Abstract
A crucial step in the immune response is the binding of antigenic peptides to MHC proteins. Class II MHC proteins present their bound peptides to CD4+T cells, helping to activate both the humoral and the cellular arms of the adaptive immune response. Peptide loading onto class II MHC proteins is regulated temporally, spatially, and developmentally in antigen-presenting cells1. To help visualize these processes, we have developed a series of novel fluorogenic probes that incorporate the environment-sensitive amino acid analogs 6-N,N-dimethylamino-2-3-naphthalimido-alanine and 4-N,N-dimethylaminophthalimido-alanine. Upon binding to class II MHC proteins these fluorophores exhibit large changes in emission spectra, quantum yield, and fluorescence lifetime. Peptides incorporating these fluorophores bind specifically to class II MHC proteins on antigen presenting cells, and can be used to follow peptide binding in vivo. Using these probes we have tracked a developmentally-regulated cell surface peptide binding activity in primary human monocyte-derived dendritic cells.
Class II MHC proteins bind peptides in an extended conformation through a combination of conserved hydrogen bonds to the peptide main chain and allele-specific pockets that accommodate some of the side chains of the bound peptide. Although peptides as short as six amino acids have been shown to bind with high affinity, naturally processed and presented peptides are longer, typically 13-25 residues, with a core 9-11 residue sequence interacting with the MHC protein and the reminder extending out to both ends of the peptide binding site2. For HLA-DR, the most abundant of the three human class II MHC proteins, the major determinant of binding is a large pocket that accommodates a hydrophobic or aromatic amino acid side chain near the amino terminus of the peptide (termed the P1 position)3,4. Additional pockets accommodate peptide side chains at the P4, P6 and P9 positions, with the fine specificity varying between allelic variants2,5. To develop fluorogenic probes for binding to HLA-DR proteins, we investigated whether solvatochromic amino acid analogs could be used to replace the aromatic amino acid that usually occupies the P1 pocket in HLA-DR binding peptides. The P1 pocket is lined by conserved aliphatic and aromatic amino acids, and represents a highly non-polar environment5. We anticipated that the binding of environment-sensitive fluorophores into such a binding pocket would induce large spectral changes.
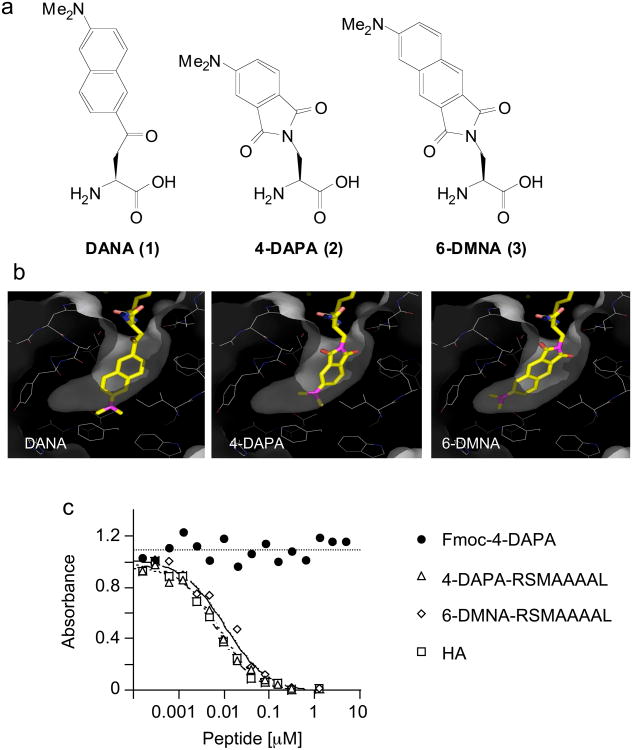
Using the crystal structure of HLA-DR1 in complex with the influenza-derived HA peptide (PKYVKQNTLKLAT)5 as a guide, we carried out molecular modeling studies to replace the tyrosine residue occupying the P1 pocket with three amino acids that integrate environment-sensitive fluorophores into the residue side chains: 6-dimethylamino-2′-naphthoyl-alanine (DANA/ALADAN)6,7 (1), 4-N,N,-dimethylaminophthalimido-alanine (4-DAPA)8 (2), and 6-N,N-dimethylamino-2,3-naphthalimido-alanine (6-DMNA)9 (3) (Fig. 1a). Peptides containing the 4-DAPA and 6-DMNA residues have been previously developed as probes for the phosphotyrosine-binding SH29 and phosphoserine/phospothreonine-binding 14-3-3 domains8, and DANA/ALADAN has been used to measure RNAase assembly7, delta-opioid antagonist binding10, and electrostatics within protein G6. In each case the amino acid side chains carry highly solvatochromic moieties as found in several environment-sensitive fluorescence probes11. Rigid body docking and torsional angle adjustment revealed that the side chain of DANA/ALADAN was too long to fit into the P1 pocket of HLA-DR1, whereas the side chains of the 4-DAPA and 6-DMNA residues could be accommodated without major distortion of the MHC protein or the bound peptide (Fig. 1b).
Figure 1. 4-DAPA and 6-DMNA can be modeled into the P1 pocket of DR1 without major distortion.
(a) Chemical structures of DANA, 4-DAPA and 6-DMNA. (b) In the crystal structure of the DR1-HA peptide complex, a tyrosine residue is buried deep into the hydrophobic P1 pocket 5. DANA, 4-DAPA and 6-DMNA were modeled in silico into this pocket in place of tyrosine. Residues shown lining the pocket are (clockwise from upper right) Pheα54, Pheα32, Trpα43 (behind), Ileα7, Trpβ153, Pheα48, Thrβ90, Valβ91, Tyrβ83, Glyβ86, and Valβ85. The side chains of Asnβ82 and Hisβ81 (upper left) form hydrogen bonds with the peptide main chain at the mouth of the P1 pocket. Other residues lining the pocket but not shown are Pheα24, Pheα26, and Pheα48 (c) Binding of fluorogenic peptides to DR1 assessed using a competitive binding assay. DR1 was incubated with biotin-HA peptide and various concentrations of unlabelled inhibitor peptides, with biotin-HA binding quantified by a sandwich ELISA assay using streptavidin alkaline phosphatase. Binding of Fmoc-(4-DAPA) was assessed to evaluate non-specific binding of the fluorophore. IC50 values for these and other peptides (Supplementary Table I online) were determined as described (Supplementary Methods online).
N-Fmoc-protected derivatives of 4-DAPA and 6-DMNA were used in the preparation of the short peptides xRSMAAAAL (where x is 4-DAPA or 6-DMNA) by solid phase peptide synthesis. The RSMAAAAL sequence includes optimal P2, P4, P6 and P9 side chains (underlined). Analogous sequences have been used previously to investigate binding preferences in HLA-DR1 and related allelic variants12. We tested the relative affinity of these peptides for recombinant soluble HLA-DR1 extracellular domains using a standard competition-binding assay (Fig.1c). (4-DAPA)-RSMAAAAL and (6-DMNA)-RSMAAAAL bound tightly to HLA-DR1, with IC50 values comparable to the tight binding influenza-derived HA 306-318 peptide13, whereas neither the protected 4-DAPA amino acid alone (Fmoc-4-DAPA) nor 4-DAPA-containing control peptide (4-DAPA-ETDKDKRS) exhibited detectable binding to HLA-DR1 (Fig.1c. and Supplementary Table 1 online).
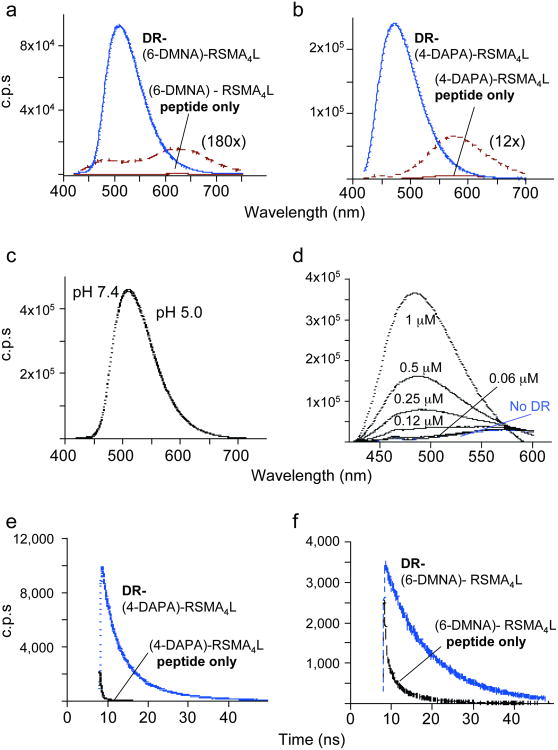
To evaluate the spectral properties of the bound fluorophores, HLA-DR1 was incubated with excess synthetic (4-DAPA)-RSMAAAAL and (6-DMNA)-RSMAAAAL peptides, and MHC-peptide complexes were isolated by gel filtration chromatography. Excitation and emission spectra of the bound peptides were compared with the free forms (Fig. 2a and Supplementary Fig.1 online). Both (6-DMNA)-RSMAAAAL and (4-DAPA)-RSMAAAAL exhibited large changes in fluorescence upon binding to HLA-DR1. For (6-DMNA)-RSMAAAAL, binding to HLA-DR1 resulted in more than a 1000-fold increase in fluorescence intensity and 115 nm blue shift of the emission maximum (Table I). For (4-DAPA)-RSMAAAAL, changes in fluorescence intensity and emission λmax were smaller but still substantial (Fig. 2b and Table I). Excitation λmax (390-400 nm) were not appreciably changed for either chromophore upon binding to HLA-DR1 (Supplemental Fig. 1 online). Similar if not such dramatic changes were observed after transfer of (6-DMNA)-RSMAAAAL to solvents of different polarity (Supplementary Table 2 online). In vivo, peptides bind to class II MHC proteins in low pH endosomal compartments1, therefore we evaluated whether the fluorogenic peptide probes would be useful under these conditions. The spectral properties of the bound complexes (as well of the free peptides) were essentially identical in physiological saline (pH 7.4) and low pH endosomal conditions (pH 5.0) (Fig. 2c). A titration of HLA-DR1 into a fixed concentration of (4-DAPA)-RSMA4L suggests an isoemissive point (Fig. 2d), indicating the possibility of ratiometric measurements of peptide binding. Finally, we evaluated whether the fluorescence lifetimes of the peptides were altered upon binding to HLA-DR1. Differential fluorescence lifetimes can be exploited to provide large increases in signal-to-noise ratio, and is increasingly being exploited in fluorescence lifetime imaging microscopy (FLIM) applications. For both (4-DAPA)-RSMAAAAL and (6-DMNA)-RSMAAAAL, binding to HLA-DR1 induced large increases in fluorescence lifetimes (Fig. 2e,f and Supplementary Table 3 online). Thus, both the fluorescence spectra and fluorescence lifetimes of the peptides exhibit large changes upon binding to HLA-DR1.
Figure 2. Spectral properties of 6-DMNA and 4-DAPA peptides free in solution and bound to DR1.
Fluorescence emission spectra of (a) (6-DMNA)-RSMA4L (20 nM) and (b) (4-DAPA)-RSMA4L (400 nM) peptides and their complexes with HLA-DR1 are shown, using 400 nm excitation. Free peptide spectra are shown also on an expanded scale. Note the shift in emission λmax upon binding of the peptide to DR and the increase in the emission intensity. Fluorescence intensities are reported as counts per second (c.p.s.). (c) The fluorescence spectrum of the purified DR-(6-DMNA) complex was recorded in citrate phosphate buffer (pH 5.0) and in PBS (pH 7.4). No difference was observed between the two pH. (d) Various concentrations of DR1 were incubated with 1 μM (4-DAPA)-RSMA4L and the emission spectra of the mixtures recorded. (f,g) Fluorescence lifetime measurements. Fluorescence lifetime is increased upon binding of the (4-DAPA)-RSMA4L (f) and (6-DMNA)-RSMA4L (g) to DR. Fluorescence lifetime values measured by time-correlation single-photon counting spectroscopy (Supplementary Table 3 online) were determined as described (Supplementary Methods online).
Table 1. Spectral properties of fluorogenic peptides in aqueous solution and bound to DR.
| Sample | λemission(nm) | Relative fluorescence intensity (c.p.s./nM) | Fold difference DR-pep / pep |
|---|---|---|---|
| DR-(6-DMNA)-RSMA4L | 505 | 4496 | -- |
| (6-DMNA)-RSMA4L alone | 505 (λmax complex) | 2.1 | 2190 |
| 620 (λmax free peptide) | 4.1 | 1100 | |
| DR-(4-DAPA)-RSMA4L | 471 | 599 | -- |
| (4-DAPA)-RSMA4L alone | 471 (λmax complex) | 1.3 | 470 |
| 575 (λmax free peptide) | 16 | 37 | |
| DR-(4-DAPA)-HA | 471 | 578 | -- |
| (4-DAPA)-HA alone | 471 (λmax complex) | 0.8 | 708 |
| 575 (λmax free peptide) | 16 | 36 |
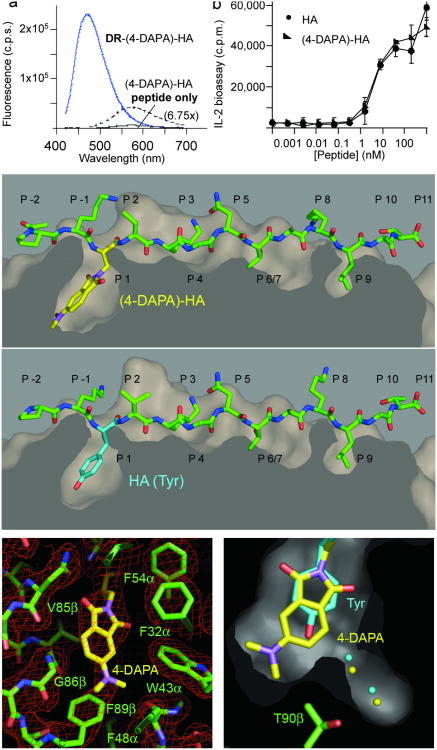
Ideally, introduction of the fluorescent reporter group would not significantly alter the structure of the MHC-peptide complex or its function in triggering T cell responses. To evaluate these issues, we introduced the 4-DAPA residue into the well-characterized HA peptide, by substituting the tyrosine residue at the P1 position with 4-DAPA. The HA peptide with tyrosine substituted by 4-DAPA (4-DAPA-HA) bound well to HLA-DR1, with an IC50 inferior to 1 nM in a standard competition binding assay. Fluorescence intensity, emission λmax, and lifetime changes upon binding to HLA-DR1 were similar to those observed for (4-DAPA)-RSMAAAAL (Fig. 3a, Table 1, and Supplementary Table 3 online). To test the biological activity of the modified peptide, HLA-DR1-positive B cells were treated with HA or (4-DAPA)-HA and used to stimulate a HLA-DR1-restricted, HA peptide-specific T cell hybridoma. HA and (4-DAPA)-HA exhibited essentially identical activation profiles, as judged by an IL-2 secretion assay (Fig. 3b). To test for any structural alterations resulting from introduction of the 4-DAPA side chain, we determined the crystal structure of the HLA-DR1:(4-DAPA)-HA complex at 2.3 Å resolution, using the bacterial superantigen SEC to promote crystallization (Supplementary Table 4 online). The HA and (4-DAPA)-HA peptides adopt essentially identical conformations bound to HLA-DR1 (RMSD 0.24 Å), with differences observed only at the termini of the flexible and solvent-exposed Lys side chains at the P-1, P2 and P8 positions (Fig. 3c). As anticipated, the side chain of the 4-DAPA residue occupies the P1 pocket, completely buried by interactions with many aromatic and hydrophobic side chains in this region (Fig. 3d). We had expected that the 4-DAPA side chain would displace one or two of the water molecules in the P1 pocket that form hydrogen bonds to the Thrβ89 hydroxyl group, however the structure reveals that 4-DAPA predominately adopts the ring-flipped orientation, placing the dimethylamino group on the other side of the ring interacting with the Thrβ89 methyl group, and not displacing the water molecules (Fig. 3e). Overall, introduction of the 4-DAPA fluorophore does not significantly alter the binding affinity, biological activity, or structure of the HA peptide.
Figure 3. Structural and functional characterization of (4-DAPA)-HA peptide.
(a) (4-DAPA)-HA was incubated with HLA-DR1 as described for (4-DAPA)-RSMA4L, and the complex formed was purified by gel filtration. Fluorescence of DR-(4-DAPA)-HA was compared with the free peptide. (b) Activation of HA1.7 TCR hybridoma by antigen presenting cells pulsed with HA (filled circles) or (4-DAPA)-HA (closed triangles) peptides. T cell activation reported as counts per minute (c.p.m.) measured in a thymidine incorporation bioassay for IL-2 as secreted by activated T cells. Error bars indicated standard deviation of triplicate measurements. (c-e) Crystal structure of (4-DAPA)-HA bound to DR1. (c) top, (4-DAPA)-HA peptide shown with surface representation of the DR1 peptide binding site, with 4-DAPA side chain shown with yellow bonds extending down into the P1 pocket; bottom, unmodified HA peptide from the crystal structure of the DR1-HA-SEC (3B2) complex (PDB ID 1JWU) shown after alignment of MHC peptide binding domain, with tyrosine side chain at the P1 position shown with magenta bonds. (d) ∣2Fo−Fc∣ omit map of the region around the P1 pocket with all residues shown removed from the model before map calculation. (e) section through the P1 pocket, showing the HA peptide tyrosine side chain and the (4-DAPA)-HA fluorophore, along with the corresponding ordered water molecules, colored as in (c). Panels (c-f) made using Pymol37.
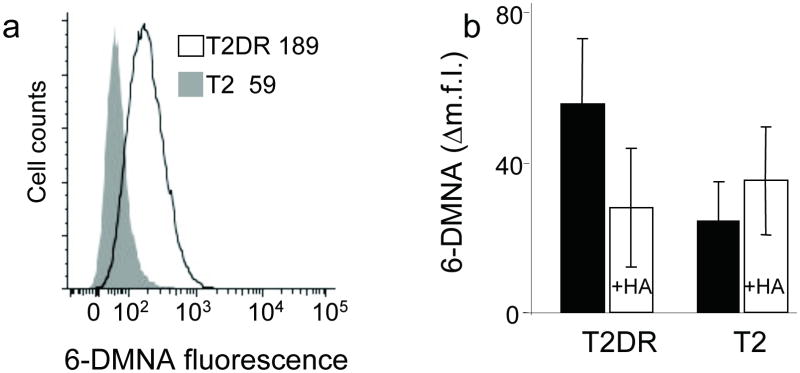
We tested the ability of the fluorogenic peptides to report on binding to class II MHC proteins in a cellular context, using human T2 cells transfected with HLA-DR1 α and β chains. We incubated (6-DMNA)-RSMAAAAL peptide with T2 or T2DR cells for 90 min at 37 °C in PBS, pH 7.4 and analyzed peptide binding by flow cytometry. Binding of the (6-DMNA)-RSMAAAAL peptide to T2DR cells was observed, with a mean fluorescence intensity approximately 3-fold above the background signal observed for binding to untransfected cells (Fig. 4a). Binding of (6-DMNA)-RSMAAAAL involved the conventional peptide-binding site on DR1, as specific binding was inhibited in the presence of excess unlabelled HA peptide (Fig. 4b). Comparable binding was observed on fixed cells (Supplementary Figure 2 online) indicating that under these conditions peptide loading occurs primarily on the cell surface (as expected for this cell line which lacks HLA-DM, the endosomal peptide exchange factor required for the efficient peptide loading14).
Figure 4. (6-DMNA)-peptide binds specifically to DR molecules expressed on live cells.
(a) T2 cells expressing HLA-DR1 (T2DR, dark line) or untransfected T2 cells (shaded profile) were incubated with (6-DMNA)-RSMA4L under conditions which inhibit endocytosis, and the fluorescence of the bound peptide was measured by flow cytometry. Numbers in inset indicate the mean fluorescence intensity values for the indicated cells. (b) Binding of (6-DMNA)-RSMA4L peptide to HLA-DR1 on these cells was reduced in the presence of excess unlabelled HA peptide. The Δm.f.i. values shown are mean fluorescence intensity of labeled cells minus background autofluorescence from the same cells, and represent the average and standard deviation of three independent experiments.
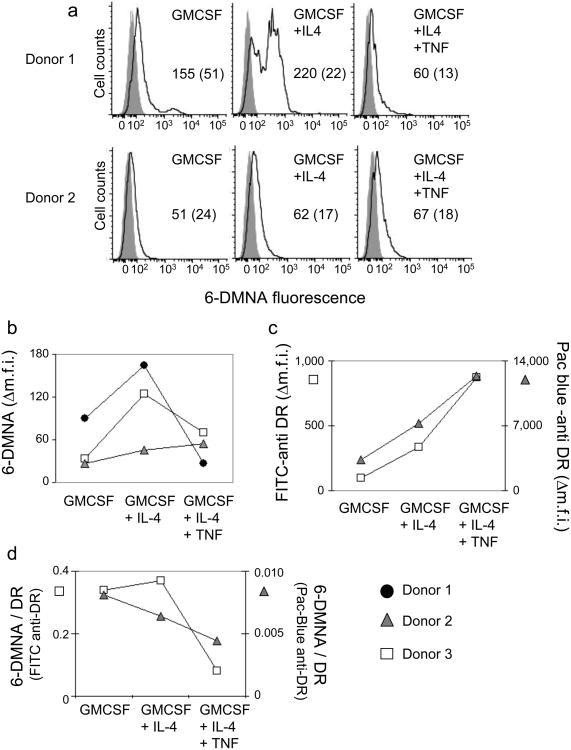
Dendritic cells (DC) play a key role in the immune response by collecting antigens throughout the body and carrying them to lymph nodes for presentation to T cells in a developmentally driven process. To evaluate the MHC-peptide binding activity of DC in various maturation states, we prepared DC from peripheral blood monocytes using a cytokine-driven in vitro differentiation protocol. Peripheral blood monocytes isolated from HLA-DR1+ individuals were cultured with GMCSF, GMCSF+IL-4, or GMSCF+IL-4 followed by TNFα, to generate preDC, immature DC, and mature DC, respectively15. These cells were then incubated with (6-DMNA)-RSMAAAAL as described above. Each of the cell types bound the fluorescent peptide, but to a varying degree, depending on the DC differentiation state and the individual donor (Fig. 5a). Despite the inter-donor variation, clear differences in the peptide-binding capacity of the various DC subsets could be observed (Fig. 5b), particularly after accounting for the number of MHC molecules on the surface of these cells, which was measured using anti-MHC antibody staining (Fig. 5c). It is apparent that pre-DC (GMCSF) and immature DC (GMCSF+IL-4) bind peptide more readily than do mature DC (GMCSF+IL-4+TNFα) (Fig. 5d).
Figure 5. (6-DMNA)-peptide binds to human monocyte-derived DC in a maturation-dependent manner.
Peptide binding to monocyte derived DC subsets isolated from HLA-DR1+ donors and prepared by an in vitro differentiation protocol (see methods) was performed essentially as described for T2DR. (a) Fluorescent peptide binding to DC subsets isolated from two different donors. Solid line, (6-DMNA)-RSMA4L, shaded curve, autofluorescence. Numbers indicate the mean fluorescence intensity for peptide treated cells (with autofluorescence intensity for the same cells shown in parenthesis). (b) Δm.f.i. values for DC subsets generated from cells obtained from three different donors. (c) Expression of HLA-DR1 was measured using murine anti human HLA-DR monoclonal antibody L243 labeled with FITC or Pacific-Blue, for DC subsets from two of the donors in (b). (d) Peptide binding expressed as the ratio of 6-DMNA fluorescence to FITC-anti-DR or Pacific-Blue-anti-DR antibody fluorescence.
The relative ability of immature and mature DC to bind peptide antigen and display the resultant MHC-peptide complexes on the surface has been controversial. Some studies have shown that immature DC are inefficient in acquiring peptide cargo and presenting MHC peptide complexes on the surface as compared to mature DC16,17, with maturation necessary even for loading of preprocessed peptides18. Alternatively, it has been suggested that immature DC can present peptides efficiently19-21 however, the resultant MHC-peptide complexes are recycled more rapidly as compared to the mature DC22. Generally these studies have used indirect T cell readouts of peptide loading, or antibodies specific for particular MHC-peptide complexes. Our results obtained using a direct binding assay fall in line with the latter observations and indicate that the immature DC, despite having lower levels of absolute MHC on their surface, can efficiently present exogenously added preprocessed peptides. The cellular location of peptide loading events in immature and mature DC, and in particular the contribution of shallow endocytic compartments as compared to loading on the surface, remains to be determined, and studies of these phenomena should lead to better understanding of the role of DC as antigen presenting cells.
A potential complication in the use of phthalimide- and naphthalimide-based based fluorophores is their propensity to undergo ring-opening reactions at extremes of pH and temperature (Supplementary Fig. 3a online). To evaluate any possible contribution from binding and signaling of the ring-opened forms, we synthesized the xRSMAAAAL peptides containing the ring-open forms of 4-DAPA and 6-DMNA. While the ring-opened 4-DAPA-peptide was less fluorescent than the closed-ring form, the ring-opened 6-DMNA-peptide exhibited increased fluorescence relative to the closed-ring form, with an emission λmax in aqueous solution similar to protein-bound (6-DMNA)-RSMAAAAL, although with much reduced quantum yield (Supplemental Fig. 3b and Supplementary Table 2 online). To ensure that the fluorescence properties described above reflected the ring-closed xRMSAAAAL peptides, we characterized MHC-peptide incubation mixtures containing the ring-open forms. HLA-DR1 was incubated with a synthetic mixture containing mostly the open form (in the case of 4-DAPA) or a mixture of approximately equal amounts of open and closed forms (in the case of 6-DMNA). After isolation of MHC-peptide complexes, only the closed form was observed by mass spectrometry, indicating that the open forms did not bind significantly to DR1. (Supplementary Fig. 3c online). Moreover, no significant amount of the ring-open form was observed after extended incubation of closed-ring (6-DMNA)-RSMAAAAL or (4-DAPA)-RSMAAAAL for 3 days at 37 °C under conditions of the peptide binding assay. Thus little ring opening was likely to occur under our experimental protocols, and any ring-opened form that might be present would not bind to HLA-DR1 or interfere with our studies.
In summary we have identified fluorophores that can be accommodated into synthetic peptides to faithfully report binding to class II MHC molecules. The large spectral shifts and increased fluorescence lifetimes observed upon binding to HLA-DR1, the high affinity and specific binding comparable to native viral antigens, and the insensitivity to pH over the physiological range, all make these fluorogenic peptides ideal sensors for monitoring peptide loading to cellular MHC proteins at the cell surface or in acidic endosomal compartments. While these probes were designed to bind to HLA-DR1 they are likely to bind to other HLA-DR allelic variants due to promiscuous nature of MHC-peptide binding interactions23. Compared to existing probes of antigen presentation, these reagents can be used with any cell type, do not require extensive washing prior to analysis, and report actual binding events rather just the presence of the peptide, and as such should be useful in studies of the in vivo regulation of peptide loading activity, for example by modulation of DM activity, differential proteolysis, pH regulation, differential compartmentalization of antigen and MHC all of which have been reported to accompany DC differentiation1. The approach described here could be employed for investigating other peptide/protein interactions, provided that a solvatochromic amino acid residue can be effectively incorporated into the protein of interest via chemical synthesis, semi-synthesis, or biosynthetic approaches. In ongoing work we are using these probes to monitor the activity of peptide-binding modules such as phosphotyrosine-binding SH2 domains9, phosphoserine-/phosphothreonine- binding 14-3-3 proteins24, C-terminal peptide-binding PDZ domains (unpublished results), and in the design of delta- and mu- selective opiod-receptor binding peptides 25.
Methods
Materials
All reagents for peptide synthesis were of analytical grade. Resin and amino acids were obtained from AnaSpec (USA). Purified recombinant human cytokines GMCSF, IL-4, and TNFα were obtained from R&D Systems. Antibodies for FACS MHC analysis were from BD Biosciences (Pharmingen) and BioLegend (San Diego,CA). Fmoc-PAL-PEG-PS resin (0.20 mmol/g) was obtained from Applied Biosystems. Amino acid and reagents were obtained from Novabiochem and Sigma-Aldrich. 4-DAPA and 6-DMNA were synthesized according to the previously reported methods8,9.
Peptide synthesis and purification
RSMAAAAL (0.68 mmole) and EDKDKSR (0.41 mmole) were synthesized on a Rink-amide resin using an automatic peptide synthesizer adopting standard Fmoc-based SPPS chemistry. The 6-DMNA and 4-DAPA were then coupled manually to 0.01 to 0.02 μmol resin-bound peptide. Two equivalents of Fmoc-amino acid were mixed with two equivalents each of DIEA /HATU in 0.30 mL DMF and reacted with the resin for 60 min. The peptides were deprotected (20% piperidine in DMF) and N-acetyl capped using a mixture of pyridine and acetic acid (0.15 M in DMF, 30 min). For peptide 4-DAPA-HA, the remaining Lys and Pro residues were coupled manually after Fmoc-4-DAPA addition and deprotection. Peptides was cleaved from the resin and ether precipitated and washed before purification by reverse phase HPLC (water/0.01% TFA using an acetonitrile/0.01% TFA gradient). The major peak was collected, dried and re-suspended in water at pH 4.0 or in water containing 100 mM urea. MALDI-TOF confirmed the identity of the peptides (Ac-4-DAPA peptide amide: expected 1089.54; observed mass 1090.69; Ac-6-DMNA peptide-amide expected mass 1139.55 and the observed mass 1140.42). Peptides were stored frozen at −80 °C until use.
MHC-peptide complex formation
Soluble recombinant DR1 extracellular domain was prepared by expression of truncated HLA-DRA*0101 and HLA-DR1*0101 genes in S2 insect cells cultured in serum-free medium followed by immunoaffinity isolation of the secreted protein as described26. For peptide loading, 7 μM of HLA-DR1 was incubated with 30 μM (4-DAPA)-RSMAAAAL or (6-DMNA)-RSMAAAAL in PBS (10 mM Na2HPO4 containing 137 mM NaCl and 2.7 mM KCl, (pH 7.4), 1mM PMSF, protease inhibitor cocktail from Sigma Aldrich, 0.02 % sodium azide) for 3 days at 37°C. MHC-peptide complexes were isolated by gel filtration using a Superdex 300 column (Pharmacia) with phosphate-buffered PBS containing 0.02% sodium azide buffer. The MHC-peptide complex eluting between 31 and 34 min was collected and concentrated. Non-bound protein tends to aggregate under the incubation conditions and is typically not found in these fractions5. The complex thus purified from free peptide was stable for several months at 4 °C. UV-Visible spectra of the purified peptides and the DR-peptide complex were measured in PBS. Fluorescence excitation and emission spectra were obtained for purified DR-peptide complex and the free peptide, with 5 nm bandpass for excitation and emission. Raman spectra of pure water obtained under the same conditions, generally were subtracted from the experimental spectra. For direct binding measurement in presence of excess peptide, 4-DAPA-peptide (1 μM) was incubated with 0.125 - 1 μM of HLA-DR1 in Buffer A for 3 days and the fluorescence measured as described above. The complex of DR-(4-DAPA)-HA and those of the synthetic mixtures containing open and closed forms of (4-DAPA)-RSMA4L (see supplementary Figure 1) were similarly prepared.
Cell culture
T2, a human B-cell/T cell hybrid cell line lacking the MHC class II regions and conventionally used as antigen presenting cells in T cell activation studies, and T2 transfected with full-length HLA-DR1 alpha and beta subunit genes (T2DR)34, were cultured in RPMI + 10% FCS at 37 °C, 5% CO2. Expression of HLA-DR1 was verified by flow cytometry using FITC-labeled anti-DR antibody L243. DC from peripheral blood monocytes were prepared and grown in the presence of GMCSF, GMCSF+IL-4 and GMCSF+IL-4+TNFα essentially as described 15. Briefly, buffy coats were prepared from blood collected from a healthy DRB1*01/DRB1*04 donor following institutional guidelines and approved protocols. Monocytes were isolated from blood using RosetteSep™ Enrichment cocktail (Stem Cell Technologies, Vancouver, Canada) Enriched monocytes (1-2 × 106/mL) were cultured in medium containing recombinant human GMCSF (25 ng/mL) alone or with IL-4 (20 ng/mL), with medium replenished every third day. After 6 days of culture, TNF-α (20 ng/mL) was added with further culture for 6 days to generate GMCSF+IL-4+TNFα DC.
Cellular peptide loading assay
T2, T2DR, and monocyte-derived DC subsets, were washed in phosphate citrate buffer (137 mM NaCl, 2.7 mM KCL, 10 mM Na2HPO4 1.14 mM citric acid adjusted to pH 7.00) supplemented with 0.1% BSA and 0.05% NaN3 / 50 μM 2-deoxyglucose to prevent endocytosis. Washed cells were incubated for 30 min on ice, washed again, and finally resuspended to 2.5 ×105 cells in a volume of 25 μL. Peptide (6 μl; final urea concentration 20 mM) was added to give 30 μM final concentration, with or without 2 mM unlabeled HA competitor peptide. The cells were incubated in peptide-containing solution at 37 °C for 90 min, and then were washed at 4 °C, followed by flow cytometry analysis using a LSR II instrument equipped with a violet 405 nm laser. 6-DMNA fluorescence was collected after passing through 505 nm long pass dichroic and 530/30 or 525/20 bandpass filters (bandpass filters characterized as wavelength of maximal transmittance / full width at half maximum transmission). Live cells were gated for analysis using 7-amino-actinomycin D. In some experiments, cell autofluorescence was subtracted using an adjacent spectral region (450/50 bandpass filter), as suggested by Alberti et al 35. Expression of MHC II molecules was measured using anti-DR antibody L243 labelled with FITC (Pharmingen) or with Pacific-blue (Biolegend), by incubation for 20 minutes on ice followed by washing and flow cytometry. Pacific-blue has excitation and emission maxima of 402 and 420 nm, respectively. Antibody staining values are reported after subtraction of appropriately-labelled isotype controls. Flow cytometry data are shown on a “logicle” sinh(×)-based scale 36
Supplementary Material
Supplemental Figure 1 Absorbance and Fluorescence Spectra
(a) UV-visible spectra of the free peptides. Peptides in a 100 μL volume of PBS pH 7.4 were scanned from 500 – 210 nm using a scan speed of 1 nm/sec (Varian). Molar extinction coefficient of 6480 M−1cm−1 for 4-DAPA (λmax.em 421nm) and 7980 M−1cm−1 for 6-DMNA (λmax.em 388nm) were used to estimate the concentration of the free peptides (b) Spectra of purified 4-DAPA peptide and 6-DMNA peptide complexes were obtained under similar conditions. Fluorescence excitation and emission spectra of the purified complex (c and d) were measured as described under methods. Emission scans were performed using 388 nm for the DR (6-DMNA)-RSMAAAAL peptide complex and 421 nm for the (4-DAPA)-RSMAAAAL peptide complex which corresponds to their UV absorption maxima. Excitation scans were obtained using 500 nm emission. The peak intensity for the 4-DAPA peptide-DR1 complex is therefore slightly lower in the excitation scan (471 nm is the λmax of emission).
Supplemental Figure 2 Peptide binding takes place primarily on the cell surface
T2 and T2DR cells were fixed in formaldehyde, washed and incubated with the fluorogenic peptide (6-DMNA)-RSMA4L in the presence or absence of excess unlabelled HA peptide.
Supplementary Fig 3 Ring-open forms
(a) Structures of the open forms of (4-DAPA)- and (4-DMNA)- peptides. The fluorophores of the 4-DAPA and 6-DMNA residues undergo spontaneous (reversible) hydrolysis leading to opening of the ring structure. (b) Spectral properties of the open forms of (6-DMNA)-RSMA4L and (4-DAPA)-RSMA4L. Fluorescence (left panels) and UV-visible absorbance (right panels) spectra of the synthetic mixtures of purified closed and open forms of (6-DMNA)-RSMA4L (top panels) and of (4-DAPA)-RSMA4L (bottom panels) were measured. Fluorescence spectra of the purified DR peptide complexes derived from the incubation with synthetic mixtures of open and closed forms were simultaneously measured. Note that although the spectral properties of the open (6-DMNA)-RSMA4L peptide is similar to that of the DR1-peptide complex it is ∼ 100 fold less fluorescent (Supplementary Table 1). The purified DR1-peptide complex from synthetic mixtures is identical to the DR-peptide complex purified from incubations with the pure closed form of the peptides. (c) Open forms of the fluorogenic peptides are largely excluded from the binding pocket of DR1. Mass spectra of pure closed (4-DAPA)-RSMA4L and (6-DMNA)- RSMA4L peptides, their synthetic peptide mixture, incubation mixtures of DR+ synthetic peptide mixtures, and the peptide eluted from purified DR bound with the synthetic mixture were obtained by MALDI TOF. The synthetic mixtures were tested for their ability to inhibit the binding of biotin-HA to DR1, with the resultant IC50 values shown. In an independent experiment, we also tested the effect of the open and closed peptides after fresh isolation over a reverse phase column using a very shallow gradient (see UV spectra in supplementary Fig 2) The peptides after two day incubation gave a mass corresponding to that of the closed peptide and the IC50 values matched those of the closed forms of the peptide (data not shown) indicating that such incubation in the presence of DR1 favors the formation of closed forms of the peptide and that the binding of the open forms is even weaker than suggested by the IC50 values.
Supplementary Table 1 Binding affinity of fluorogenic peptides for HLA-DR1
Supplementary Table 2 (6-DMNA)-RSMA4L fluorescence intensity in different solvents and bound to HLA-DR1
Supplementary Table 3 Fluorescence lifetimes for free and DR-bound peptides (76 Kb)
Supplementary Table 4 Data collection and refinement statistics (molecular replacement) (72 Kb)
Supplementary Methods Preparation and analysis of ring open forms; crystal structure; MHC-peptide binding assay; T cell activation; fluorescence lifetime measurement (96 Kb)
Acknowledgments
We would like to thank Mauricio Calvo-Calle for performing T cell activation assays, Richard Konz for assistance with flow cytometry experimentation and data interpretation, Elvedin Lukovic and Corrie Painter for peptide synthesis, Iwona Strug for characterization of eluted peptides, Eric Schreiter and Zarixia Zavala-Ruiz for helpful discussions, Liying Lu for preparation of HLA-DR, Jerome Bill for the HA1.7 hybridoma, and the staff at the National Synchrotron Light Source (NSLS). Diffraction data for this study were measured at beamline ×25 of the NSLS, support for which comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. This work was supported by NIH-AI38996 (LJS) and NSF CHE-0414243 (BI).
Footnotes
P.V. performed the biochemical, cellular, and spectroscopic studies, M.S. synthesized the fluorogenic amino acids and performed the chemical stability studies, T.N. determined the crystal structure, O.S. and S.C. performed biophysical and immunological assays, and P.V., M.S., B.I., and L.J.S wrote the manuscript
References
- 1.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 2.McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med Res Rev. 2002;22:168–203. doi: 10.1002/med.10006. [DOI] [PubMed] [Google Scholar]
- 3.Jardetzky TS, et al. Peptide binding to HLA-DR1: a peptide with most residues substituted to alanine retains MHC binding. Embo J. 1990;9:1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato AK, et al. Determinants of the peptide-induced conformational change in the human class II major histocompatibility complex protein HLA-DR1. J Biol Chem. 2000;275:2165–2173. doi: 10.1074/jbc.275.3.2165. [DOI] [PubMed] [Google Scholar]
- 5.Stern LJ, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 6.Cohen BE, et al. Probing protein electrostatics with a synthetic fluorescent amino acid. Science. 2002;296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 7.Nitz M, Mezo AR, Ali MH, Imperiali B. Enantioselective synthesis and application of the highly fluorescent and environment-sensitive amino acid 6-(2-dimethylaminonaphthoyl) alanine (DANA) Chem Commun (Camb) 2002:1912–1913. doi: 10.1039/b205224e. [DOI] [PubMed] [Google Scholar]
- 8.Vazquez ME, Rothman DM, Imperiali B. A new environment-sensitive fluorescent amino acid for Fmoc-based solid phase peptide synthesis. Org Biomol Chem. 2004;2:1965–1966. doi: 10.1039/b408001g. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez ME, Blanco JB, Imperiali B. Photophysics and biological applications of the environment-sensitive fluorophore 6-N,N-dimethylamino-2,3-naphthalimide. J Am Chem Soc. 2005;127:1300–1306. doi: 10.1021/ja0449168. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, et al. [Aladan3]TIPP: a fluorescent delta-opioid antagonist with high delta-receptor binding affinity and delta selectivity. Biopolymers. 2005;80:325–331. doi: 10.1002/bip.20200. [DOI] [PubMed] [Google Scholar]
- 11.Saroja G, Soujanya T, Ramachandram B, Samanta A. 4-Aminophthalimide derivatives as environment-sensitive probes. J Fluorescence. 1998;8:405–410. [Google Scholar]
- 12.Hammer J, et al. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci U S A. 1994;91:4456–4460. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J Immunol. 1990;144:1849–1856. [PubMed] [Google Scholar]
- 14.Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 15.Potolicchio I, et al. Conformational variation of surface class II MHC proteins during myeloid dendritic cell differentiation accompanies structural changes in lysosomal MIIC. J Immunol. 2005;175:4935–4947. doi: 10.4049/jimmunol.175.8.4935. [DOI] [PubMed] [Google Scholar]
- 16.Pierre P, Mellman I. Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Cell. 1998;93:1135–1145. doi: 10.1016/s0092-8674(00)81458-0. [DOI] [PubMed] [Google Scholar]
- 17.Turley SJ, et al. Transport of peptide-MHC class II complexes in developing dendritic cells. Science. 2000;288:522–527. doi: 10.1126/science.288.5465.522. [DOI] [PubMed] [Google Scholar]
- 18.Inaba K, et al. The formation of immunogenic major histocompatibility complex class II-peptide ligands in lysosomal compartments of dendritic cells is regulated by inflammatory stimuli. J Exp Med. 2000;191:927–936. doi: 10.1084/jem.191.6.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cella M, Engering A, Pinet V, Pieters J, Lanzavecchia A. Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature. 1997;388:782–787. doi: 10.1038/42030. [DOI] [PubMed] [Google Scholar]
- 20.Colledge L, Bennett CL, Reay PA, Blackburn CC. Rapid constitutive generation of a specific peptide-MHC class II complex from intact exogenous protein in immature murine dendritic cells. Eur J Immunol. 2002;32:3246–3255. doi: 10.1002/1521-4141(200211)32:11<3246::AID-IMMU3246>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 21.Veeraswamy RK, Cella M, Colonna M, Unanue ER. Dendritic cells process and present antigens across a range of maturation states. J Immunol. 2003;170:5367–5372. doi: 10.4049/jimmunol.170.11.5367. [DOI] [PubMed] [Google Scholar]
- 22.Wilson NS, El-Sukkari D, Villadangos JA. Dendritic cells constitutively present self antigens in their immature state in vivo and regulate antigen presentation by controlling the rates of MHC class II synthesis and endocytosis. Blood. 2004;103:2187–2195. doi: 10.1182/blood-2003-08-2729. [DOI] [PubMed] [Google Scholar]
- 23.Southwood S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 24.Vazquez ME, Nitz M, Stehn J, Yaffe MB, Imperiali B. Fluorescent caged phosphoserine peptides as probes to investigate phosphorylation-dependent protein associations. J Am Chem Soc. 2003;125:10150–10151. doi: 10.1021/ja0351847. [DOI] [PubMed] [Google Scholar]
- 25.Vazquez ME, et al. 6-N,N-dimethylamino-2,3-naphthalimide: a new environment-sensitive fluorescent probe in delta- and mu-selective opioid peptides. J Med Chem. 2006;49:3653–3658. doi: 10.1021/jm060343t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty alpha beta heterodimers in the absence of antigenic peptide. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 27.Zavala-Ruiz Z, Strug I, Anderson MW, Gorski J, Stern LJ. A polymorphic pocket at the P10 position contributes to peptide binding specificity in class II MHC proteins. Chem Biol. 2004;11:1395–1402. doi: 10.1016/j.chembiol.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Frayser M, Sato AK, Xu L, Stern LJ. Empty and peptide-loaded class II major histocompatibility complex proteins produced by expression in Escherichia coli and folding in vitro. Protein Expr Purif. 1999;15:105–114. doi: 10.1006/prep.1998.0987. [DOI] [PubMed] [Google Scholar]
- 29.Andersen PS, et al. Role of the T cell receptor alpha chain in stabilizing TCR-superantigen-MHC class II complexes. Immunity. 1999;10:473–483. doi: 10.1016/s1074-7613(00)80047-3. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski z, Minor w. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 34.Riberdy JM, Cresswell P. The antigen-processing mutant T2 suggests a role for MHC-linked genes in class II antigen presentation. J Immunol. 1992;148:2586–2590. [PubMed] [Google Scholar]
- 35.Alberti S, Parks DR, Herzenberg LA. A single laser method for subtraction of cell autofluorescence in flow cytometry. Cytometry. 1987;8:114–119. doi: 10.1002/cyto.990080203. [DOI] [PubMed] [Google Scholar]
- 36.Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69:541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 37.DeLano WL. The PyMOL molecular graphics system. 2002 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Absorbance and Fluorescence Spectra
(a) UV-visible spectra of the free peptides. Peptides in a 100 μL volume of PBS pH 7.4 were scanned from 500 – 210 nm using a scan speed of 1 nm/sec (Varian). Molar extinction coefficient of 6480 M−1cm−1 for 4-DAPA (λmax.em 421nm) and 7980 M−1cm−1 for 6-DMNA (λmax.em 388nm) were used to estimate the concentration of the free peptides (b) Spectra of purified 4-DAPA peptide and 6-DMNA peptide complexes were obtained under similar conditions. Fluorescence excitation and emission spectra of the purified complex (c and d) were measured as described under methods. Emission scans were performed using 388 nm for the DR (6-DMNA)-RSMAAAAL peptide complex and 421 nm for the (4-DAPA)-RSMAAAAL peptide complex which corresponds to their UV absorption maxima. Excitation scans were obtained using 500 nm emission. The peak intensity for the 4-DAPA peptide-DR1 complex is therefore slightly lower in the excitation scan (471 nm is the λmax of emission).
Supplemental Figure 2 Peptide binding takes place primarily on the cell surface
T2 and T2DR cells were fixed in formaldehyde, washed and incubated with the fluorogenic peptide (6-DMNA)-RSMA4L in the presence or absence of excess unlabelled HA peptide.
Supplementary Fig 3 Ring-open forms
(a) Structures of the open forms of (4-DAPA)- and (4-DMNA)- peptides. The fluorophores of the 4-DAPA and 6-DMNA residues undergo spontaneous (reversible) hydrolysis leading to opening of the ring structure. (b) Spectral properties of the open forms of (6-DMNA)-RSMA4L and (4-DAPA)-RSMA4L. Fluorescence (left panels) and UV-visible absorbance (right panels) spectra of the synthetic mixtures of purified closed and open forms of (6-DMNA)-RSMA4L (top panels) and of (4-DAPA)-RSMA4L (bottom panels) were measured. Fluorescence spectra of the purified DR peptide complexes derived from the incubation with synthetic mixtures of open and closed forms were simultaneously measured. Note that although the spectral properties of the open (6-DMNA)-RSMA4L peptide is similar to that of the DR1-peptide complex it is ∼ 100 fold less fluorescent (Supplementary Table 1). The purified DR1-peptide complex from synthetic mixtures is identical to the DR-peptide complex purified from incubations with the pure closed form of the peptides. (c) Open forms of the fluorogenic peptides are largely excluded from the binding pocket of DR1. Mass spectra of pure closed (4-DAPA)-RSMA4L and (6-DMNA)- RSMA4L peptides, their synthetic peptide mixture, incubation mixtures of DR+ synthetic peptide mixtures, and the peptide eluted from purified DR bound with the synthetic mixture were obtained by MALDI TOF. The synthetic mixtures were tested for their ability to inhibit the binding of biotin-HA to DR1, with the resultant IC50 values shown. In an independent experiment, we also tested the effect of the open and closed peptides after fresh isolation over a reverse phase column using a very shallow gradient (see UV spectra in supplementary Fig 2) The peptides after two day incubation gave a mass corresponding to that of the closed peptide and the IC50 values matched those of the closed forms of the peptide (data not shown) indicating that such incubation in the presence of DR1 favors the formation of closed forms of the peptide and that the binding of the open forms is even weaker than suggested by the IC50 values.
Supplementary Table 1 Binding affinity of fluorogenic peptides for HLA-DR1
Supplementary Table 2 (6-DMNA)-RSMA4L fluorescence intensity in different solvents and bound to HLA-DR1
Supplementary Table 3 Fluorescence lifetimes for free and DR-bound peptides (76 Kb)
Supplementary Table 4 Data collection and refinement statistics (molecular replacement) (72 Kb)
Supplementary Methods Preparation and analysis of ring open forms; crystal structure; MHC-peptide binding assay; T cell activation; fluorescence lifetime measurement (96 Kb)