Summary
Biochemical evidence implicates the death-domain (DD) protein PIDD as a molecular switch capable of signaling cell survival or death in response to genotoxic stress. PIDD activity is determined by binding-partner selection at its DD: whereas recruitment of RIP1 triggers prosurvival NF-κB signaling, recruitment of RAIDD activates proapoptotic caspase-2 via PIDDosome formation. However, it remains unclear how interactor selection, and thus fate decision, are regulated at the PIDD platform. We show that the PIDDosome functions in the ‘Chk1-suppressed’ apoptotic response to DNA damage, a conserved ATM/ATR–caspase-2 pathway antagonized by Chk1. In this pathway, ATM phosphorylates PIDD on Thr788 within the DD. This phosphorylation is necessary and sufficient for RAIDD binding and caspase-2 activation. Conversely, nonphosphorylatable PIDD fails to bind RAIDD or activate caspase-2, and recruits prosurvival RIP1 instead. Thus, ATM phosphorylation of the PIDD DD enables a binary switch through which cells elect to survive or die upon DNA injury.
Introduction
In response to DNA damage, cells can either repair and survive the lesions, or remove the damaged genome altogether through apoptotic cell death. An incorrect decision and genomic instability can ensue, stimulating cancer development (Ciccia and Elledge, 2010; Lowe et al., 2004). Despite the radical nature and significance of fate choice after DNA damage, the mechanisms by which cells elect to survive or die upon DNA injury remain unclear.
PIDD (p53-inducible protein with a death domain; LRDD) is a scaffold protein that interacts with prosurvival and prodeath signaling factors after DNA damage (Lin et al., 2000; Telliez et al., 2000). PIDD is composed of seven N-terminal leucine-rich repeats (LRRs), two ZO-1 and Unc5-like (ZU-5) domains, a putative oligomerization domain (UPA; uncharacterized protein domain in UNC5, PIDD and Ankyrin), and a C-terminal death domain (DD). The ZU-5 domains bind PCNA following UV irradiation, implicating PIDD in polη-mediated translesion synthesis (Logette et al., 2011). The PIDD DD serves as a docking site for other DD-containing proteins, including the caspase-2 adaptor protein RAIDD (RIP-associated Ich-1/CED homologous protein with death domain; CRADD) (Duan and Dixit, 1997). Recruitment of RAIDD to PIDD results in the assembly of a PIDD-RAIDD-caspase-2 complex designated PIDDosome (Tinel and Tschopp, 2004), reminiscent of the apoptosome (cyt-c-APAF1-caspase-9) and death-inducing signaling complex (DISC; FAS-FADD-caspase-8). Indeed, the PIDDosome acts as a caspase-2 activation platform in vitro or when overexpressed in vivo, by enabling autocatalytic caspase-2 activation through induced proximity (Berube et al., 2005; Bouchier-Hayes et al., 2009; Tinel and Tschopp, 2004).
PIDD can also participate in a prosurvival signaling complex by recruiting receptor-interacting protein kinase 1 (RIP1) in place of RAIDD (Janssens et al., 2005). Docking of RIP1 onto the PIDD DD mobilizes an alternative PIDDosome comprising PIDD, RIP1, and NEMO (NF-κB essential modulator; IKK-γ). Formation of this complex triggers NEMO sumoylation, initiating a series of events that culminate in NEMO-induced phosphorylation and degradation of IκB, release of NF-κB into the nucleus, and transcription of prosurvival NF-κB target genes (Hayden and Ghosh, 2008; Janssens et al., 2005). Thus, two distinct PIDDosomes can be assembled from PIDD, PIDD-RAIDD-caspase-2 and PIDD-RIP1-NEMO, with opposing effects on cell fate.
Both PIDDosomes have been implicated in the DNA damage response. Topoisomerase II inhibitors readily stimulate recruitment of RIP1 to PIDD, and PIDD is necessary for etoposide-induced NF-κB activation (Janssens et al., 2005). In addition, enforced assembly of the PIDD-RAIDD-caspase-2 complex via overexpression sensitizes cells to genotoxic stress-induced apoptosis (Tinel and Tschopp, 2004). These observations have led to the ‘switch hitter’ model for PIDD function, whereby PIDD acts upon DNA damage to dictate cell survival or death by mobilizing antagonistic PIDDosomes (Janssens et al., 2005; Tinel et al., 2007; Wu et al., 2005).
Thus, a key question with regard to PIDD switch function is the mechanism by which PIDD discriminates between prodeath RAIDD and prosurvival RIP1 after DNA damage. The full-length PIDD protein (100 kDa) is autoproteolytically processed in cells to generate three fragments. PIDD-N (48 kDa) contains the N-terminal LRRs and the proximal ZU-5 domain; PIDD-C (51 kDa) comprises the distal ZU-5 domain, UPA and DD; and PIDD-CC (37 kDa), which lacks the ZU-5 domain but retains an intact UPA and DD (Tinel et al., 2007). Interestingly, whereas PIDD-C preferentially binds RIP1, PIDD-CC primarily interacts with RAIDD. Furthermore, the relative abundances of PIDD-C and PIDD-CC can be influenced by DNA damage, with increasing doses favoring PIDD-CC maturation (Tinel et al., 2007). Thus the selection of RAIDD or RIP1 by PIDD might occur as a function of DNA damage severity. However, endogenous PIDD-FL is often constitutively processed into PIDD-C and PIDD-CC in cells (Logette et al., 2011; Manzl et al., 2009; Tinel et al., 2007), suggesting additional regulatory mechanisms.
Efforts to elucidate the physiologic role of PIDD after DNA damage have yielded contradictory findings (Baptiste-Okoh et al., 2008; Kim et al., 2009; Lin et al., 2000; Manzl et al., 2009; Manzl et al., 2012; Vakifahmetoglu et al., 2006). A caveat has been the lack of a clearly defined experimental setting for endogenous PIDDosome assembly and caspase-2 function after DNA damage (Kitevska et al., 2009; Kumar, 2009; Vakifahmetoglu-Norberg and Zhivotovsky, 2010).
We have recently shown that caspase-2 plays an essential and evolutionarily conserved role in γ-irradiation (IR)-induced apoptosis in zebrafish and human cells, and that this role can be uncovered upon concurrent inhibition of checkpoint kinase 1 (Chk1) (Sidi et al., 2008). In this ‘Chk1-suppressed’ (CS) pathway of apoptotic cell death, caspase-2 becomes activated downstream of ATM and ATR through an as yet unknown mechanism. The selective activation and essential role of caspase-2 in the CS pathway have been further documented in human cancer cells and Caspase-2−/− mouse embryonic fibroblasts (MEFs), respectively (Ho et al., 2009; Myers et al., 2009; Pan et al., 2009).
Here we show that PIDD and RAIDD are responsible for caspase-2 activation in the CS pathway, via PIDDosome formation. We then identify the mechanism by which PIDD discriminates between RAIDD and RIP1 to decide cell fate after DNA damage.
Results
Caspase-2 processing in the CS pathway requires PIDD and RAIDD, but not Apaf-1 or FADD
Three major protein complexes have been implicated in caspase-2 activation. The Apaf-1-based apoptosome and FADD-based DISC can indirectly promote caspase-2 processing via the caspase-9/-8—caspase-3/-6/-7 amplification cascade (Inoue et al., 2009; Lavrik et al., 2006; Manzl et al., 2009; Olsson et al., 2009). In contrast, the PIDD and RAIDD-based PIDDosome can directly activate caspase-2 in vitro or upon overexpression in vivo (Park et al., 2007; Tinel and Tschopp, 2004).
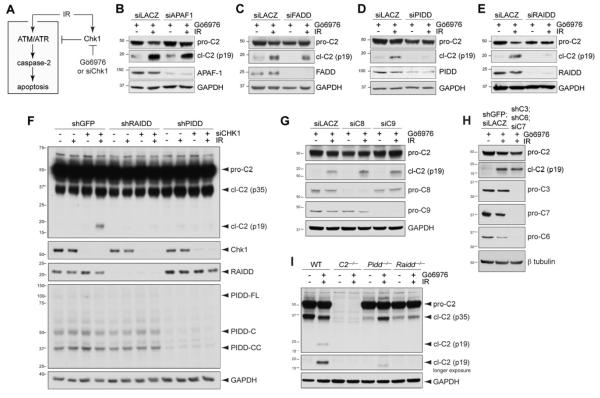
To test the endogenous contributions of these platforms to caspase-2 activation during CS apoptosis, we depleted their core components by RNAi and assessed the effects on caspase-2 processing 24 hours after initiating the CS pathway by combined exposure of HeLa cells to ionizing radiation (IR) and the Chk1 inhibitor, Gö6976 (Figure 1A-E and Supplemental Figure S1A,B) (Sidi et al., 2008). Whereas siRNAs against APAF1 or FADD had no effect (Figure 1C,D), siRNAs to PIDD or RAIDD blocked caspase-2 processing in the CS pathway (Figure 1E,F). This was also observed in p53−/− HCT116 cells (Figure S1B). Consistent with the non-involvement of Apaf-1 or FADD, depletions of CASP9, CASP8, or all downstream executioner caspase activity in CASP3;CASP6;CASP7 triple-knockdown cells, had no effect on caspase-2 cleavage (Figure 1G,H). This also alleviated concerns that caspase-2 cleavage reflects a byproduct of DNA damage-induced caspase cascades, a common caveat in the field (Inoue et al., 2009; Manzl et al., 2009).
Figure 1. PIDD and RAIDD are required for caspase-2 activation in the CS pathway.
(A) Diagram of the CS pathway. Pathway activation after IR is restrained by Chk1. Combined delivery of IR and Chk1 inhibitor (Gö6976) or siRNA activates the pathway. IR and Chk1 inhibition are individually insufficient for pathway activation (Sidi et al., 2008).
(B-E) HeLa cells transfected with the indicated siRNAs were treated with Gö6976 (1 μM) with or without IR (10 Gy) and harvested 24 hr post IR. Lysates were analyzed by western blot. pro-C2, procaspase-2; cl-C2 (p19), cleaved caspase-2, p19 fragment (mature cleavage product). cl-C2 images are longer exposures of the same membrane.
(F) HeLa cells stably expressing the indicated shRNAs were transfected with CHK1 siRNA (+) or LACZ siRNA (-), treated with or without IR (10 Gy) 16 hr after transfection, and harvested 16 hr post IR. Lysates were analyzed by western blot.
(G) HeLa cells transfected with the indicated siRNAs were treated with Gö6976 (1 μM) with or without IR (10 Gy) and harvested 24 hr post IR. Lysates were analyzed by western blot. C8, caspase-8; C9, caspase-9.
(H) HeLa cells stably expressing the indicated shRNAs and transfected with the indicated siRNAs were treated with Gö6976 (1 μM) with or without IR (10 Gy), and harvested 24 hr post IR. Lysates were analyzed by western blot. C3, caspase-3; C6, caspase-6; C7, caspase-7.
(I) SV40-transformed MEFs of indicated genotypes were treated with Gö6976 (1 μM) with or without IR (10 Gy) and harvested 24 hr post IR. Lysates were analyzed by western blot.
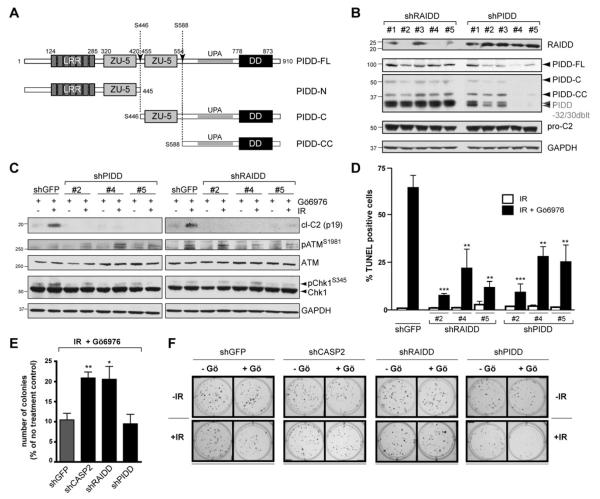
Further depletions of PIDD or RAIDD by shRNAs in stably transduced cell lines also blocked caspase-2 activation after IR+Chk1 inhibitor (Figure 2B,C) or IR+Chk1 siRNA (Figure 1F), but not after heat shock (Figure S1C) (Bouchier-Hayes et al., 2009; Tu et al., 2006). Finally, Pidd−/− and Raidd−/− MEFs (Berube et al., 2005; Manzl et al., 2009) also failed to trigger caspase-2 processing after IR+Chk1 inhibition (Chk1i) (Figures 1I and S1D). Thus PIDD and RAIDD, but not the apoptosome or DISC, are specifically required for caspase-2 activation during CS apoptosis. Additionally, PIDD and RAIDD can be placed between ATM/ATR and caspase-2 in the CS pathway, because none of the PIDD or RAIDD hairpins affected ATM activation, as assessed by ATM autophosphorylation on Ser1981, and with the sole exception of shPIDD#5 (which presumably mediated off-target effects), none of the shRNAs affected ATR activity, as assessed by Chk1 phosphorylation on Ser345 (Figure 2C).
Figure 2. PIDD and RAIDD are required for CS apoptosis.
(A) Schematic diagram of PIDD maturation. Autoproteolytic cleavage sites are indicated (arrows).
(B) HeLa cells stably expressing the indicated shRNAs were analyzed by western blot. PIDD-32/30 doublet: PIDD specific bands running at ~30 kDa, presumably resulting from autoproteolysis of shorter PIDD isoforms (Cuenin et al., 2008).
(C) HeLa cells stably expressing the indicated shRNAs were treated with Gö6976 (1 μM) with or without IR (10 Gy), and harvested 24 hr post IR. Lysates were analyzed by western blot.
(D) HeLa cells stably expressing the indicated shRNAs were treated with 10 Gy IR with or without Gö6976 (1 μM) (black and white bars, respectively), and were analyzed by TUNEL staining at 48 hr post IR. Data are means +/− SEM. **p < 0.01; ***p < 0.001 two-tailed Student’s t-test.
(E) HeLa cells stably expressing the indicated shRNAs were treated with Gö6976 (1 μM) and IR (5 Gy) and colony numbers were recorded 14 days post IR. Data are means +/− SEM. *p < 0.05; **p < 0.01 two-tailed Student’s t-test.
(F) Representative images of a clonogenic assay. HeLa cells stably expressing the indicated shRNAs were treated with or without Gö6976 (1 μM) or IR (2 Gy) and stained with crystal violet 14 days after IR.
PIDD and RAIDD are required for CS apoptosis
Depletions of PIDD or RAIDD markedly impaired CS pathway-induced apoptosis, as demonstrated by 65-85% reductions in TUNEL staining 48 hours after IR+Chk1i (Figure 2D). These phenotypes were similar to those observed in shCASP2 cells (Sidi et al., 2008) or Caspase-2−/− MEFs (Ho et al., 2009). Consistent with these observations, depletion of CASP2 or RAIDD protected against the long-term cytotoxic effects of IR+Chk1i, with a two fold increase in colony survival 14 days after treatment compared to control cells (Figure 2E,F). While PIDD knockdown failed to phenocopy CASP2 or RAIDD depletion in the long-term assay, this is likely due to removal of the prosurvival functions of PIDD mediated by PIDD-RIP1-NEMO—NF-κB and/or PIDD-PCNA—polη pathways (Janssens et al., 2005; Logette et al., 2011). Indeed, shPIDD cells, but not shCASP2 or shRAIDD cells, failed to trigger IκBα degradation after IR+Chk1i (Figure S2), consistent with previous results with doxorubicin (Janssens et al., 2005). Despite differential effects on NF-κB signaling and 14-day colony survival, PIDD deficiency nevertheless removes caspase-2 processing and TUNEL reactivity after IR+Chk1i (Figures 1D,F,I, 2C-D, and S1A,B,D), demonstrating that PIDD, like RAIDD and caspase-2, is critical for CS apoptotic signaling.
CS apoptosis requires PIDDosome assembly
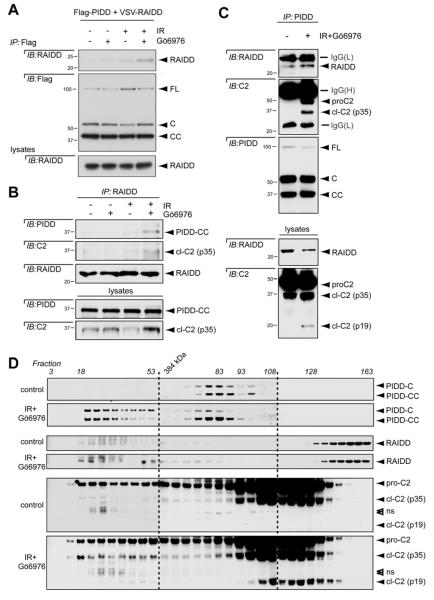
The genetic requirement for PIDD and RAIDD indicated that the CS pathway likely operates through the PIDDosome. Thus far, however, there has been no published account of PIDDosome assembly from endogenous components that results in caspase-2 activation after DNA damage. Starting our analyses with tagged PIDD and RAIDD constructs, we observed that PIDD and RAIDD weakly interacted in unstimulated cells or in cells treated with IR or Chk1i alone. However, the interaction was substantially enhanced in double-treated cells (Figure 3A). This provided initial support for PIDDosome formation during CS apoptosis.
Figure 3. The CS pathway triggers PIDDosome assembly.
(A) HeLa cells transfected with the indicated constructs were treated with or without Gö6976 (1 μM) or 10 Gy IR and harvested 24 hr post IR. Anti-Flag immunoprecipitates were analyzed by western blot. PIDD-FL is constitutively autoprocessed in cells into -C and -CC fragments (Tinel et al., 2007).
(B) HeLa cells treated with or without IR (10 Gy) or Gö6976 (1 μM) were harvested 24 hr post IR. Lysates were immunoprecipated with anti-RAIDD antibody and analyzed by western blot.
(C) HeLa cells treated with or without IR (10 Gy) or Gö6976 (1 μM) were harvested 24 hr post IR. Lysates were immunoprecipated with anti-PIDD antibody and analyzed by western blot.
(D) HeLa cells treated with or without IR (10 Gy) and Gö6976 (1 μM) were lysed 24 hr post IR and loaded on a S400 HiPrep 16/60 Sephacryl column (1 ml/min). An aliquot of each fraction was analyzed by SDS-PAGE with the indicated antibodies. Empty arrowheads indicate non-specific bands.
We then investigated complex assembly at the endogenous level. Both PIDD-CC and caspase-2 (p35 subunit) were successfully detected in RAIDD immunoprecipitates from IR+Chk1i–treated cells, but not unstimulated or single-treated cells (Figure 3B). Reciprocally, both RAIDD and caspase-2 (full-length and p35 subunit) were readily detected in PIDD immunoprecipitates from cells undergoing CS apoptosis but not control cells (Figure 3C). Furthermore, size exclusion chromatography demonstrated the recruitment of PIDD (C and CC fragments), RAIDD, and caspase-2 to a large molecular complex in cells undergoing CS apoptosis, but not control cells (Figure 3D). PIDD-CC, RAIDD and procaspase-2 consistently co-eluted in high molecular weight fractions, and notably, fractions 49-53 also contained both caspase-2 cleavage products, altogether indicative of bona fide functional PIDDosomes. Finally, the physical interaction between PIDD and RAIDD is essential for caspase-2 activation in the CS pathway (see Figure 5 below). Collectively, the genetic and biochemical data show that both caspase-2 cleavage and apoptotic signaling through the CS pathway require PIDDosome assembly and function.
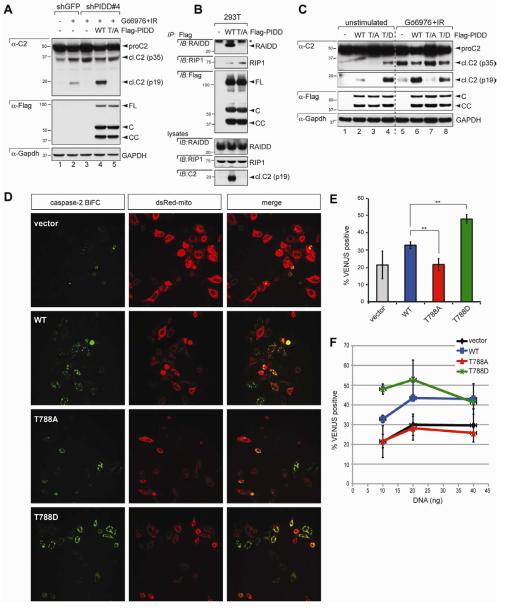
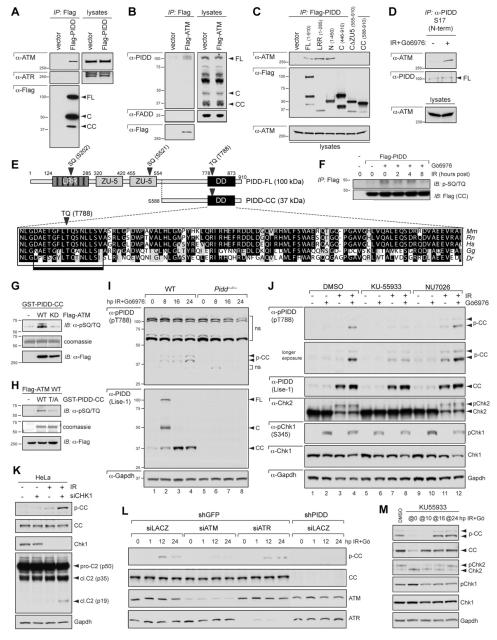
Figure 5. PIDD phosphorylation on T788 is necessary and sufficient for caspase-2 activation.
(A) HeLa cells stably expressing the indicated shRNAs were transfected with the indicated C-terminally Flag-tagged, shRNA-resistant PIDD constructs, treated with or without Gö6976 (1 μM) or IR (10 Gy), and harvested 24 hr post IR. Lysates were analyzed by western blot. WT, wild-type PIDD; T/A, T788A mutant.
(B) 293T cells transfected with the indicated C-terminally Flag-tagged PIDD constructs were harvested at 24 hr post transfection. Protein extracts were immunoprecipitated with anti-Flag antibodies. Immunoprecipitates and whole-cell lysates were analyzed by western blot. Asterisk indicates IgG light chain.
(C) HeLa cells transfected with the indicated C-terminally Flag-tagged PIDD constructs were either left untreated or treated with Gö6976 (1 μM) and IR (10 Gy), and harvested 24 hr post IR. Lysates were analyzed by western blot. T/D, T788D mutant.
(D) HeLa-Bcl-XL cells were transfected with 5 ng of C2-CARD-VC and C2-CARD-VN pBiFC plasmids, encoding the C- and N-terminal moieties of the Venus protein, respectively, fused to the caspase recruitment domain (CARD) of caspase-2. Cells were co-transfected with the indicated PIDD constructs (10 ng) and dsRed-mito (10 ng, used as transfection reporter), and imaged 24 hr post-transfection by confocal microscopy. Caspase-2 bifluorescence complementation (BiFC; left column) is the Venus fluorescence signal that occurs when N- and C-terminal Venus moieties are brought in proximity as a result of caspase-2 recruitment to the PIDDosome.
(E) HeLa cells transfected and imaged as in (D) were scored for Venus positivity. Data from 3 independent experiments (>100 cells each) are shown as means +/− SEM. **p < 0.01, two-tailed Student’s t-test.
(F) HeLa cells transfected as in (D) but with incremental amounts of the indicated PIDD constructs (10, 20, or 40 ng) were imaged as in (D) and scored for Venus positivity. Data from 3 independent experiments (>100 cells each) are shown as means +/− SEM.
PIDDosome formation in the CS pathway does not correlate with an increase in PIDD-CC maturation
We next sought to determine the mechanism of PIDDosome assembly in the CS pathway. PIDD interacts with RAIDD primarily through the PIDD-CC fragment (Tinel et al., 2007). PIDD-CC accumulates in cells exposed to IR (Cuenin et al., 2008; Tinel et al., 2007) [Figures S3 and 4J (lanes 1-4)]. Because initiation of the CS pathway via IR+Chk1i leads to yet greater levels of DNA damage than that seen after IR alone (Sidi et al., 2008), a rise in PIDD-CC might account for the enhanced RAIDD recruitment and caspase-2 activation observed in this context. However, we did not detect any evident increase, whether net, relative, nuclear, or cytoplasmic, in PIDD-CC levels after IR+Chk1i compared to that observed after IR alone, whether in MEFs or HeLa cells [Figures S3, 4J (lanes 1-4), and 4K]. These results pointed to a different mechanism of PIDDosome assembly after DNA damage.
Figure 4. ATM phosphorylates PIDD on T788 during CS apoptosis.
(A) HeLa cells transfected with Flag-tagged PIDD were lysed, immunoprecipitated with anti-Flag antibodies and analyzed by western blot.
(B) HeLa cells transfected with Flag-ATM were lysed, immunoprecipitated with anti-Flag antibodies and analyzed by western blot.
(C) HeLa cells transfected with the indicated Flag-tagged PIDD deletion constructs were lysed 24 hr post transfection, immunoprecipitated with anti-Flag antibodies and analyzed by western blot.
(D) HeLa cells either untreated or treated with IR (10 Gy) and Gö6976 (1 μM) were harvested 24 hr post IR. Nuclear extracts were immunoprecipated with an anti-PIDD antibody targeted to the PIDD N-terminus. Immunoprecipitates were analyzed by western blot.
(E) Diagram of PIDD-FL highlighting three candidate ATM phosphorylation sites (bold arrowheads). Blow up shows a clustal alignment of the DD amino acid sequences of PIDD proteins from the indicated species. Mm, Mus musculus; Rn, Rattus norvergicus; Hs, Homo sapiens; Gg, Gallus gallus (chicken); Dr, Danio rerio (zebrafish). The predicted full ATM target sequence (box) and the target TQ motif (bold arrowhead) are indicated.
(F) HeLa cells transfected with empty vector or C-terminally Flag-tagged PIDD were treated with IR (10 Gy) with or without Gö6976 (1 μM) and harvested at the indicated time points after IR. Protein extracts were immunoprecipitated with anti-Flag antibodies and analyzed by western blot.
(G) Recombinant GST-PIDD-CC proteins were incubated with wild-type (WT) or kinase-dead (KD) Flag-ATM for an in vitro kinase assay (IVK). Reactions were analyzed by coomassie staining and western blot.
(H) Recombinant wild-type (WT) or T788A mutant (T/A) GST-PIDD-CC proteins were incubated with Flag-ATMWT for an IVK. Reactions were analyzed by coomassie staining and western blot. Residual pSQ/TQ immunoreactivity in the T/A sample suggests additional specific or non-specific phosphorylation events occurring in the reaction.
(I) SV40 MEFs of indicated genotypes were treated with IR (10 Gy) and Gö6976 (1 μM) and harvested at the indicated time points after IR. Extracts were analyzed by western blot. p-CC, phospho-PIDD-CC; upper arrowhead marks p-CC harboring an additional post-translational modification (see text). ns, non-specific bands.
(J) SV40 WT MEFs treated with or without IR (10 Gy) or Gö6976 (1 μM) were additionally treated with ATM inhibitor KU55933 or DNA-PKcs inhibitor NU7026 (10 μM each) at 8 hpIR. Cells were harvested at 24 hpIR and analyzed by western blot.
(K) HeLa cells transfected with CHK1 siRNA (+) or LACZ siRNA (-) were treated with or without IR (10 Gy) or Gö6976 (1 μM) at 16 hr post-transfection. Cells were harvested at 16 hpIR and analyzed by western blot.
(L) HeLa cells stably expressing the indicated shRNAs were transfected with the indicated siRNAs and treated with or without Gö6976 (1 μM) or IR (10 Gy) 36 hr post-transfection. Cells were harvested at the indicated time points after IR and analyzed by western blot.
(M) SV40 WT MEFs treated with IR (10 Gy) and Gö6976 (1 μM) were exposed to ATM inhibitor KU55933 added at (‘@’) the indicated time points after stimulus. Cells were harvested at 24 hr after IR+Gö6976 treatment and analyzed by western blot.
PIDD interacts with ATM through the LRR domain
The fact that PIDD acts downstream of ATM and ATR in the CS pathway (Figure 2C) suggested that ATM and/or ATR might engage PIDD directly, plausibly through phosphorylation. This might in turn serve as the trigger for the PIDD-RAIDD interaction shown in Figure 3.
We first asked whether ATM and ATR are PIDD interactors. Flag-PIDD immunoprecipitated from transiently transfected HeLa cells was found to associate with ATM, but not ATR (Figure 4A). Reciprocally, Flag-ATM interacted with PIDD-FL, but not FADD (Figure 4B). As shown in Figure 4C, ATM bound PIDD-N, but not PIDD-C or PIDD-CC. A construct comprising solely the LRR domain interacted with ATM with similar affinity as PIDD-N (Figure 4C). These results indicated that ATM binds PIDD on the LRR domain.
At the endogenous level, ATM barely interacted with PIDD in untreated cells. By contrast, initiating the CS pathway via IR+Chk1i triggered a robust interaction between both molecules (Figure 4D). Therefore, ATM recruits PIDD during CS apoptosis, correlating with PIDDosome assembly. Intriguingly, ATM also interacted with PIDD after IR alone (Figure S4A). This contrasts with the PIDD-RAIDD interaction, which occurs specifically after IR+Chk1i (Figure 3A,B). Therefore, Chk1 must antagonize PIDDosome assembly downstream of the ATM-PIDD interaction but upstream of the PIDD-RAIDD interaction. This positions Chk1 antagonism of CS apoptosis at the level of the PIDD platform (see Discussion).
ATM phosphorylates PIDD on T788 within the DD
The interaction between ATM and PIDD suggested that ATM might play a direct role in PIDDosome assembly during CS apoptosis. A Scansite (http://scansite.mit.edu) analysis of PIDD-FL identified three candidate ATM phosphorylation sites: SQ(202/203), located in the fourth LRR; SQ(521/522), located in the distal ZU-5 domain; and TQ(788/789), located in the DD (Figure 4E). Because the DD, but not the LRRs or ZU-5 domains, provides the docking site for RAIDD (Tinel and Tschopp, 2004), we focused on T788.
Sequence alignments revealed that the TQ(788/789) motif is conserved in vertebrates (Figure 4E). In addition, Flag-PIDD immunoprecipitated from cells undergoing CS apoptosis, but not control cells, was reactive to antibodies raised against phosphorylated ATM/ATR substrate peptides (phospho-SQ/TQ antibodies). The RAIDD-interacting PIDD-CC fragment, which contains TQ(788/789) but not SQ(202/203) or SQ(521/522), became reactive to phospho-SQ/TQ antibodies shortly after IR+Chk1i (Figure 4F). These observations indicated that PIDD might indeed be an ATM substrate.
To test this directly, we first performed kinase assays. A recombinant GST-PIDD-CC substrate incubated with wild-type ATM, but not kinase-dead ATM, showed a marked increase in phospho-SQ/TQ immunoreactivity (Figure 4G). This increase was dependent on an intact T788 residue, because a nonphosphorylatable GST-PIDD-CCT788A substrate was significantly less phosphorylated than wild-type GST-PIDD-CC (Figure 4H). These results showed that ATM can phosphorylate PIDD on T788 in vitro.
To test whether ATM phosphorylates endogenous PIDD during CS apoptosis, we generated an antibody against PIDD phosphorylated on T788 [α-pPIDD (pT788); see Supplemental Experimental Procedures]. The target epitope, ETGFLpT788QSNLL, is identical in humans and mice (Figure 4F). This enabled us to demonstrate the specificity of the antibody in both shPIDD cells and Pidd−/− MEFs (see Figures 4I,L below). In WT MEFs treated with IR+Chk1i, the antibody detected phospho-PIDD-CC (p-CC) as a band running ~1-2 kDa above native PIDD-CC (38/39 kDa vs. 37 kDa, respectively), whose intensity peaked at 24 hours post stimulus, and was absent in Pidd−/− lysates (Figure 4I). The antibody did not recognize native PIDD-CC but detected an additional, weaker band running ~5kDa above p-CC (Figures 4I and S4B). Because this band was also undetectable in Pidd−/− lysates (Figure 4I), it likely reflects a phosphorylation-dependent secondary modification of PIDD-CC.
p-CC was first detectable 8-16 hours after IR+Chk1i and peaked at 24 hours (lanes 1-4 in Figure 4I; Figures S3A and S4B), correlating with the kinetics of caspase-2 activation during CS apoptosis (Figures S3B-C, 4K, and S4B). In contrast, p-CC was barely detectable after IR or Chk1i alone [Figures 4J (lanes 1-4), 4K and S3A), or during heat shock-induced caspase-2 activation (Figure S1C). These observations indicate that PIDD phosphorylation is specific to CS apoptosis. Most importantly, inhibition or depletion of ATM, but not ATR or DNA-PK, blocked endogenous PIDD phosphorylation during CS apoptosis in both MEFs (Figure 4J,M) and HeLa cells (Figure 4L). In summary, ATM: (i) interacts with PIDD via the LRR domain; (ii) phosphorylates PIDD directly on T788 in vitro; and (iii) is required for phosphorylation of this residue in live cells treated with IR+Chk1i. These observations show that ATM phosphorylates PIDD on T788 during CS apoptosis.
T788 phosphorylation triggers RAIDD recruitment and caspase-2 activation
We next sought to determine the functional consequences of PIDD phosphorylation by ATM. As shown earlier (Figure 1F and 2C), PIDD-depleted cells fail to cleave caspase-2 after IR+Chk1i (Figure 5A, compare lanes 2 and 3). Introduction of a shRNA-resistant PIDDWT construct into this line was sufficient to restore caspase-2 processing (compare lanes 3 and 4). In contrast, a shRNA-resistant but nonphosphorylatable PIDDT788A construct failed to rescue caspase-2 processing in shPIDD cells (compare lanes 3, 4 and 5). This was not due to indirect effects of the T788A mutation on PIDD stability or maturation, because the PIDDWT and PIDDT788A constructs generate equivalent amounts of the FL, C, and CC fragments in transfected cells (Figure 5A). These results therefore indicate that T788 phosphorylation is specifically required for caspase-2 activation in the CS pathway.
We then investigated the mechanism by which PIDD phosphorylation leads to caspase-2 activation. Because T788 lies within the PIDD DD, which serves as the docking site for RAIDD (Tinel and Tschopp, 2004), we analyzed the effects of the T788A mutation on the PIDD-RAIDD interaction. Whereas PIDDWT robustly interacted with endogenous RAIDD in HEK293T and HeLa cells, RAIDD recruitment was abolished by the T788A mutation (Figures 5B and 6A). As a consequence, PIDDT788A shows a complete loss of function for caspase-2 activation, as assessed by immunoblotting (Figure 5B,C) and caspase-2 bifluorescence complementation (BiFC) microscopy (Figure 5D-F; see legend for details) (Bouchier-Hayes et al., 2009). The inability of PIDDT788A to recruit RAIDD or activate caspase-2 did not result from indirect effects of the mutation on PIDD maturation (Figure 5A-C), nor was it caused by a general alteration of the DD that would globally affect its ability to interact with other proteins (see RIP1 interaction data below). Increasing doses of transfected PIDDT788A failed to induce even marginal levels of caspase-2 BiFC, showing that the mutant behaves as a true function-null, as opposed to hypomorphic, allele (Figure 5F).
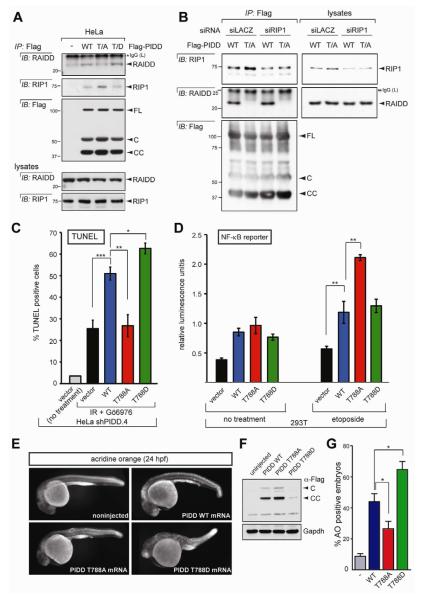
Figure 6. T788 phosphorylation determines RAIDD versus RIP1 selection and fate decision by PIDD.
(A) HeLa cells transfected with the indicated C-terminally Flag-tagged PIDD constructs were harvested at 24 hr post transfection. Protein extracts were immunoprecipitated with anti-Flag antibodies and analyzed by western blot.
(B) HeLa cells co-transfected with the indicated siRNAs and C-terminally Flag-tagged PIDD constructs were harvested at 36 hr post siRNA transfection. Protein extracts were
immunoprecipitated with anti-Flag antibodies and analyzed by western blot. Asterisk indicates IgG light chain.
(C) shPIDD.4 HeLa cells transfected with the indicated PIDD constructs were treated with or without 10 Gy IR and Gö6976 (1 μM) and analyzed by TUNEL staining at 48 hr post IR. Data are means +/− SEM. *p < 0.05; **p < 0.01; ***p < 0.001, two-tailed Student’s t-test.
(D) HEK293T cells transiently transfected with the indicated PIDD constructs were treated with or without etoposide (40 μM) and analyzed with a NF-κB reporter. **p < 0.01, two-tailed Student’s t-test.
(E) One-cell stage wild-type zebrafish embryos were injected with the indicated synthetic Flag-PIDD mRNAs (100 pg each), stained with the cell-death marker acridine orange at 24 hr post-fertilization (hpf), and imaged with a fluorescence microscope. Representative images are shown, anterior to the left.
(F) Whole-body protein extracts from 24-hpf embryos as in (E) were analyzed by western blot.
(G) Quantification of acridine-orange stains such as in (E). Data collected from six independent experiments (at least 30 embryos per construct in each) are represented as means +/− SEM. *p < 0.05, two-tailed Student’s t-test.
In contrast with nonphosphorylatable PIDD, a phosphomimetic variant, PIDDT788D, showed greater affinity for RAIDD compared to PIDDWT (Figure 6A), as well as an enhanced ability to activate caspase-2 (Figures 5C and S5, compare lanes 2 and 4 in each panel; see also BiFC data in Figure 5D-F). Notably, this hypermorphic activity of PIDDT788D in unstimulated cells matched that produced by PIDDWT in cells undergoing CS apoptosis (Figure 5C, compare lanes 4 and 6), a context where T788 is physiologically phosphorylated by ATM (Figure 4J-N). Altogether, these results indicate that phosphorylation of PIDD on T788 is both necessary and sufficient for PIDDosome assembly and caspase-2 activation in the CS pathway.
T788 phosphorylation status determines RAIDD versus RIP1 recruitment and fate choice by PIDD
The PIDD DD not only interacts with RAIDD but can also bind prosurvival RIP1 after DNA damage (Janssens et al., 2005). However, the mechanism by which PIDD discriminates between RAIDD and RIP1 has remained unclear. We investigated the role of T788 phosphorylation in this process. Whereas the T788D phosphomimetic improved the affinity of PIDD for RAIDD, it impaired RIP1 binding (Figure 6A). In contrast, while the T788A mutation eliminated the ability of PIDD to recruit RAIDD, it stimulated the recruitment of RIP1 (Figures 5B and 6A). The inability of PIDDT788A to interact with RAIDD was not due to a failure to dissociate from RIP1, because removing RIP1 via RNAi failed to restore RAIDD recruitment to PIDDT788A (Figure 6B). None of the mutations affected the stability of PIDD-C (RIP1-recruiting PIDD fragment) or PIDD-CC (RAIDD-recruiting fragment). Collectively, these data indicate that the choice of RAIDD or RIP1 by PIDD is determined by the phosphorylation status of T788 in the docking site.
If T788 phosphorylation determines the selection of proapoptotic RAIDD versus prosurvival RIP1 at the PIDD DD, then T788 phosphorylation should also impact fate decisions by PIDD. Consistent with its decreased affinity for RAIDD but increased affinity for RIP1, non-phosphorylatable PIDDT788A was incapable of inducing apoptosis in HeLa cells or live zebrafish embryos (Figure 6C,E-G), but showed a markedly enhanced ability to stimulate etoposide-induced NF-κB signaling (~two-fold increase compared to WT PIDD, P<0.01; Figure 6D). Conversely, the PIDDT788D phosphomimetic, which is hypermorphic for RAIDD binding but hypomorphic for RIP1 binding, failed to stimulate NF-κB signaling (Figure 6D) but showed enhanced apoptosis-inducing capacity compared to WT, both in vitro (25-30% increase, P=0.0256; Figure 6C), and in vivo (~50% increase, P<0.05; Figure 6E-G). Collectively, these experiments indicate that T788 phosphorylation is a central determinant of cell fate specification by PIDD.
Discussion
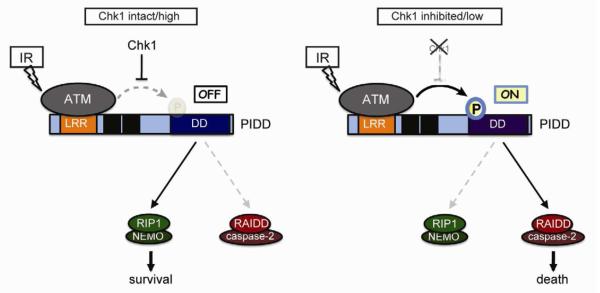
Here we show that the DNA damage response kinase, ATM, controls a phosphorylation switch within the PIDD death domain which, in the OFF position, dictates recruitment of prosurvival RIP1, and in the ON position, triggers RAIDD binding, caspase-2 activation, and apoptotic cell death (Figure 7). This phosphorylation switch is regulated by the Chk1 kinase, a core effector of cell cycle arrest and DNA repair after DNA injury. Thus, PIDD encodes a genome surveillance platform that integrates both DNA integrity (via ATM) and genome-maintenance potential (via Chk1) to decide cell survival or death in the face of genotoxic stress (Figure 7).
Figure 7. Mechanism of life vs. death decisions at the PIDD docking site.
See text for details.
Assigning the PIDDosome to a defined apoptotic pathway
Recent efforts to elucidate the PIDDosome‘s physiologic significance have yielded contradictory results (Baptiste-Okoh et al., 2008; Berube et al., 2005; Kim et al., 2009; Lin et al., 2000; Manzl et al., 2009; Manzl et al., 2012; Niizuma et al., 2008; Read et al., 2002; Ribe et al., 2012; Vakifahmetoglu et al., 2006). Here, we identify the CS pathway as an apoptotic context in which the PIDDosome both assembles and is required for caspase-2 activation after genotoxic stress. Surprisingly, whereas IR is a potent trigger of PIDDosome assembly in the absence of Chk1, such lesions are by themselves poor inducers of complex formation (Figure 3A). This may explain why caspase-2 is only marginally required for apoptosis induction in HeLa cells or MEFs exposed to IR alone (Ho et al., 2009; Sidi et al., 2008), and why neither PIDD, RAIDD, nor caspase-2 are required for IR-induced apoptosis in mouse thymocytes (Kim et al., 2009; Manzl et al., 2009). Altogether, it appears that a cell’s engagement of the PIDDosome after DNA damage is tightly controlled by Chk1 signaling, which acts to restrain complex assembly.
Mechanism of PIDD switch function
PIDD has been implicated as a cell-fate switch that integrates DNA damage signals and orchestrates an appropriate, survival or death response. PIDD switch activity centers on its ability to interact with a prosurvival factor, RIP1, or a prodeath molecule, RAIDD, both of which compete for access to the PIDD DD (Janssens et al., 2005; Tinel and Tschopp, 2004). However, how PIDD senses DNA injury and in turn properly discriminates between RIP1 and RAIDD had remained unclear.
Tinel et al. proposed that DNA damage impacts on the autoproteolytic cleavage events that generate the RIP1- and RAIDD-interacting forms of PIDD (Cuenin et al., 2008; Tinel et al., 2007). However, we could not detect any striking accumulation of PIDD-CC in cells undergoing CS apoptosis. Alternatively, a DNA-damage signaling molecule might directly regulate binding-partner selection at the PIDD DD (Janssens et al., 2005). Here, we identify this molecule as the ATM kinase, a core sensor and transducer of DNA double-strand breaks (Ciccia and Elledge, 2010; Lowe et al., 2004). ATM phosphorylates PIDD on T788 within the DD, an event that appears sufficient to decide between RAIDD or RIP1 recruitment. T788 is located in the N-terminal half of helix 1 in the PIDD DD, which comprises several residues involved in homotypic interactions that help build the PIDDosome core complex (Park et al., 2007). Phosphorylation of T788 by ATM might induce a conformational change that exposes these residues at the DD:DD interfaces. Alternatively, T788 phosphorylation might open up the N-terminal tail of PIDD-CC to unmask the PIDD DD, a possibility that awaits structural resolution of full-length PIDD-CC (H. Wu, personal communication).
Refining the mechanism by which Chk1 suppresses CS apoptosis
We previously proposed that Chk1 antagonizes the CS pathway via a negative feedback loop that targets ATM/ATR. This was based on the fact that IR activates ATM and ATR to a much lesser extent than IR+Chk1i in HeLa cells (Sidi et al., 2008). However, our present observations in MEFs challenge this model, as we find that IR activates ATM as efficiently as IR+Chk1i in these cells (pChk2 blot in Figure 4J, compare lanes 3 and 4). Why then does ATM phosphorylate PIDD only when Chk1 is inhibited?
Given the data, Chk1 must act downstream of active ATM but upstream of PIDD phosphorylation. Because ATM binds the PIDD LRRs after IR regardless of Chk1 activity (Figures 4D and S4A), Chk1 must block PIDD phosphorylation by ATM following substrate recognition by the kinase (Figure 7). Chk1 might directly phosphorylate PIDD to induce a conformational change that, while compatible with ATM binding, prevents ATM from phosphorylating T788. Alternatively, a Chk1 substrate might interact with PIDD or ATM to interfere with T788 phosphorylation.
Significance of PIDD switch activity
Our data suggest that PIDD integrates signals from the DNA damage-sensing and checkpoint-surveillance machineries (via ATM and Chk1, respectively) to decide between cellular life and death (Figure 7). The death decision, which is favored by ATM, only takes place if Chk1 signaling is minimal, as occurs during checkpoint termination. This suggests that the PIDDosome acts to eliminate damaged cells that have violated checkpoint surveillance. The PIDDosome might therefore constitute a potent tumor-suppressive device. Consistent with this notion, caspase-2 has recently emerged as a tumor suppressor in mice (Ho et al., 2009; Ren et al., 2012), and it has been suggested that apoptosis through the CS pathway may contribute to this function (Kumar, 2009). While lymphoma suppression by caspase-2 does not depend on PIDD (Manzl et al., 2012), other tumor types may be suppressed by PIDDosome signaling, likely as a function of ATM and Chk1 levels, and extent of genomic instability. Future studies should help further clarify the biologic contexts in which cells engage the PIDDosome, rather than other caspase-2 activation pathways, to trigger apoptosis. Our identification of the CS pathway as a tractable setting for PIDDosome signaling provides a molecular entry point into PIDDosome biology after DNA damage.
Experimental Procedures
Cell Culture and Reagents
HeLa, HEK293T, and TP53—/— HCT116 cells were cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS). Caspase2—/—, Raidd—/—, and Pidd—/— SV40- or E1A/Ras-transformed MEFs, kindly provided by Andreas Villunger and Douglas Green, were cultured as described previously (Manzl et al., 2009). Unless otherwise specified, cells seeded in 6 cm dishes were grown to 50-80% confluence for treatment with DMSO or Gö6976 (1 μM final; Calbiochem) one hour before IR (0 or 10 Gy, Gammacell 1000 137Cs -irradiator) or etoposide treatment (40 μM, Enzo Life Sciences). KU-55933 and NU7026 were from TOCRIS.
RNAi and DNA transfection
Lentiviral shRNA transduction, and siRNA and DNA transfections were essentially performed as previously described (Sidi et al., 2008). See Supplemental Experimental for further details and siRNA and shRNA sequences.
TUNEL and NF-κB Reporter Assays
TUNEL assays were performed using the APO-BRDU kit (BD Biosciences, Franklin Lakes, NJ, USA) as described previously (Sidi et al., 2008). NF-κB reporter assays were performed with the Cignal Reporter kit (QIAGEN) according to the manufacturer’s instructions.
Size Exclusion Chromatography (Gel Filtration)
SEC was performed in a DuoFlow BioLogic System according to the manufacturer’s manual (BioRad). Cells were washed twice with PBS and resuspended in hypotonic buffer (20 mM Hepes-KOH, 10 mM KCl, 1 mM MgCl2, 1mM EDTA, 1 mM EGTA, 1 mM DTT, pH 7.5) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (PhosSTOP, Roche). Resuspended cells were subjected to three rounds of freeze thawing in liquid nitrogen. Debris was removed by centrifugation at 10,000xg for 20 min at 4 °C, followed by filtration at 0.2 m. The column was equilibrated with gel filtration buffer (150 mM NaCl, 20 mM Hepes-KOH, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, pH 7.5). Whole cell lysates (5 mg) were applied to a S400 (HiPrep 16/60 Sephacryl) gel filtration column (Amersham Biosciences). Samples were eluted at 1 ml/min and monitored with an online detector at 280 nm.
In Vitro Kinase Assays
HEK293T cells were transfected with pcDNA3-FLAG-ATM plasmids using X-tremeGENE HP (Roche). 48 hr post transfection, cells were extracted in 0.1% NP-40 lysis buffer (0.1% NP-40, 50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 5mM EDTA) supplemented with protease and phosphatase inhibitors (Roche). Lysates (~10 mg) were immunoprecipitated with 20 μl protein A beads (Bio-Rad) coupled to anti-FLAG M2 antibody (Sigma). Recombinant GST-tagged PIDD-CC was expressed in DH5α (Invitrogen) and purified using glutathione beads (GE Healthcare). Flag-ATM and GST-PIDD-CC beads were mixed together and resuspended in kinase reaction buffer with 200 μM ATP (Cell Signaling) for 30 min at 30°C.
BiFC Assay and Confocal Microscopy
5 × 104 Hela cells were plated on dishes containing coverslips (Mattek) coated with fibronectin (1mg/ml; Millipore) 24 hr before transfection. Cells were transfected with pBiFC.C2 CARD-VC155, pBiFC.C2 CARD-VN173, dsRedmito (10 ng; Clontech), and PIDD plasmids using lipofectamine 2000 (Invitrogen). qVD-OPH (5 M; MP Biomedicals) was included to prevent cell detachment upon apoptosis. 24 hr post transfection, cells were analyzed by fluorescence microscopy. Red cells were counted and the percentage of Venus+ cells against total transfected cells was calculated from a minimum of 300 cells per well. Images were taken with a spinning disk confocal microscope (Zeiss) with a 20X objective and analyzed with ImageJ software (see Supplemental Experimental Procedures for details).
mRNA Injections and Acridine Orange Labeling in Zebrafish Embryos
Synthetic mRNAs were in vitro transcribed from pCSDest-subcloned PIDD templates using the Ambion mMESSAGE mMACHINE kit and injected (100 ng/μl) into one-cell stage embryos as previously described (Sidi et al., 2008). 24 hr embryos were stained with acridine orange (10 μg/ml final) and lyzed for western blot analysis as previously described (Sidi et al., 2008), and imaged with a Nikon SMZ 1500 fluorescence microscope.
Statistics
Statistical significance was analyzed by two-tailed Student’s t tests. Data are represented as mean ± SEM.
Supplementary Material
Highlights.
The PIDDosome is physiologically required for ‘Chk1-suppressed’ apoptosis
PIDD is an ATM substrate; phosphorylation occurs on T788 within the death domain
T788 phosphorylation is necessary and sufficient for RAIDD recruitment
T788 status dictates RIP1 vs RAIDD binding, coupling DNA damage to fate decision
Acknowledgements
We gratefully acknowledge Jennifer Pascual, Alexander Mir and Junjun Ding for technical assistance, Andreas Villunger, Douglas Green, William Hahn, Michael Kastan and Bert Vogelstein for reagents, John R. Gilbert for editorial review, and Hao Wu, Patricia Vinciguerra and Antoine Tinel for helpful discussions. This work was supported by NIH grants HL88664 (S.S, A.T.L), GM095942 (J.W), and awards from the Claudia Adams Barr Foundation, Tisch Cancer Institute, JJR Foundation, and Searle Scholars Program to S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data Supplemental Data include Supplemental Experimental Procedures and five figures.
References
- Baptiste-Okoh N, Barsotti AM, Prives C. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proc Natl Acad Sci U S A. 2008;105:1937–1942. doi: 10.1073/pnas.0711800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, Yeh WC, Mak TW, Benchimol S. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci U S A. 2005;102:14314–14320. doi: 10.1073/pnas.0506475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchier-Hayes L, Oberst A, McStay GP, Connell S, Tait SW, Dillon CP, Flanagan JM, Beere HM, Green DR. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell. 2009;35:830–840. doi: 10.1016/j.molcel.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenin S, Tinel A, Janssens S, Tschopp J. p53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-kappaB and caspase-2 in response to genotoxic stress. Oncogene. 2008;27:387–396. doi: 10.1038/sj.onc.1210635. [DOI] [PubMed] [Google Scholar]
- Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci U S A. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Browne G, Melino G, Cohen GM. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ. 2009;16:1053–1061. doi: 10.1038/cdd.2009.29. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Kim IR, Murakami K, Chen NJ, Saibil SD, Matysiak-Zablocki E, Elford AR, Bonnard M, Benchimol S, Jurisicova A, Yeh WC, et al. DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis. 2009;14:1039–1049. doi: 10.1007/s10495-009-0375-1. [DOI] [PubMed] [Google Scholar]
- Kitevska T, Spencer DM, Hawkins CJ. Caspase-2: controversial killer or checkpoint controller? Apoptosis. 2009;14:829–848. doi: 10.1007/s10495-009-0365-3. [DOI] [PubMed] [Google Scholar]
- Kumar S. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more? Nat Rev Cancer. 2009;9:897–903. doi: 10.1038/nrc2745. [DOI] [PubMed] [Google Scholar]
- Lavrik IN, Golks A, Baumann S, Krammer PH. Caspase-2 is activated at the CD95 death-inducing signaling complex in the course of CD95-induced apoptosis. Blood. 2006;108:559–565. doi: 10.1182/blood-2005-07-007096. [DOI] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- Logette E, Schuepbach-Mallepell S, Eckert MJ, Leo XH, Jaccard B, Manzl C, Tardivel A, Villunger A, Quadroni M, Gaide O, et al. PIDD orchestrates translesion DNA synthesis in response to UV irradiation. Cell Death Differ. 2011;18:1036–1045. doi: 10.1038/cdd.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, Logette E, Tschopp J, Villunger A. Caspase-2 activation in the absence of PIDDosome formation. The Journal of cell biology. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Peintner L, Krumschnabel G, Bock F, Labi V, Drach M, Newbold A, Johnstone R, Villunger A. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ. 2012 doi: 10.1038/cdd.2012.54. doi: 10.1038/cdd.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niizuma K, Endo H, Nito C, Myer DJ, Kim GS, Chan PH. The PIDDosome mediates delayed death of hippocampal CA1 neurons after transient global cerebral ischemia in rats. Proc Natl Acad Sci U S A. 2008;105:16368–16373. doi: 10.1073/pnas.0806222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Vakifahmetoglu H, Abruzzo PM, Hogstrand K, Grandien A, Zhivotovsky B. DISC-mediated activation of caspase-2 in DNA damage-induced apoptosis. Oncogene. 2009;28:1949–1959. doi: 10.1038/onc.2009.36. [DOI] [PubMed] [Google Scholar]
- Pan Y, Ren KH, He HW, Shao RG. Knockdown of Chk1 sensitizes human colon carcinoma HCT116 cells in a p53-dependent manner to lidamycin through abrogation of a G2/M checkpoint and induction of apoptosis. Cancer Biol Ther. 2009;8:1559–1566. doi: 10.4161/cbt.8.16.8955. [DOI] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. The Journal of cell biology. 2002;159:739–745. doi: 10.1083/jcb.200209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren K, Lu J, Porollo A, Du C. Tumor-suppressing Function of Caspase-2 Requires Catalytic Site Cys-320 and Site Ser-139 in Mice. J Biol Chem. 2012;287:14792–14802. doi: 10.1074/jbc.M112.347625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribe EM, Jean YY, Goldstein RL, Manzl C, Stefanis L, Villunger A, Troy CM. Neuronal caspase 2 activity and function requires RAIDD, but not PIDD. Biochem J. 2012;444:591–599. doi: 10.1042/BJ20111588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telliez JB, Bean KM, Lin LL. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta. 2000;1478:280–288. doi: 10.1016/s0167-4838(00)00029-7. [DOI] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. Embo J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Tu S, McStay GP, Boucher LM, Mak T, Beere HM, Green DR. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat Cell Biol. 2006;8:72–77. doi: 10.1038/ncb1340. [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene. 2006;25:5683–5692. doi: 10.1038/sj.onc.1209569. [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu-Norberg H, Zhivotovsky B. The unpredictable caspase-2: what can it do? Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Mabb A, Miyamoto S. PIDD: a switch hitter. Cell. 2005;123:980–982. doi: 10.1016/j.cell.2005.11.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.