Abstract
The inflammatory bowel diseases are considered an abnormal host immune response to an environmental stimulus. Evidence suggests a role for intestinal bacteria in initiating and/or providing an ongoing stimulus for inflammation in inflammatory bowel disease. Helicobacter pylori is the major cause of active chronic gastritis and peptic ulcers in humans and has been linked to gastric carcinoma and lymphoma. Studies in various animal models, particularly mice, have identified enterohepatic Helicobacter species that are capable of causing hepatitis and enterocolitis. We hypothesize that Helicobacter species may have a role in maintaining inflammation in humans with inflammatory bowel disease. In order to investigate this, biopsy specimens were obtained from patients with and without inflammatory bowel disease. DNA was extracted from the tissues and subjected to PCR with primers designed to detect the ribosomal DNA of members of the Helicobacter species. DNA from six biopsy samples from 60 inflammatory bowel disease patients tested positive. This included 5 of 33 ulcerative colitis patients that were positive compared to 0 of 29 age-matched controls (P < 0.04). Sequencing of the bands produced by PCR amplification revealed ≥99% homology with H. pylori. These results indicate that a member of the Helicobacter species may be involved in some cases of ulcerative colitis.
Inflammatory bowel diseases such as ulcerative colitis and Crohn's disease are chronic diseases characterized by exacerbations and remissions of mucosal inflammation and ulceration. These diseases cause significant morbidity and are associated with an increased risk of colon cancer (4, 17). The cause(s) of inflammatory bowel disease is not known. The current hypothesis is that genetically susceptible individuals develop an abnormal immune response to an environmental stimulus or agent (5, 19, 25). The factor(s) that initiates the inflammation or contributes to exacerbations is not known. A number of clinical observations suggest that bacteria or gut flora play a role in these diseases; however, the data linking specific infectious agents remains inconclusive (8, 11, 15, 20, 21, 25). On the other hand, studies in rodent models of inflammatory bowel disease have suggested that elements of the normal gut flora may be involved in the pathogenesis of the intestinal inflammation (10, 26).
Gastric infection by Helicobacter pylori is a well-recognized cause of chronic active gastritis and gastric and duodenal ulcers and is linked to the development of gastric malignancies (14). Studies in mice have suggested that persistent intestinal infection by related organisms such as Helicobacter hepaticus and Helicobacter bilis is associated with the development of chronic inflammation and enterocolitis (6, 7, 12, 13, 18). These findings raise the possibility that intestinal infection by Helicobacter species may be involved in the pathogenesis of the intestinal inflammation in patients with inflammatory bowel disease. To date, no evidence is available that confirms a relationship between Helicobacter species and the development of human inflammatory bowel disease. In this report, primers which amplify Helicobacter species-specific sequences of ribosomal DNA were used for PCR analysis to search for the presence of Helicobacter DNA in endoscopically obtained biopsy tissue.
MATERIALS AND METHODS
Colonic biopsy samples were obtained from patients enrolled in a population-based case-control study of inflammatory bowel disease. In developing an administrative definition of inflammatory bowel disease, subjects identified through the administrative database of Manitoba Health, the single health insurer for the province, were mailed questionnaires regarding their histories of inflammatory bowel disease and willingness to participate in future studies. Sixty percent of the subjects responded and mailed back the questionnaires (3). This group was used to develop the population-based University of Manitoba Inflammatory Bowel Disease Epidemiology Database (3). Those who returned questionnaires and thereby consented to be known to the researchers were logged in the University of Manitoba Inflammatory Bowel Disease Research Registry. At the beginning of the present study, there were 2,890 subjects in the research registry. Persons in this research registry who agreed to participate in future studies furnished a mailing address and a telephone number to facilitate contact. We accessed the registry in search of subjects under the age of 50 years and mailed them information sheets and questionnaires for a case-control study examining possible etiologic risk factors for inflammatory bowel disease.
A population-based set of controls was developed from the Manitoba Health (MH) population registry. The Manitoba Health population registry contains demographic information on all persons registered with the Manitoba Health public health insurance system. The registry is regularly updated with vital registrations and information from medical and hospital transactions and closely matches population estimates derived from the Canadian census (Statistics Canada) (22). A random sample of registered persons was selected with stratification for age (5-year intervals) and gender to achieve balance with the case series for those two variables. With the specified stratification, Manitoba Health's Information Services generated a mailing list of eligible controls and sent an information package prepared by the investigators explaining the study and requesting participation. The investigators did not know the identity of the controls unless they received a mailed response. For a second control group we asked patients with inflammatory bowel disease to refer us to one or more siblings.
All cases and controls completed a questionnaire and consented to provide a blood sample. Controls were invited to participate in colonoscopy plus biopsies, and those agreeing were paid an honorarium. Approximately 10% of controls who were enrolled in our study collecting questionnaire data and blood agreed to participate in the colonoscopy plus biopsy study. Cases were asked to contact the study personnel when they were to undergo their next colonoscopy for clinical reasons. All patients who were to undergo colonoscopies consented to providing extra biopsy tissue for study purposes.
Each of the cases and controls provided eight biopsy samples from the cecum and eight biopsy samples from the rectum at colonoscopy. In subjects with a previous cecal resection, biopsy samples were obtained from the right colon distal to the ileocolonic anastomosis. All biopsy samples were snap frozen in liquid nitrogen and stored at −70°C. These studies were approved by the research ethics board of the University of Manitoba
Nucleic acid purification and amplification.
Nucleic acids were extracted from tissue samples essentially as previously described (16), except that tissues were homogenized in only 1 ml of lysis buffer in 10-ml polypropylene snap cap tubes. The tissue lysis buffer that was used in this study is a modification (16) of the commonly used Chomczynski lysis buffer (9). Specifically, 0.2 M sodium acetate was used at pH 7.0 instead of at pH 4.0. DNA was quantified by spectrophotometry at 260 nm.
The Helicobacter genus-specific primers 5′TATGACGGGTATCC GGC-3′ and 5′-ATTCCACCTACCTCCTCCCA-3′ were designed to amplify a 375-bp sequence within an area of the 16S rRNA gene conserved among members of the Helicobacter genus (2). An NCBI BLAST analysis confirmed that these primers identify the target sequence within the genome of known Helicobacter species including H. pylori, H. felis, H. muridarum, H. bilis, and H. hepaticus strains. PCR primers were constructed at the Institute for Molecular Biology and Medicine (Mobix Laboratory, McMaster University, Hamilton).
PCRs were prepared with 2.8 μM each primer, 3 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 5 μl of Amplitaq Gold Buffer (Applied Biosystems), 1.25 μg of template DNA, and 1.6 U of Taq polymerase (Applied Biosystems) plus distilled water to a total volume of 50 μl. PCR mixtures were heated to 94°C for 10 min, followed by 45 cycles of denaturation at 94°C for 30 s, primer annealing at 53°C for 30 s, and extension at 72°C for 45 s, followed by 10 min at 72°C in a PTC 200 DNA Engine (MJ Research, Waltham, Mass.). PCR products were subjected to electrophoresis on a 2% agarose gel containing ethidium bromide, and the size of the product was confirmed by using DNA molecular size standards.
The specificity of the PCR primers designed for these studies was evaluated. The primer set amplified DNA extracted from cultured H. pylori, H. hepaticus, and H. felis (data not shown). No bands were observed with these Helicobacter PCR primers with DNA extracts from Escherichia coli or Campylobacter jejuni. PCR analysis detected bands in mouse gastric tissue known to be infected with H. pylori (data not shown). To confirm that the method of extracting DNA allowed identification of bacteria adherent to the mucosa, human gastric tissue with histologically identifiable H. pylori infection was obtained; DNA extracted from this tissue had a strong positive band for Helicobacter DNA (data not shown).
To assess the sensitivity of the PCR for the detection of H. pylori DNA, serially diluted samples of H. pylori DNA were subjected to the PCR. Strong bands were detected in samples diluted down to 0.000125 μg of H. pylori DNA (data not shown)
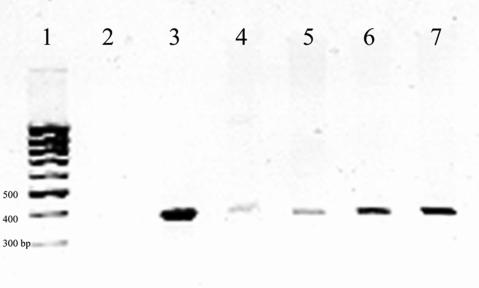
To ensure that the DNA extraction method would allow extraction of bacterial DNA from tissue, tissue known to be negative for Helicobacter was combined with cultured H. pylori bacteria in decreasing amounts from 108 to 100 bacteria (±101 bacteria in each sample). After DNA extraction, PCR analysis of the DNA from the bacteria-spiked tissues detected H. pylori DNA with bacterial concentrations of between 10 and 100 bacteria per sample (Fig. 1).
FIG. 1.
Assessment of tissue extraction efficiency for the detection of Helicobacter rRNA gene sequences. DNA extracts from tissues treated with serially diluted Helicobacter pylori bacteria were amplified with pan-Helicobacter species PCR primers, followed by electrophoresis of the PCR product on an ethidium bromide-containing agarose gel. Lane 1, 1-kb DNA ladder; lane 2, negative control; lane 3, positive control; lane 4, DNA from tissue with 10 ± 10 bacteria present; lane 5, DNA from tissue with 100 ± 10 bacteria present; lane 6, DNA from tissue with 1,000 ± 100 bacteria present; lane 7, DNA from tissue with 10,000 ± 1,000 bacteria present.
Controls were included in each set of PCR amplifications of patient sample DNA. A negative control lacking DNA template was used as well as a positive control containing DNA extracted from cultured H. pylori Sydney strain. In addition, each specimen was run with and without a DNA spike consisting of the addition of 1 μg of H. pylori DNA to the PCR mixture to exclude the presence of PCR inhibitors. H. pylori DNA was used for this in preference to human gene internal controls as some inhibitors may be specific for Helicobacter DNA. PCR products from tissue specimens that produced a positive band were sequenced by a fluorescence-based DNA sequencing method (Mobix Laboratory, McMaster University, Hamilton). The sequences were compared to that of known members of the Helicobacter genus with the Blast program at the National Center for Biotechnology Information.
Fisher's exact test was used to compare proportions of positive results between cases and controls. Student's t test was used to compare clinical information between groups.
Nucleotide sequence accession numbers. Sequences M28, M36, M37, and M38 were submitted to GenBank and assigned accession numbers AY426557, AY426558, AY426559, and AY426560, respectively.
RESULTS
Biopsy specimens were obtained from 29 age-matched control patients and 60 patients with inflammatory bowel disease, of which 33 had ulcerative colitis, 25 had Crohn's disease, and 2 had indeterminate colitis. The mean age of the ulcerative colitis patients was 43 years, that of the Crohn's disease patients was 38.8 years, and that of the indeterminate colitis patients was 31.5 years, compared to a mean age of 42.0 years for the controls (Table 1). Information on all previous antibiotic treatment was not available for these patients, although no subjects had used antibiotics within 1 week of tissue collection.
TABLE 1.
Clinical information for subjects undergoing biopsy
| Group (n) | Mean age (yr) ± SD | Sex (M:F)a | Mean duration of disease (yr) ± SD |
|---|---|---|---|
| Crohn's disease (25) | 38.8 ± 10.9 | 14:11 | 10.2 ± 5.9 |
| Ulcerative colitis (33) | 43.3 ± 13.6 | 18:13 | 14.4 ± 9.0 |
| Indeterminate colitis (2) | 31.5 ± 14.0 | 0:2 | 7.0 ± 7.1 |
| Controls (29) | 42.0 ± 8.1 | 14:15 |
M, male; F, female.
None of the cases or control specimens contained inhibitors of the PCR, as evaluated by the presence of a band on the spiked sample. The 29 control subjects were all negative for Helicobacter DNA. Within the inflammatory bowel disease group, 60 patients were tested. On PCR amplification, the DNA extracted from six patients produced bands of the size appropriate for extracted species (Table 2, Fig. 2). Five of these positive results were from subjects with ulcerative colitis and one was from a patient with active Crohn's disease. The PCR amplification product from each of these patients was sequenced. Four (M28, M36, M37, and M38) of the six sequences showed >95% identity compared to the corresponding 16S rRNA gene sequence of the H. pylori type strain ATCC 43504 and the sequenced strains, J99 and 26695. The partial 16S rRNA gene fragments M28 (345 bases), M36 (342 bases), M37 (339 bases), and M38 (342 bases) all showed one base change at nucleotide 296 (A to G) compared to that of strains ATCC 43504, J99, and 26695.
TABLE 2.
Clinical information for patients with and without PCR evidence of Helicobacter DNA
| Patient group | No. positive for Helicobacter | Diagnosisa (n) | Mean duration of disease (yr) ± SD | Mean age (yr) ± SD | Sex (M:F)b | Disease activity (no. noted) |
|---|---|---|---|---|---|---|
| PCR positive | 6 | UC (5), CD (1) | 19.7 ± 9.8 | 52 ± 9.6 | 2:4 | Active (2) |
| PCR negative | 54 | UC (28), CD (24), I (2) | 11.8 ± 7.7 | 39.8 ± 12.5 | 30:24 | Active (12) |
UC, ulcerative colitis; CD, Crohn's disease; I, indeterminate colitis.
M, male; F, female.
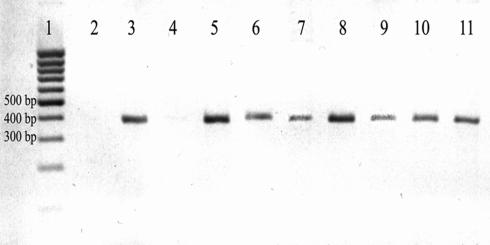
FIG. 2.
Assessment of DNA extracted from endoscopic biopsy samples taken during evaluation of the intestines of patients with and without inflammatory bowel disease: PCR products obtained from amplification of DNA with pan-Helicobacter species PCR primers. Lanes: 1, 1-kb DNA ladder; 2, negative control; 3, positive control; 4, ulcerative colitis subject negative for extracted DNA; 5, ulcerative colitis subject from lane 4 with H. pylori DNA spike; 6, positive subject with ulcerative colitis; 7, positive subject with ulcerative colitis; 8, positive subject with active ulcerative colitis; 9, positive subject with active Crohn's disease; 10, positive subject with ulcerative colitis; 11, positive subject with ulcerative colitis.
When compared to the 16S rRNA gene sequence of H. hepaticus ATCC 51449 (sequenced strain) and H. bilis (accession number U51873), these four sequences all showed ≤95% identity. The remaining two PCR sequences also showed the highest match (85% identity) to H. pylori 16S rRNA gene sequences, but with the limited DNA obtained from the biopsy samples, we were unable to obtain a good clean sequence for submission to GenBank.
The proportion of ulcerative colitis patients showing Helicobacter sp. DNA was greater than the proportion of controls (P < 0.04 by Fisher's exact test). The patients who tested positive were significantly older than the patients who were negative for Helicobacter DNA (mean age, 52 years, versus 39.8 years for the negative ulcerative colitis patients; P = 0.024) and had a significantly longer duration of disease (mean, 19.7 years, versus a mean of 11.8 for Helicobacter-negative patients; P = 0.025). Patients with ulcerative colitis who were positive for Helicobacter DNA also tended to have inactive disease (four of six positive patients), although this was not statistically significant.
[13C]urea breath testing and serology for Helicobacter were not performed at the time the biopsy samples were taken: after these results were obtained, the patients who tested positive for Helicobacter DNA were contacted and requested to undergo further testing. Five of the six patients (all of the ulcerative colitis patients) agreed to urea breath testing, which was performed approximately 1 year postendoscopy. Only one of these patients tested positive, suggesting active gastric H. pylori infection.
DISCUSSION
H. pylori is well recognized as the major cause of chronic active gastritis, gastric and duodenal ulcers, and gastric malignancies. Other members of the Helicobacter family have been identified in various species, where they reside throughout the gastrointestinal tract. Some of these have been termed the enterohepatic helicobacters for their abilities to persistently colonize and cause inflammation within the liver, small intestine, and colon. In particular, H. hepaticus and H. bilis have been linked to chronic colitis in a variety of mouse models (6, 7, 24, 28). These observations suggest a possible link to human inflammatory bowel disease.
One previous study searched for microbial agents such as Helicobacter in biopsy samples taken from normal or inflamed mucosa of 11 patients with Crohn's disease. Of the five biopsy samples from inflamed mucosa, three contained numerous bacteria, including Helicobacter, as assessed by broad-range and genus-specific hybridization analysis of the PCR products obtained by amplification of fragments of the 16S rRNA gene (27). However, species-specific primers were unsuccessful at identifying which members of the extracted genus were present. The authors suggested that, considering the mixture of bacterial species present in inflamed tissue, it was unlikely that Helicobacter as a single agent would be responsible for Crohn's disease in these patients. However, their failure to confirm the presence of Helicobacter extracted by species-specific primers raises questions about the specificity of the original genus-specific primer set. It is possible that the primers were detecting a closely related bacterium such as Campylobacter. It should be noted that none of the patients included in this study had ulcerative colitis.
We examined biopsy samples from a more extensive population-based cohort of patients with both active and inactive inflammatory bowel disease. For this study, we used a PCR primer set designed to amplify an area of the 16S rRNA gene common to Helicobacter species identified on Blast search. We defined precisely the specificity and sensitivity of this primer set to ensure that the PCR amplification method would detect only Helicobacter species and that DNA from as few as 10 helicobacters in 100 μg of tissue was detectible. We also tested each sample for inhibitors that might interfere with PCR amplification, to ensure that each negative was truly devoid of Helicobacter DNA.
The lack of Helicobacter sp. DNA in the control patient groups is of interest. Considering the age of the control population, a proportion (approximately 20%) of these patients might be expected to have gastric infection with H. pylori (23), which might have been detectable by PCR testing of feces. However, patients undergoing colonoscopy have a bowel cleansing preparation prior to endoscopy, and this would minimize fecal material in the biopsy samples taken for DNA isolation. It may be that this would prevent detection of any Helicobacter species not adherent to the mucosa.
Our studies identified six patients who had Helicobacter DNA present in their colonic biopsy samples. Five of these had ulcerative colitis and one had Crohn's disease. Statistically, there was a significant difference between the ulcerative colitis group and controls (P < 0.04). Though the sequencing of the amplified fragments from the inflammatory bowel disease patients showed homology with H. pylori, this does not rule out the possibility of the presence of a member of the Helicobacter family with significant sequence homology to H. pylori. Gastric H. pylori status for all subjects at the time of colonoscopy was not known. However, only one of the ulcerative colitis patients whose tissue contained Helicobacter DNA had a positive urea breath test approximately 1 year postendoscopy, suggesting that the presence of gastric Helicobacter pylori was not a significant factor in these patients.
In conclusion, H. pylori-like DNA sequences were identified in biopsy samples from 5 of 33 patients with ulcerative colitis and in 1 of 25 with Crohn's disease, while all 29 age-matched controls were negative. These patients with positive findings had a longer history of disease and tended to have inactive disease. Though sequencing of the PCR product indicated the DNA detected showed homology to H. pylori, it remains entirely possible that this organism is distinct from gastric H. pylori. While the DNA in these tissues may be due to bacteria or bacterial DNA in transit from the stomach, these results raise the possibility that a proportion of inflammatory bowel disease patients, particularly those with ulcerative colitis, have transient infection or colonization of the lower gastrointestinal tract with a Helicobacter sp. which may contribute to the persistence of colonic inflammation.
Acknowledgments
We thank Rajka Borojevic and Michael Sargent for invaluable laboratory assistance.
This work was funded in part by the Crohn's and Colitis Foundation of Canada. Cathy Streutker was supported by a CAG/Abbott/CIHR fellowship award. Charles Bernstein is the recipient of a Crohn's and Colitis Foundation of Canada Research Scientist Award and a Canadian Institutes of Health Research Investigator Award. Kenneth Croitoru is the recipient of a Crohn's and Colitis Foundation of Canada Research Scientist Award.
REFERENCES
- 1.Amati, L., L. Caradonna, E. Jirillo, and D. Caccavo. 1999. Immunological disorders in inflammatory bowel disease and immunotherapeutic implications. Ital. J. Gastroenterol. Hepatol. 31:313-325. [PubMed] [Google Scholar]
- 2.Beckwith, C. S., C. L. Franklin, R. R. Hook, Jr., C. L. Besch-Williford, and L. K. Riley. 1997. Fecal PCR assay for diagnosis of Helicobacter infection in laboratory rodents. J. Clin. Microbiol. 35:1620-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, C. N., J. F. Blanchard, P. Rawsthorne, and A. Wajda. 1997. The epidemiology of Crohn's disease and ulcerative colitis in a central Canadian province: a population based study. Am. J. Epidemiol. 149:916-924. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, C. N., E. Kliewer, A. Wajda, and J. F. Blanchard. 2001. The incidence of cancer among patients with inflammatory bowel disease: a population-based study. Cancer 91:854-862. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg, R. S., L. J. Saubermann, and W. Strober. 1999. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr. Opin. Immunol. 11:648-656. [DOI] [PubMed] [Google Scholar]
- 6.Burich, A., R. Hershberg, K. Waggie, W. Zeng, T. Brabb, G. Westrich, J. L. Viney, and L. Maggio-Price. 2001. Helicobacter-induced inflammatory bowel disease in IL-10- and T cell-deficient mice. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G764-G778. [DOI] [PubMed] [Google Scholar]
- 7.Cahill, R. J., C. J. Foltz, J. G. Fox, C. A. Dangler, F. Powrie, and D. B. Schauer. 1997. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect. Immun. 65:3126-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadwick, N., I. J. Bruce, S. Schepelmann, R. E. Pounder, and A. J. Wakefield. 1998. Measles virus RNA is not detected in inflammatory bowel disease with hybrid capture and reverse transcription followed by the polymerase chain reaction. J. Med. Virol. 55:305-311. [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Dianda, L., A. M. Hanby, N. A. Wright, A. Sebesteny, A. C. Hayday, and M. J. Owen. 1997. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am. J. Pathol. 150:91-97. [PMC free article] [PubMed] [Google Scholar]
- 11.Duclos, P., and B. J. Ward. 1998. Measles vaccines: a review of adverse events. Drug Safety 19:435-454. [DOI] [PubMed] [Google Scholar]
- 12.Foltz, C. J., J. G. Fox, R. Cahill, J. C. Murphy, L. Yan, B. Shames, and D. B. Schauer. 1998. Spontaneous inflammatory bowel disease in multiple mutant mouse lines: association with colonization by Helicobacter hepaticus. Helicobacter 2:69-78. [DOI] [PubMed] [Google Scholar]
- 13.Fox, J. G., L. Yan, B. Shames, J. Campbell, J. C. Murphy, and X. Li. 1996. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect. Immun. 64:3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guindi, M. 1999. Role of Helicobacter pylori in the pathogenesis of gastric carcinoma and progression of lymphoid nodules to lymphoma. Can. J. Gastroenterol. 13:224-227. [DOI] [PubMed] [Google Scholar]
- 15.Haga, Y. O., Funakoshi, K. Kuroe, K. Kanazawa, H. Nakajima, H. Saito, Y. Murata, A. Munakata, and Y. Yoshida. 1996. Absence of measles viral genomic sequence in intestinal tissues from Crohn's disease by nested polymerase chain reaction. Gut 38:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamel, A. L., L. Lin, C. Sachvie, E. Grudenski, and G. P. S. Nayar. 2000. PCR detection and characterization of type-2 porcine circovirus. Can. J. Vet. Res. 64:44-52. [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis, J. D., J. J. Deren, and G. R. Lichtenstein. 1999. Cancer risk in patients with inflammatory bowel disease. Gastroenterol. Clin. North Am. 28:459-477. [DOI] [PubMed] [Google Scholar]
- 18.Maloy, K. J., L. Salaun, R. Cahill, G. Dougan, N. J. Saunders, and F. Powrie. 2003. CD4+CD25+T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 197:111-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merger, M., and K. Croitoru. 1998. Infections in the immunopathogenesis of chronic inflammatory bowel disease. Semin. Immunol. 10:69-78. [DOI] [PubMed] [Google Scholar]
- 20.Papadakis, K. A., and S. R. Targan. 1999. Current theories on the causes of inflammatory bowel disease. Gastroenterol. Clin. North Am. 28:283-296. [DOI] [PubMed] [Google Scholar]
- 21.Prantera, C., and M. L. Scribano. 1999. Crohn's disease: the case for bacteria. Ital. J. Gastroenterol. Hepatol. 3:244-246. [PubMed] [Google Scholar]
- 22.Roos, L. L., C. A. Mustard, and J. P. Nicol. 1993. Registries and administrative data: organization and accuracy. Med. Care 3:201-212. [DOI] [PubMed] [Google Scholar]
- 23.Roosendaal, R., E. J. Kuipers, J. Buitenwerf, C. van Uffelen, S. G. Meuwissen, G. J. van Kamp, and C. M. Vandenbroucke-Grauls. 1997. Helicobacter pylori and the birth cohort effect: evidence of a continuous decrease of infection rates in childhood. Am. J. Gastroenterol. 92:1480-1482. [PubMed] [Google Scholar]
- 24.Shomer, N. H., C. A. Dangler, M. D. Schrenzel, and J. G. Fox. 1997. Helicobacter bilis-induced inflammatory bowel disease in scid mice with defined flora. Infect. Immun. 65:4858-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strober, W., I. J. Fuss, and R. S. Blumberg. 2002. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 20:495-549. [DOI] [PubMed] [Google Scholar]
- 26.Taurog, J. D., J. A. Richardson, J. T. Croft, W. A. Simmons, M. Zhou, J. L. Fernandez-Sueiro, E. Balish, and R. E. Hammer. 1994. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 180:2359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiveljung, A., J. D. Soderholm, G. Olaison, J. Jonasson, and H. J. Monstein. 1999. Presence of eubacteria in biopsies from Crohn's disease inflammatory lesions as determined by 16S rRNA gene-based PCR. J. Med. Microbiol. 3:263-268. [DOI] [PubMed] [Google Scholar]
- 28.Ward, J. M., M. R. Anver, D. C. Haines, J. M. Melhorn, P. Gorelick, L. Yan, and J. G. Fox. 1996. Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab. Anim. Sci. 1:15-20. [PubMed] [Google Scholar]