., T. Ichimiya, T. Yamasaki, and M. Nasu. 1998. The in vitro effect of macrolides on the interaction of human polymorphonuclear leukocytes with Pseudomonas aeruginosa in biofilm. Chemotherapy 44:190-197. [DOI] [PubMed] [Google Scholar]
- 59.Terry, J. M., S. E. Pina, and S. J. Mattingly. 1992. Role of energy metabolism in conversion of nonmucoid Pseudomonas aeruginosa to the mucoid phenotype. Infect. Immun. 60:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Delden, C., E. C. Pesci, J. P. Pearson, and B. H. Iglewski. 1998. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect. Immun. 66:4499-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 64.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, A. Heydorn, K. Mathee, C. Moser, L. Eberl, S. Molin, N. Hoiby, and M. Givskov. 2000. Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology 146:2481-2493. [DOI] [PubMed] [Google Scholar]
- 65.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHAO. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]