Abstract
Four seronegative foals aged 6 to 7 months were exposed to an aerosol of influenza strain A/Equi/2/Kildare/89 at 106 50% egg infective doses (EID50)/ml. Nasopharyngeal swabs were collected for 10 consecutive days after challenge. Virus isolation was performed in embryonated eggs, and the EID50 was determined for all positive samples. The 50% tissue culture infective dose was determined using Madin-Darby canine kidney (MDCK) cells. Samples were also tested by an in vitro enzyme immunoassay test, Directigen Flu A, and by reverse transcription-PCR (RT-PCR) using nested primers from the nucleoprotein gene and a single set of primers from the matrix gene. RT-PCR using the matrix primers and virus isolation in embryonated eggs proved to be the most sensitive methods for the detection of virus. The Directigen Flu A test was the least sensitive method. The inclusion of 2% fetal calf serum in the viral transport medium inhibited the growth of virus from undiluted samples in MDCK cells but was essential for the maintenance of the virus titer in samples subjected to repeated freeze-thaw cycles.
Equine influenza is considered to be the most important respiratory disease of the horse in the majority of countries where the breeding and racing of horses is a major industry. There are two subtypes of equine influenza virus, which is an orthomyxovirus: A/Equi 1/H7N7, first isolated in 1956 (20), and A/Equi 2/H3N8, first isolated in 1963 (24). Both subtypes have caused disease. However, it is generally accepted that A/Equi 1/H7N7 has not been isolated since 1979 and may be extinct (22, 26). In contrast, A/Equi 2/H3N8 continues to circulate worldwide with the exception of a small number of island countries, such as Australia, New Zealand, and Iceland, where equine influenza has never been recorded. Infection with this subtype appears to be enzootic in North America and Europe, where two separate virus lineages have evolved and outbreaks frequently occur despite the mandatory vaccination of some horse populations (2, 10, 16a). However, it is immunologically naïve populations that are most at risk from equine influenza. Influenza is highly contagious, and the introduction of a single infected horse can result in explosive virus spread in unprotected horses over a wide geographical area. In South Africa in 1986, the introduction of the virus into the country for the first time resulted in thousands of horses suffering severe respiratory disease and necessitated the cancellation of racing for 5 months (5, 18).
The risk associated with the increase in the international movement of horses by air transport for racing and breeding purposes means that it is imperative that there are sensitive virus detection systems available for the rapid diagnosis of equine influenza (21). Traditionally, equine influenza virus is diagnosed by the isolation of virus from nasopharyngeal swabs in embryonated hen eggs or by the detection of a fourfold-or-greater rise in antibody titer in paired sera by hemagglutination inhibition (16).
The main objective of this study was to compare the sensitivity of virus isolation using embryonated eggs and tissue culture with that of more rapid detection systems such as the immunoassay Directigen Flu A (DFA) and reverse transcription-PCR (RT-PCR) followed by gel electrophoresis. We also examined the potential negative effects of repeat freeze-thawing of samples on all four detection systems and evaluated the benefit of inclusion of fetal calf serum (FCS) in virus transport medium (VTM).
MATERIALS AND METHODS
Virus isolation and quantification.
Four seronegative foals aged 6 to 7 months were exposed to an aerosol of 10 ml of A/Equi/2/Kildare/89 at 106 50% egg infective doses (EID50)/ml as described previously by Mumford et al. (9). Nasopharyngeal swabs were collected for 10 consecutive days after challenge and immersed in chilled VTM consisting of phosphate-buffered saline with 100 U of penicillin, 100 μg of streptomycin, and 5 μg of amphotericin B per ml. Within 24 h of collection, 100 μl of each sample was inoculated into the allantoic cavity of three 9- to 12-day-old embryonated hen eggs. The allantoic fluid was harvested after 2 days of incubation at 34°C and tested by hemagglutination using 1% hen blood (16). If hemagglutination was observed, the virus isolate was typed by hemagglutination inhibition using type-specific ferret antisera supplied by the National Institute of Biological Standards, London, England.
Madin-Darby canine kidney (MDCK) cells were propagated at 34°C in an atmosphere of 5% CO2 in Eagle's minimum essential medium supplemented with FCS (10% [vol/vol]). For virus isolation and quantification, the cells were maintained in serum-free medium and in the presence of 1.25 μg of beef pancreas trypsin (Sigma-Aldrich)/ml. Samples were cultured for 7 days at 34°C in an atmosphere of 5% CO2, and the supernatant medium was tested daily by hemagglutination (16).
Quantification assays to determine the EID50 and the 50% tissue culture infective dose (TCID50) were carried out on samples that were frozen within 24 h of collection and had subsequently been thawed once (7).
Enzyme-linked immunosorbent assay.
DFA, an in vitro enzyme immunoassay membrane test manufactured by Becton Dickinson and Co. (Paramus, N.J.), was used in accordance with the manufacturer's instructions. Extracted nasopharyngeal samples (125 μl) were added to the test device, and any influenza A antigen that bound to the membrane surface was detected using enzyme-conjugated monoclonal antibodies against the nucleoprotein. Each DFA test device has an H1N1 antigen spot in the center of the membrane which develops as a purple dot and indicates the integrity of the test. The development of a purple triangle surrounding the dot is indicative of a positive reaction. In this study, the degree of positive reaction was scored as + (outline of triangle), ++ (lightly colored triangle), and +++ (dark-purple triangle).
PCR and gel electrophoresis.
Primers from the nucleoprotein genes NPF (5′-AGCAAAAGCAGGGTAGATAA-3′), NPR (5′-TCCTTGCATCAGAGAGCACA-3′), NPFI (5′-GCAGGGTAGATAATCACTCA-3′), and NPRI (5′-AGTACCATCCTTTCTATTGT-3′), designed by Oxburgh and Hagstrom (17), were synthesized by Genosys Biotechnologies, Inc., (Cambridge, United Kingdom). The PCR was performed essentially as described previously (17), except that the thermal cycle programs used were denaturation for 5 min at 95°C and then 30 cycles of 95°C for 1 min, 52°C for 1 min 30 s, and 72°C for 2 min 30 s, with a final extension time of 7 min at 72°C.
Primers M52C (5′-CTTCTAACCGAGGTCGAAACG-3′) and M253R (5′-AGGGCATTTTGGACAAAG/TCGTCTA-3′), designed by Fouchier et al. (3) from highly conserved regions of the matrix gene, were synthesized by Eurogentec (Seraing, Belgium). The reverse transcription and PCR were performed as described previously (3), except that the cycling conditions were 30 min at 42°C and 2 min at 94°C once and then 40 cycles of 15 s at 94°C, 30 s at 45°C, and 1 min at 72°C, with a final extension of 3 min at 72°C.
PCR products of 552 bp (outer nucleoprotein primers), 241 bp (inner nucleoprotein primers), and 244 bp (matrix primers) were visualized on 1.2 or 1% agarose gels stained with ethidium bromide.
Assessment of sensitivities of assays after freezing of samples.
Three dilutions of A/Equi/2/Kildare/89 (107.5 EID50/ml [sample A], 106 EID50/ml [sample B], and 102.5 EID50/ml [sample C]) were prepared in VTM with or without FCS at a final concentration of 2% (vol/vol). The samples were subjected to repeat freeze (−70°C)-thaw (37°C) cycles and were examined for the presence of virus by isolation in embryonated eggs and MDCK cells, by DFA, and by RT-PCR after each cycle.
Statistical analysis.
Microsoft SPSS Base 10.0 was the statistical software used for formal testing, and the significance level was set at 0.05. The paired t test was used for a two-way comparison of means, and Pearson's correlation was used for comparing quantitative tests.
RESULTS AND DISCUSSION
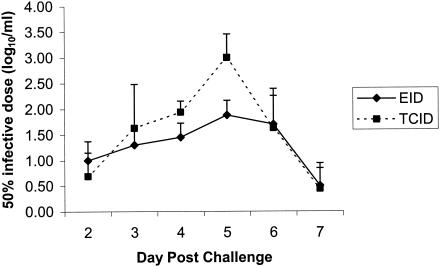
Virus was isolated from the four horses postchallenge. The results of the virus isolation and titration in both embryonated eggs and tissue culture are summarized in Table 1. Virus was first detected on day 2 postchallenge and persisted for a mean period of 5.5 days. Peak titers in the embryonated eggs ranged from 101.5 EID50/ml to 102.8 EID50/ml. Peak titers in the MDCK cells ranged from 102.5 TCID50/ml to 104 TCID50/ml. Pearson's correlation showed a significant (P = 0.014) positive correlation between the two methods of quantification of virus load on each day postchallenge (Fig. 1), but the tissue culture system was slightly less sensitive (85%) than the embryonated eggs. The results suggest that for virus A/Equi/2/Kildare/89, which is very closely related to A/Equi/2/Suffolk/89 (14), the present prototype of the European lineage, embryonated eggs is the quantification system of choice. Virus quantification is necessary for vaccine efficacy studies where there is a regulatory requirement to monitor the quantity of virus shed by vaccinated and unvaccinated horses after experimental challenge. At present, such quantification is usually carried out in embryonated eggs (11, 12). However, it has proven more difficult to isolate virus in eggs during some of the disease outbreaks that have occurred since 1989 (16a) and the comparative sensitivities of virus isolation in tissue culture and in eggs need to be evaluated for other isolates, particularly those of American lineage.
TABLE 1.
Detection of equine influenza virus by virus isolation, antigen detection, and RT-PCR in foals infected experimentally with A/Equi/2/Kildare/89a
| Horse and day postchallenge | VI | EID50/ml | TCID50/ml | DFAb | PCR O | PCR I | PCR M |
|---|---|---|---|---|---|---|---|
| Horse 1 | |||||||
| 1 | − | − | − | − | − | ||
| 2 | + | 101.5 | 101.25 | − | − | + | + |
| 3 | + | 101.5 | 100.75 | − | − | + | + |
| 4 | + | 101.1 | 101.75 | − | − | + | + |
| 5 | + | 101.2 | 102.5 | − | − | + | + |
| 6 | + | − | − | − | − | − | + |
| 7 | − | − | − | − | + | ||
| 8 | − | − | − | − | − | ||
| 9 | − | − | − | − | − | ||
| 10 | − | − | − | − | − | ||
| Horse 2 | |||||||
| 1 | − | − | − | − | − | ||
| 2 | + | 101 | − | − | − | − | + |
| 3 | + | 101.8 | 103 | + | + | + | NAc |
| 4 | + | 101.7 | 102.25 | ++ | − | + | + |
| 5 | + | 101.8 | 103.25 | +++ | + | + | + |
| 6 | + | 102.2 | 102.25 | +++ | − | + | + |
| 7 | + | 100.8 | − | − | − | − | + |
| 8 | − | − | − | − | − | ||
| 9 | − | − | − | − | − | ||
| 10 | − | − | − | − | − | ||
| Horse 3 | |||||||
| 1 | − | − | − | − | − | ||
| 2 | + | 101.2 | 101.5 | + | − | + | + |
| 3 | + | 101.3 | 102.75 | ++ | − | + | + |
| 4 | + | 102 | 102.25 | − | − | + | + |
| 5 | + | 102.2 | 104 | − | + | + | + |
| 6 | + | 101.8 | 102.25 | ++ | + | + | + |
| 7 | + | 101.2 | 101.75 | − | − | + | + |
| 8 | − | − | − | − | − | ||
| 9 | − | − | − | − | − | ||
| 10 | − | − | − | − | − | ||
| Horse 4 | |||||||
| 1 | − | − | − | − | + | ||
| 2 | + | − | − | − | − | − | + |
| 3 | + | 100.6 | − | − | − | − | + |
| 4 | + | 101 | 101.5 | − | + | + | + |
| 5 | + | 102.3 | 102.25 | − | + | + | + |
| 6 | + | 102.8 | 102 | − | − | + | + |
| 7 | − | − | − | − | − | ||
| 8 | − | − | − | − | − | ||
| 9 | − | − | − | − | − | ||
| 10 | − | − | − | − | − |
VI, virus isolation (in embryonated eggs) from the nasal secretions prior to freezing; PCR O, PCR using the NPF and NPR primers; PCR 1, nested PCR using NPF, NPR, NPFI, and NPRI; PCR M, PCR using M52C and M253R. All aliquots assayed by EID50, TCID50, DFA, and PCR had undergone a single freeze-thaw cycle.
For DFA scores, see Materials and Methods.
NA, not applicable.
FIG. 1.
The mean EID50 and the mean TCID50 of equine influenza virus in nasal secretions of foals from days 2 to 7 postchallenge.
Virus culture in eggs or cell monolayers takes several days. Infection is usually identified by hemagglutination, and virus identity has to be confirmed by hemagglutination inhibition with specific antisera. DFA takes approximately 15 min and is simple to perform. However, published studies have demonstrated a wide range of sensitivities for the detection of both human influenza (6, 15, 25) and equine influenza (1, 8, 23, 27) by DFA when compared to virus culture. In this study, the sensitivity of the DFA test was 32% of that of virus isolation in eggs from the unfrozen samples. The results are shown in Table 1. No virus was detected in the samples collected from horses 1 and 4, despite the fact that they shed virus for 4 days. Virus was detected in the samples collected from horse 2 on 4 of the 6 days of shedding. DFA was positive for samples collected from horse 3 on days 2, 3, and 6 but failed to detect virus on days 4 and 5 when shedding peaked. It has been suggested that DFA is most useful at the peak of infection but less sensitive early or late in infection when low levels of virus are shed (13). However, in this study neither the positive results nor the strength of the reactions correlated with the quantity of virus shed (Table 1). These data are consistent with those of Ryan-Poirier et al. (19), who found that the intensity of positive reaction in specimens from human patients did not correlate with the amount of virus in the specimen. They found that DFA detects cell-associated antigens more readily than free virus and speculated that an increase in the epithelial cells or cell debris in the specimen may increase the sensitivity. This may explain why in some investigations of equine influenza outbreaks and experimental challenge studies DFA has been more sensitive than virus isolation (8, 23, 27), while in others, such as the present study, it has been significantly less sensitive (1).
RT-PCR requires a longer time to get a result than DFA but is considerably faster than virus isolation. In this study, the nasopharyngeal samples that had been thawed once after storage at −70°C were subjected to nested PCR with primers from the nucleoprotein gene (17) and a single set of primers from the matrix gene (3). The results are presented in Table 1. No virus was detected in the samples from horse 1 by use of the outer nucleoprotein primers. However, PCR product was detected in the samples collected on days 2, 3, 4, and 5 when the nested primers were used. Virus was detected in these samples and in the samples collected on days 6 and 7 with the primers from the matrix gene. Virus was isolated in embryonated eggs from the day 6 sample prior to freezing but not after one freeze-thaw cycle. Virus was not detected by any other method in the sample collected on day 7. This sensitivity pattern was reproduced in horses 2 and 4, i.e., nested PCR using the nucleoprotein primers was more sensitive than that using the outer primers on their own and the single pair of matrix primers were in turn more sensitive than the nested nucleoprotein primers. Virus was detected by using the matrix primers in the samples collected from horse 4 on days 1 to 6 inclusive. Virus was not detected in the sample collected on day 1 by any other method and was only detected in the sample collected on day 2 by virus isolation prior to freezing. For horse 3, the sensitivity of the matrix primers was equal to that of the nested primers. Overall the detection limit of PCR using the nucleoprotein primers in our study was consistent with that reported previously (17), i.e., EID50/30 μl (corresponding to 33 EID50/ml). However, PCR using the single set of primers from the matrix gene proved more sensitive and is less prone to contamination than nested PCR. In a recent retrospective study of an outbreak of equine influenza in a riding school in The Netherlands, RT-PCR using these primers proved to be more sensitive than virus isolation or two different enzyme immunoassays (23). Furthermore, these primers were originally designed to detect all known subtypes of influenza A viruses from multiple species (3). It appears that avian influenza viruses can occasionally become infective for horses (4), and given the zoonotic potential of avian influenza viruses, it is not only advantageous to the equine industry but also important for public health to have a diagnostic assay that is capable of detecting genotypically diverse viruses.
In this experimental challenge study, the quantification assays were carried out on samples that were frozen within 24 h of collection and had subsequently been thawed once. This single freeze-thaw resulted in a reduction in the sensitivity of virus detection by isolation in embryonated eggs, i.e., no virus was detected in three samples that were positive on initial screening of the unfrozen samples. The samples concerned were those collected from horse 1 on day 6 and horse 4 on days 2 and 3. The EID50 was repeated by using different aliquots that had undergone one freeze-thaw cycle, and virus was isolated from one sample but at a low titer (100.6 EID50/ml). The other two samples were negative. FCS was omitted from the VTM in this study as it inhibits the growth of equine influenza virus in tissue culture. The effects of freeze-thawing on the viability of virus in VTM in the presence or absence of 2% FCS was examined and the results are summarized in Table 2. There was significantly greater decline in EID50 in samples A, B, and C when FCS was omitted from the VTM (P = 0.034, 0.032, and 0.014, respectively) as determined by the paired t test. A similar effect was observed when the virus was quantified in MDCK cells.
TABLE 2.
Detection of equine influenza virus by virus isolation, antigen detection, and RT-PCR after repeat freeze-thaw cycles in samples with or without FCS
| No. of freeze-thaw cycles | Samplea | EID50/ml | TCID50/ml | DFA | RT-PCR Ob | RT-PCR Ic |
|---|---|---|---|---|---|---|
| 0 | A (F) | 107.3 | ≥107.5 | +++ | + | + |
| A | 107.5 | ≥107.5 | ++ | + | + | |
| B (F) | 106 | 105.25 | − | + | + | |
| B | 105.83 | 105.75 | − | + | + | |
| C (F) | 102.5 | 102.75 | − | − | + | |
| C | 102.3 | 103 | − | − | + | |
| 1 | A (F) | 107.16 | 107 | +++ | + | + |
| A | 105.5 | 106.75 | ++ | + | + | |
| B (F) | 104.6 | 105.25 | − | + | + | |
| B | 103.6 | 103.75 | − | + | + | |
| C (F) | 102 | 102 | − | − | + | |
| C | 101 | − | − | − | + | |
| 2 | A (F) | 106.5 | 106.5 | +++ | + | + |
| A | 105.3 | 105.5 | ++ | + | + | |
| B (F) | 104 | 104.5 | − | + | + | |
| B | 101.16 | 101.25 | − | + | + | |
| C (F) | 101.6 | 101.75 | − | − | + | |
| C | − | − | − | − | + | |
| 3 | A (F) | 106.3 | 106.75 | +++ | + | + |
| A | 104.16 | 104.5 | ++ | + | + | |
| B (F) | 103.6 | 104.25 | − | + | + | |
| B | − | − | − | + | + | |
| C (F) | 101 | − | − | − | + | |
| C | − | − | − | − | + | |
| 4 | A (F) | 106.16 | 106.25 | +++ | + | + |
| A | 103.83 | 103.5 | ++ | + | + | |
| B (F) | 103.3 | 103.5 | − | + | + | |
| B | − | − | − | + | + | |
| C (F) | 100.83 | − | − | − | + | |
| C | − | − | − | − | − |
F, with FCS. Other samples are without FCS.
PCR O, PCR using the NPF and NPR primers.
PCR I, nested PCR using NPF, NPR, NPFI, and NPRI.
The relative sensitivities of DFA and RT-PCR were also evaluated in this experiment (Table 2). Only virus in test item A was detected by DFA, but virus was detected after each of the freeze-thaw cycles irrespective of the inclusion or omission of FCS from the VTM. Virus was detected in samples A and B after each freeze-thaw cycle by PCR using only the outer primers NPF and NPR as well as by nested PCR. Virus was only detected in sample C by nested PCR. There was no decrease in sensitivity over the course of the experiment with the exception of the fourth freeze-thaw cycle in the sample with no FCS. Virus was not detected in this sample by virus culture or by DFA.
The results indicate that to ensure maximum sensitivity it is advisable to attempt virus isolation from samples as soon as possible after collection and prior to freezing. The omission of FCS from the VTM decreases the viability of the virus on freeze-thawing. An inhibitory effect was observed with FCS in undiluted samples in tissue culture but was not evident in diluted samples. Thus, samples that are to be tested by virus isolation after freeze-thaw should have an alternative cryoprotectant or stabilizer included in the VTM, or if FCS is used, they should be diluted in serum-free medium prior to attempting isolation in tissue culture. This is less important if the samples are to be tested by DFA or PCR, assays that do not require viable virus.
In conclusion, this is the first report of a comparison of equine influenza virus isolation and quantification in embryonated eggs and tissue culture and virus detection using a commercial kit (DFA) and RT-PCR using primers from the matrix and the nucleoprotein gene. The RT-PCR using the matrix primers (3) was the most sensitive technique. It has recently been introduced as a routine assay in our diagnostic laboratory. We find the DFA to be a useful supplementary diagnostic test, but a negative result does not preclude the possibility of influenza virus infection. Virus isolation in embryonated eggs is carried out routinely as it is essential for surveillance and vaccine strain selection.
Acknowledgments
We thank Susan Fisher and Rosaline Cullinane for assistance with the preparation of the manuscript.
Support was provided by the Department of Agriculture, Food and Rural Development under the National Development Plan. Michelle Quinlivan is a PhD student funded by the Irish Research Council for Science, Engineering and Technology.
REFERENCES
- 1.Chambers, T. M., K. F. Shortridge, P. H. Li, G. Powell, and K. L. Watkins. 1994. Rapid diagnosis of equine influenza by the Directigen flu-A enzyme immunoassay. Vet. Rec. 135:275-279. [DOI] [PubMed] [Google Scholar]
- 2.Daly, J. M., A. C. K. Lai, M. M. Binns, T. M. Chambers, M. Barrandeguy, and J. A. Mumford. 1996. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J. Gen. Virol. 77:661-671. [DOI] [PubMed] [Google Scholar]
- 3.Fouchier, R. A. M., T. M. Bestebroer, S. Herfst, L. Van Der Kemp, G. F. Rimmelzwaan, and A. D. M. E. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo, Y., M. Wang, Y. Kawaoka, O. Gorman, T. Ito, T. Saito, and R. G. Webster. 1992. Characterisation of a new avian-like influenza A virus from horses in China. Virology 188:245-1255. [DOI] [PubMed] [Google Scholar]
- 5.Guthrie, A. J., K. B. Stevens, and P. P. Bosman. 1999. The circumstances surrounding the outbreak and spread of equine influenza in South Africa. Rev. Sci. Tech. 18:179-185. [DOI] [PubMed] [Google Scholar]
- 6.Johnston, S. L. G., and H. Bloy. 1993. Evaluation of a rapid enzyme immunoassay for detection of influenza A virus. J. Clin. Microbiol. 31:142-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karber, G. 1931. 50% end point calculation. Arch. Exp. Pathol. Pharmakol. 162:480-483. [Google Scholar]
- 8.Morley, P. S., J. R. Bogdan, H. G. G. Townsend, and D. M. Haines. 1995. Evaluation of Directigen Flu A assay for detection of influenza antigen in nasal secretions of horses. Equine Vet. J. 27:131-134. [DOI] [PubMed] [Google Scholar]
- 9.Mumford, J. A., D. Hannant, and D. M. Jessett. 1990. Experimental infection of ponies with equine influenza (H3N8) viruses by intranasal inoculation or exposure to aerosols. Equine Vet. J. 22:93-98. [DOI] [PubMed] [Google Scholar]
- 10.Mumford, J. A., and J. M. Wood. 1993. WHO/OIE meeting: consultation on newly emerging strains of equine influenza. Vaccine 11:1172-1175. [DOI] [PubMed] [Google Scholar]
- 11.Mumford, J. A., D. M. Jessett, E. A. Rollinson, D. Hannant, and M. E. Draper. 1994. Duration of protective efficacy of equine influenza immunostimulating complex/tetanus vaccines. Vet. Rec. 134:158-162. [DOI] [PubMed] [Google Scholar]
- 12.Mumford, J. A., H. Wilson, D. Hannant, and D. M. Jessett. 1994. Antigenicity and immunogenicity of equine influenza vaccines containing a Carbomer adjuvant. Epidemiol. Infect. 112:421-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mumford, J. A. 1995. Virus infections regarding the respiratory tract. Proceedings of the 4th Congress of Equine Medicine and Surgery 1-12th December 1995, Geneva, Switzerland. Swiss-Vet 11-5:66-69.
- 14.Nelly, M. 1996. Characterisation of equine influenza virus isolates from the 1989 and 1992 influenza outbreaks in Ireland by nucleotide sequence determination of the haemagglutinin molecule. M.Sc. thesis. National University of Ireland, Dublin.
- 15.Newton, D. W., C. F. Mellen, B. D. Baxter, R. L. Atmar, and M. A. Menegus. 2002. Practical and sensitive screening strategy for detection of influenza virus. J. Clin. Microbiol. 40:4353-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Office International des Epizooties (OIE). 2000. Equine influenza, p. 546-557. In Manual of standards for diagnostic tests and vaccines. OIE, Paris, France.
- 16a.Office International des Epizooties (OIE). 1996. Conclusions and recommendations from the consultation meeting of OIE and WHO experts on equine influenza, Newmarket, United Kingdom, Septemer 18-19, 1995. Office Int. Epiz. Bull. 108:482-484.
- 17.Oxburgh, L., and A. Hagstrom. 1999. A PCR based method for the identification of equine influenza virus from clinical samples. Vet. Microbiol. 67:161-174. [DOI] [PubMed] [Google Scholar]
- 18.Rogers, A. L. 1988. A/equi-2 influenza in horses in the Republic of South Africa. J. S. Afr. Vet. Assoc. 59:123-125. [PubMed]
- 19.Ryan-Poirier, K. A., J. M. Katz, R. B. Webster, and Y. Kawaoka. 1992. Application of Directigen FLU-A for the detection of influenza A virus in human and nonhuman specimens. J. Clin. Microbiol. 30:1072-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sovinova, O., B. Tumova, F. Pouska, and J. Nemec. 1958. Isolation of a virus causing respiratory disease in horses. Acta Virol. 2:52-61. [PubMed] [Google Scholar]
- 21.Timoney, P. J. 1996. Equine influenza. Comp. Immunol. Microbiol. Infect. Dis. 19:205-211. [DOI] [PubMed] [Google Scholar]
- 22.van Maanen, C., and A. Cullinane. 2002. Equine influenza virus infections: an update. Vet. Q. 24:79-94. [DOI] [PubMed] [Google Scholar]
- 23.van Maanen, C., G. J. van Essen, J. Minke, J. M. Daly, and P. J. Yates. 2003. Diagnostic methods applied to analysis of an outbreak of equine influenza in a riding school in which vaccine failure occurred. Vet. Microbiol. 93:291-306. [DOI] [PubMed] [Google Scholar]
- 24.Wadell, G. H., M. B. Teigland, and M. M. Sigel. 1963. A new influenza virus associated with equine respiratory disease. J. Am. Vet. Assoc. 143:587-590. [PubMed] [Google Scholar]
- 25.Waner, J. L., S. J. Todd, H. Shalaby, P. Murphy, and L. V. Wall. 1991. Comparison of Directigen FLU-A with viral isolation and direct immunofluorescence for the rapid detection and identification of influenza A virus. J. Clin. Microbiol. 29:479-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webster, R. G. 1993. Are equine 1 influenza viruses still present in horses? Equine Vet. J. 25:537-538. [DOI] [PubMed] [Google Scholar]
- 27.Wernery, R., P. J. Yates, U. Wernery, and J. A. Mumford. 1999. An equine influenza outbreak in a polo club in Dubai, United Arab Emirates in 1995/96, p. 342-346. In U. Wernery, J. F. Wade, J. A. Mumford, and O. R. Kaaden (ed.), Equine infectious diseases VIII. R&W Publications Ltd., Newmarket, United Kingdom.