Abstract
West Nile virus (WNV) is an arthropod-borne virus with a world wide distribution that causes neurologic disease and death. Autophagy is a cellular homeostatic mechanism involved in antiviral responses but can be subverted to support viral growth as well. We show that autophagy is induced by WNV infection in cell culture and in primary neuron cultures. Following WNV infection, lysosomes co-localize with autophagosomes resulting in LC3B-II turnover and autolysosomal acidification. However, activation or inhibition of autophagy has no significant effect on WNV growth but pharmacologic inhibition of PI3 kinases associated with autophagy reduce WNV growth. Basal levels of p62/sequestesome1(SQSTM1) do not significantly change following WNV-induced autophagy activation, but p62 is turned over or degraded by autophagy activation implying that p62 expression is increased following WNV-infection. These data show that WNV-induces autophagy but viral growth is independent of autophagy activation suggesting that WNV-specific interactions with autophagy have diverged from other flaviviruses.
Keywords: Flavivirus, West Nile virus, autophagy, neuron, viral encephalitis, p62
Introduction
West Nile virus (WNV) is a single-stranded, positive-sense RNA virus in the family Flaviviridae. First isolated in the West Nile province of Uganda in 1937, WNV is maintained in an enzootic cycle between birds and mosquitoes and has emerged as a significant cause of neurologic infectious disease on six continents. Since its emergence in New York City in 1999, WNV has caused significant morbidity and mortality in the United States with greater than 30,000 cases reported and over 1200 deaths (www.cdc.gov/westnile). A member of the Japanese encephalitis serocomplex, WNV shares significant genetic identity with a number of other medically important flaviviruses including St. Louis encephalitis virus, Murray Valley encephalitis virus, and Japanese encephalitis virus. To date, there is no specific therapy of proven benefit for WNV infection highlighting the need to understand WNV molecular pathogenesis and identify potential new targets for therapy.
Macroautophagy (referred to as ‘autophagy’ hereafter) is an evolutionarily conserved process by which undesired intracellular components are sequestered into double membrane vesicles and delivered to lysosomes for degradation and recycling to maintain cellular homeostasis (Mizushima, 2010). Autophagy is vital for multiple fundamental cellular processes including response to starvation, development, aging, neurodegenerative diseases, cancer, and regulation of innate and adaptive immunity (Deretic, 2010; Levine and Kroemer, 2008; Mizushima, 2010). Autophagy is highly conserved in eukaryotic cells and has undergone significant co-evolution with intracellular pathogens such as bacteria, mycobacteria, and viruses (Deretic, 2010; Levine et al., 2011).
Autophagy can function as a host cell defense mechanism or can be subverted for the benefit of the invading pathogen. In experimental models of viral encephalitis using Sindbis virus and Herpes simplex type-1 virus (HSV), autophagy acts as an antiviral response in neurons. In a mouse model of Sindbis virus encephalitis, autophagy activation protected neurons from infection, and inhibition of autophagy increased cell death and mortality (Liang et al., 1998; Orvedahl et al., 2010). Similarly, inhibition of autophagy by the HSV-1 neurovirulence factor ICP34.5 allowed HSV-1 to evade host immunity and cause fatal encephalitis in a mouse model. Suppression of ICP34.5 expression attenuated the neuroinvasiveness of HSV (Orvedahl et al., 2007).
While autophagy functions as an antiviral response for some viruses, others viral infections subvert autophagy machinery to support viral growth. Members of the family Picornaviridae, such as poliovirus and coxsackieviruses, exhibit significantly reduced viral replication when autophagy is inhibited (Jackson et al., 2005; Taylor and Kirkegaard, 2008; Yoon et al., 2008). Poliovirus utilizes autophagosome lipid membrane structures to support assembly of replication complexes and uses autophagy-related double-membrane vesicles to deliver cytoplasmic virions to the extracellular milieu (Taylor and Kirkegaard, 2008). Viruses in the family Flaviviridae such as Dengue and hepatitis C virus (HCV) subvert autophagy to support viral replication (Dreux et al., 2009; Heaton and Randall, 2010; Lee et al., 2008; Li et al., 2011; McLean et al., 2011). Flavivirus NS4A from Dengue-2 and Modoc virus was associated with activation of autophagy which supported viral replication and prevented epithelial cell death (McLean et al., 2011). Additionally, Japanese encephalitis virus growth is dependent on autophagy activation during early stages of infection (Li et al., 2011).
Autophagy is regulated by a series of complex cellular signaling events that include the activation of the class III PI3 kinase, hVps34, in association with Beclin1 and inactivation of mammalian target of rapamycin (mTOR). These signaling pathways promote autophagy by activating hVps34-induced formation of phosphatidylinositol 3-phosphate (PI3) on lipids resulting in the formation of isolation membranes and early steps to initiate autophagosome formation (Kudchodkar and Levine, 2009). Over 30 autophagy (Atg) genes control autophagosome initiation, formation and completion. After initiation of the autophagosome, vesicle elongation and completion are mediated by ubiquitin-like conjugation systems. Atg7, an E1-like enzyme, and atg10, an E2-like enzyme, promote covalent association of Atg12 and Atg5 along with Atg16 to form a heterotrimeric complex that mediates growth of the autophagosome (Kudchodkar and Levine, 2009; Xie and Klionsky, 2007). A second ubiquitin-like conjugation system that includes Atg7 and Atg4 cleaves a mammalian homolog of yeast Atg8, microtubule-associated protein, light chain 3 (LC3). After cleavage, the E2-like enzyme Atg3 adds phosphatidylethanolamine (PE) to the C-terminus of the cleaved LC3 to create a species known as LC3-II (Kudchodkar and Levine, 2009). LC3 is soluble and dispersed in the cytoplasm but after cleavage and lipidation with PE, LC3-II associates with the outer and inner membranes of the autophagosome and is commonly used as a marker for autophagosomes (Klionsky et al., 2008). Once the autophagosome is completed, it docks and fuses with lysosomes to form autolysosomes resulting in degradation of the inner autophagosome contents (Klionsky et al., 2008).
The mechanisms required to target cytoplasmic cargo to the autophagosome are just now beginning to be characterized. The cellular protein p62/sequestosome1(SQSTM1) can function as an adaptor to target ubiquinated protein aggregates to the autophagosome by binding both ubiquitin and LC3 (Johansen and Lamark, 2011). Following binding of p62 to LC3-II, the autophagosome completes and fuses with lysosomes resulting in degradation of both p62 and LC3-II. Thus p62 degradation can be used as another marker of autophagy flux (Klionsky et al., 2008).
We used models of WNV-infection in cell culture, primary neuron cultures, and ex vivo organotypic brain slice cultures (BSC) to demonstrate that WNV infection activates autophagy in a wide range of cell types, but WNV growth occurs independent of autophagy activation. We also show that WNV induces LC3BII turnover but steady state levels of p62/SQSTM1 are maintained during infection. To our knowledge, this is the first report to show that WNV activates autophagy and that steady state levels of p62/SQSTM1 are not altered by virus-induced autophagy. We extend these data and show that autophagy has no significant effect on WNV growth, which distinguishes WNV from Dengue and JEV.
Results
West Nile virus infection induces LC3B lipidation
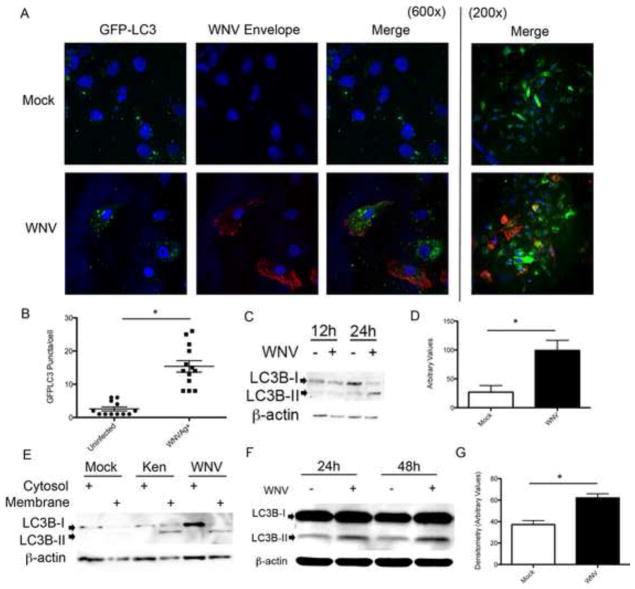
We first evaluated the formation of microtubule-associated protein 1 light chain 3B (LC3B) lipidation products formed by phosphatidylethanolamine (PE) conjugation following WNV infection. Vero cells were inoculated with WNV (MOI 3) following transfection with a green fluorescent protein (GFP)-LC3 (Cell Biolabs, CBA401) expressing plasmid as previously described (Mizushima et al., 2004). At 24 hours post-inoculation with WNV NY99, cells were fixed, labeled for WNV envelope antigen (ATCC; cy3-red), and DAPI stained for nuclei (Fig. 1A). LC3-positive puncta in antigen positive cells transfected with GFP-LC3 were counted. Based on cell counts, 73% of cells were transfected with the GFP-LC3 label. Puncta counts revealed a significant increase in GFP-LC3 puncta in WNV-infected Vero cells (16.4 puncta/cell ±1.7) compared to uninfected infected cells (2.6 puncta/cell ± 0.6, p<0.0001; Fig. 1B). We verified our imaging data by inoculating Vero cells with WNV (MOI 3), harvesting cells for whole cell lysate at 24 hour post-infection, and performing western blot analysis using antibodies specific for LC3B (Fig. 1C). Four replicate experiments demonstrated an increase in the lipidated LC3B product (LC3B-II) compared to mock-infected cells and densitometry revealed a significant increase in band density of the LC3B-II lipidation product in WNV-infected cells (Fig. 1D, p=0.018). Cytosolic expression of LC3B-I was variable throughout our studies and not significantly different between treatment groups. To verify that LC3B-II was lipidated and inserted into lipid membranes following WNV infection, Vero cells were infected with two strains of WNV [clone-derived NY99 isolate of WNV (WNV) or a clone-derived WNV isolate from Kenya (Ken)] at a MOI of 3 and harvested at 24 hours post-infection. The Kenyan isolate was used to evaluate a WNV isolate previously shown to have decreased virulence in birds compared to the NY99 strain (Brault et al., 2007). Cells were harvested and cytoplasmic and membrane fractions were analyzed by western blot. Three replicate experiments demonstrated that LC3B-II was lipidated and enriched in the membrane fraction following infection with both clone-derived WNV strains (Fig. 1E). In a model of ex vivo brain infection with WNV using organotypic brain slice cultures (BSCs) prepared as previously described (Dionne et al., 2011a), we found increased LC3B-II in WNV-infected BSCs at 24 and 48 hours post-infection (Fig. 1F). Densitometry values of the LC3B-II band corrected for β–actin in 4 replicate experiments show a significant increase in LC3B-II band density in WNV-infected BSCs compared to mock infected BSCs (*p=0.005, Fig. 1G). BSCs are a mixed population of cells that contain populations of neurons labeled green (FITC) with an antibody to NeuN and support WNV growth similar to levels seen in primary mouse cortical cultures (MCC) (Supp. Fig. A–D).
Figure 1.
LC3B is lipidated following WNV Infection. Vero cells were transfected with GFP-LC3 expression plasmid followed by A) mock or WNV (MOI 3) inoculation, harvested at 24 hours post-infection, and labeled with antibody to WNV envelope antigen (cy3:red) and nuclei stained with DAPI. Images shown at 600x and 200x magnification. B) GFP-LC3 positive puncta were counted in WNV antigen negative (uninfected) and antigen positive cells (WNVAg+, *p<0.0001). C) Vero cells were mock or WNV (MOI 3) inoculated and cells were harvested at the specified time points post-infection and analyzed by western blot analysis for the LC3B-II. D) Relative LC3B-II band densities of 4 replicate experiments were measured with Image-J software and corrected for β-actin band density (*p=0.018). E) Vero cells were mock inoculated or inoculated with clone derived WNV or a Kenyan strain (Ken) of WNV (MOI 3) and cells harvested 24 hours post-infection, membrane-cytosol fractions were resolved by SDS-PAGE and analyzed for the presence of LC3B-II in the membrane fraction. F&G) BSCs were inoculated with WNV (104 pfu/well) and tissue harvested at specified time points post-infection, processed for whole-tissue lysate and analyzed for LC3B-II with western blot. 4 replicate experiments were analyzed for relative band density at 24 hrs post-infection as above (*p=0.006).
West Nile virus-induced LC3 lipidation is dependent on autophagy activation
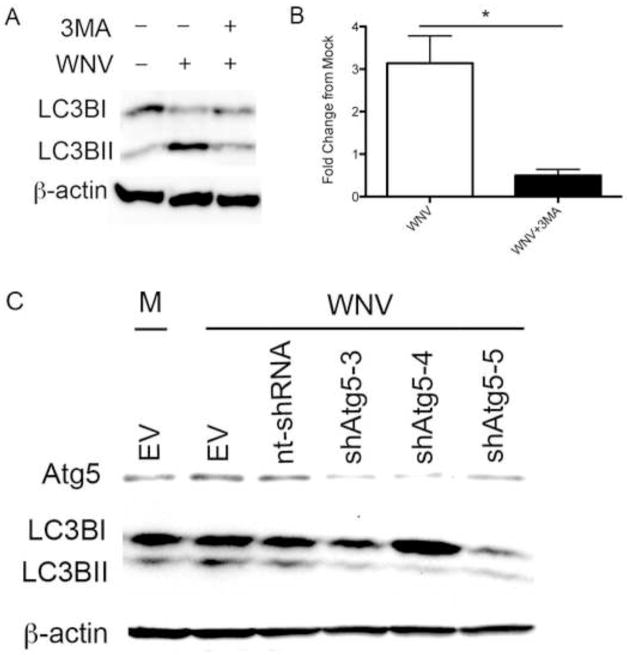
LC3 lipidation is associated with the formation of autophagosomes but is also associated with single membrane structures including phagosomes, macroendosomes, and entotic vacuoles (Florey et al., 2011; Sanjuan et al., 2007). To determine the role of autophagy in WNV-induced LC3B-II formation, we inhibited autophagy and evaluated the effect on WNV-induced LC3B-II. Vero cells were inoculated with WNV (MOI 3) followed by treatment with vehicle control or 3-methyladenine (3MA, 10mM) at 0 hours post-infection, and cells were harvested for western blot analysis at 24 hours post-infection. 3MA is an inhibitor of the class III PI3 kinase complex required for initiation of autophagy. Treatment with 3MA reduced WNV-induced LC3B lipidation in Vero cells (Fig. 2A). Western blot band densitometry of the LC3B-II band corrected for β-actin from 5 replicate experiments revealed a significant decrease in WNV-induced LC3B-II band density following treatment with 3MA compared to WNV-infected Vero cells treated with vehicle control (Fig. 2B; p=0.015). Next, we used a gene-specific knockdown of autophagy signaling to evaluate the role of Atg5 on WNV-induced autophagy. Atg5 is an important initiator of autophagy, and inhibition of Atg5 results in decreased autophagy (Hara et al., 2006). BHK cells were transduced with shRNA Atg5 using a lentivirus vector for 48 hours. Cells were mock infected or infected with WNV, harvested at 24 hours post-infection for western blot analysis. Inhibition of Atg5 with shRNA expression significantly inhibited expression of cellular Atg5 protein and inhibited WNV-induced LC3B-II compared to cells transduced with a scambled shRNA construct (Fig. 2C). This effect was consistent for 3 separate shRNA Atg5 constructs. This data indicates that Atg5 inhibition prevents WNV-induced LC3B lipidation and that LC3B-II accumulation is secondary to WNV-induced autophagy.
Figure 2.
WNV-induced LC3B-II expression is due to autophagy activation. A) Mock and WNV (MOI 3) inoculated Vero cells were treated with 3MA (10mM) or vehicle control at time 0 and analyzed with western blot at 24 hours post-infection for evidence of LC3B-II. B) 5 replicate experiments were analyzed for relative LC3B-II band density showing a significant decrease in LC3B-II band density in 3MA-treated, WNV-infected cells (*p=0.016). C) BHK cells were transduced (MOI 3) with lentivirus expressing 3 different shRNA Atg5 constructs (shATG5-3, shATG5-4, shATG5-5), a non-targeting shRNA (nt-shRNA), or empty vector (EV) controls. At 48 hours post-transduction, cells were inoculated with mock (M) or WNV and harvested at 24 hours post-infection for western blot analysis of Atg5, LC3B, and β-actin protein expression.
West Nile virus induces LC3BII turnover but has no effect basal p62/SQSTM1 levels
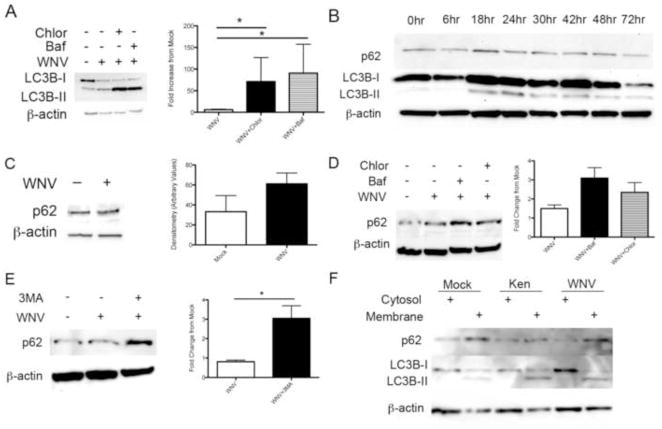
Our data show that LC3B is lipidated following WNV infection, and that WNV-induced LC3 lipidation is dependent on autophagy activation and the autophagy gene Atg5. Accumulation of LC3B-II can result from increased production following autophagy activation or decreased turnover of LC3B-II from inhibition of autophagosome breakdown. To determine the nature of WNV-induced LC3B-II accumulation, we completed a series of ‘autophagy flux’ assays by inhibiting autophagosome acidification using bafilomycin or chloroquine. If LC3B-II accumulates from WNV-induced autophagy activation, then inhibition of autophagosome acidification will result in a further increase in LC3B-II. Vero cells were mock infected or inoculated with WNV (MOI 3) followed by treatment with vehicle control, bafilomycin (50nM), or chloroquine (50μM) at time 0 and 20 hours post-infection with media changes. All treatment groups were harvested at 24 hours post-infection and western blot analysis of whole cell lystes revealed increased LC3B-II in bafilomycin and chloroquine treated, WNV-inoculated cells compared to mock infected and WNV-infected, vehicle treated cells (Fig. 3A). Densitometry values of the LC3B-II band corrected for β-actin expression in 5 replicate experiments revealed a significant increase in LC3B-II in chloroquine and bafilomycin treated, WNV-infected cells compared to WNV-infected cells treated with vehicle (Fig. 3A, p=0.03). These data show that WNV-induced LC3B-II degradation is dependent on acidification of the autophagosome.
Figure 3.
WNV infection activates LC3B turnover but basal p62/SQSTM1 levels are unchanged. A) Vero cells were mock or WNV inoculated, followed by treatment with vehicle control, chloroquine (50μM), or bafilomycin (50nM). Whole cell lysates were evaluated for LC3B-II with western blot and relative LC3B-II band density analysis of 7 replicate experiments revealed a significant increase in LC3B-II following treatment with chloroquine and bafilomycin (*p=0.03). B) Western blot analysis of 4 replicate experiments of WNV-infected MCCs (MOI 3) at indicated time points post-infection for p62 and LC3B-II protein expression. C) Mock and WNV-inoculated Vero cells were harvested and western blot analysis for p62/SQSTM1 of 7 replicate experiments by densitometry show no evidence of altered p62 levels. D) Mock and WNV-inoculated Vero cells were treated with vehicle, chloroquine (50μM) or bafilomycin (50nM). Western blot analysis of p62 relative band density from 4 replicate experiments shows increased p62 with chloroquin and bafilomycin treatment. E) Mock and WNV-inoculated Vero cells were treated with vehicle or 3MA (10mM) and western blot analysis of relative p62 band density following 3MA treatment of WNV-infected cells in 8 replicate experiments is shown (*p=0.011). F) Vero cells were inoculated with mock, WNV clone, or a Kenyan (Ken) clone of WNV. Cells were harvested and cytosol-membrane fractions were analyzed for LC3B-II and p62 using western blot analysis. 6 replicate experiments were completed. All experiments were completed at 24 hours post-infection unless otherwise indicated. Densitometry is expressed as arbitrary units.
To understand whether activation of autophagy differed between non-replicating, terminally differentiated neurons and cell lines following WNV infection, we repeated the above treatments and infections in primary mouse cortical cultures (MCCs). MCCs were harvested from E18 mouse embryos and inoculated with WNV (MOI 3) or mock inoculum at day 7 in vitro. Following one hour of inoculation, virus was removed, neurons were fed with fresh media and whole cell lysates were subjected to western blot analysis at the indicated time points post-infection (Fig. 3B). We found a robust increase in WNV-induced autophagy induction indicated by increased LC3B-II at 18 and 24 hours post-infection (Fig. 3B). However, immunoblot for p62/SQSTM1 showed no evidence of turnover despite activation of autophagy. This data show that WNV activates autophagy in the absence of significant changes in basal p62 expression levels.
P62/SQSTM1 is a cellular protein involved in selective autophagy by targeting ubiquitinated cellular substrates to the autophagosome resulting in p62 and substrate degradation following activation of autophagy and acidification of the autophagosome (Johansen and Lamark, 2011). We next determined the role of p62/SQSTM1 following infection. Vero cells were mock infected or inoculated with WNV (MOI 3) and harvested at 24 hours post-infection. We found no significant change in p62/SQSTM1 levels at 24 hours post-infection with WNV compared to mock infected cells (Fig. 3C) and densitometry of 7 replicate experiments verified these findings with evidence of a trend for increased p62/SQSTM1 levels following WNV-induced autophagy (Fig. 3C, p=0.2).
Next, we determined if levels of p62/SQSTM1 changed with inhibitors of autophagosome acidification following WNV-infection. We infected MCCs with mock inoculation or WNV (MOI 3) followed by treatment with chloroquine (50mM), bafilomycin (50nM) or vehicle control as described above. In 4 replicate experiments, we found evidence of increased p62/SQSTM1 levels in chloroquine and bafilomycin treated MCCs following WNV infection implying that p62/SQSTM1 turnover occurs following WNV infection (Fig. 3D). We next wanted to determine if p62/SQSTM1 turnover was dependent on autophagy. We infected MCCs with mock or WNV inoculum (MOI 3) and treated neurons with vehicle control or 3MA (10mM) at time 0 post-infection. We found that p62/SQSTM1 levels were significantly increased (Fig. 3E, p=0.011) following treatment with the autophagy inhibitor, 3MA, suggesting p62/SQSTM1 is eliminated by autophagy. Since WNV-induced autophagy had no effect on p62/SQSTM1 levels but p62 turnover was clearly controlled by autophagy and autophagosome acidification, we determined whether WNV infection interfered with p62/SQSTM1 delivery from the cytosol to the membrane compartment. We inoculated Vero cells with mock or WNV (MOI 3) clones (NY99 clone or Kenyan (Ken) clone) and harvested cells at 24 hours post-infection. Cells were subjected to subcellular fractionation into cytoplasmic and membrane fractions and analyzed with Western Blot analysis. WNV infection with either clone of WNV had no effect on p62/SQSTM1 localization to the membrane compared to mock infected cells (Fig. 3F). These data show that WNV infection has no effect on steady-state p62 levels, that p62 is turned over by autophagy activation, and that WNV has no effect on p62 membrane localization and degradation.
WNV-infection increases Lysosomal binding to the Autophagosome
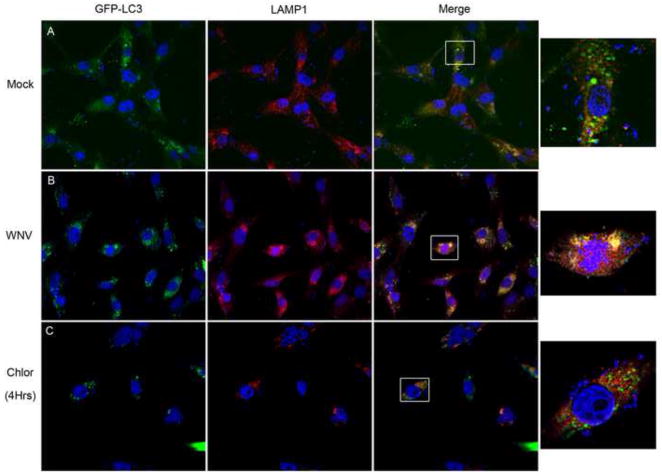
Our data show that WNV-induces autophagy and LC3B turnover, but WNV-induced autophagy has no effect on basal p62/SQSTM1 levels. Because of these findings, we next determined whether WNV infection induces lysosomal fusion to the autophagosome and acidification of the autophagosome. We transfected Vero cells with a GFP-LC3 expressing plasmid followed by mock or WNV (MOI 3) inoculation and harvested cells for immunocytochemistry at 24 hours post-infection. Following fixation (4% paraformaldehyde (PFA)), lysosomes were labeled with a primary antibody to LAMP1 and a secondary, Cy3-conjugated, antibody to rabbit IgG followed by DAPI staining. In three replicate experiments, WNV infection increased GFP-LC3 puncta co-localization with the lysosome marker LAMP1 (Fig. 4B). Mock-infected cells exhibited a lesser degree of GFP-LC3 co-localization with LAMP1 (Fig. 4A) compared to the WNV-infected cells. As a control experiment, Vero cells treated with chloroquine (10μM; 4hours) rarely exhibit co-localization of GFP-LC3 puncta with LAMP1 labeled lysosomes (Fig 4C). These data show that WNV infection activates autophagy resulting in an increase in lysosome co-localization with autophagosomes.
Figure 4. WNV-induced autophagosomes co-localize with lysosomes.
Vero cells were transfected with a GFP-LC3 expressing plasmid and then A) mock inoculated or B) inoculated with WNV (MOI 3). C) As a control, Vero cells were treated with chloroquine (50μM) following transfection with GFP-LC3 expressing plasmid, harvested at 4 hours post-treatment. All cells were fixed and labeled with an antibody to LAMP1 (Cy3:red). Boxes in the merged images were digitally magnified (1000x) at the end of each row. Images are representative of 3 replicate experiments. Experiments were completed at 24 hours post-infection and images shown at 600x original magnification.
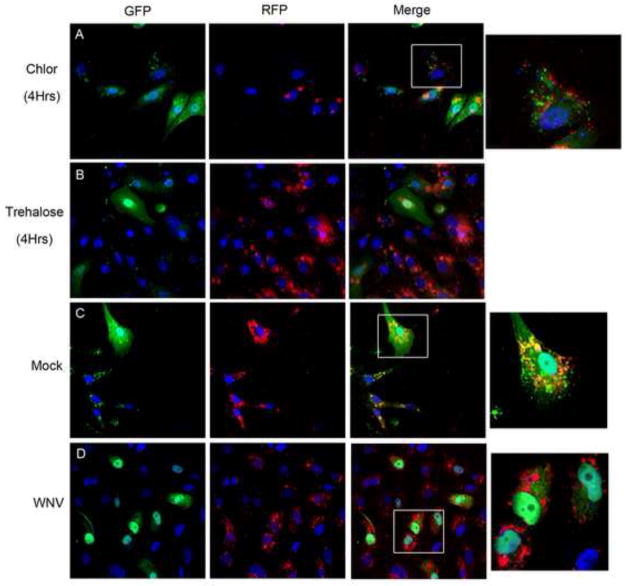
To determine whether lysosomal colocalization with the autophagosome resulted in autophagsome acidification, we used a GFP-RFP-LC3 reporter plasmid (ptfLC3; Addgene) as previously described (Kimura et al., 2007). When both fluorescent proteins are intact in the autophagosome, yellow or green signal are apparent upon imaging and as acidification of the autophagosome progresses, the GFP signal is degraded faster than the more acid resistant RFP signal resulting in a shift to red signal with autophagosome acidification. Following transfection with ptfLC3, cells were mock or WNV inoculated (MOI 3) and harvested at 24 hours post-infection followed by 4% PFA fixation for fluorescence analysis in three replicate experiments. Controls were completed in parallel in three replicate experiments with chloroquine (10μM) treatment, trehalose (10mM) treatment, and empty vector controls. Trehalose is a disaccharide synthesized in fungi, plants, and invertebrates that activates autophagy independent of mTOR in cell culture and in neurons (Rodriguez-Navarro et al., 2010; Sarkar et al., 2007). At 4 hours, chloroquine treatment resulted in a shift to green fluorescence (Fig. 5A) and trehalose treatment resulted in a shift toward red fluorescence (Fig. 5B) consistent with the properties of acidification inhibition and autophagy activation, respectively. Mock infection of ptfLC3 transfected cells resulted in predominantly yellow signal suggesting decreased autophagosome turnover with persistent green signal causing a shift to yellow (Fig. 5C), and WNV infection of ptfLC3 transfected cells resulted in a shift to red fluorescence indicating autophagosome acidification (Fig. 5D). These data indicate that WNV infection causes lysosomal-dependent autophagosome acidification.
Figure 5. WNV infection results in acidification of autophagosomes.
Vero cells were transfected with a plasmid expressing GFP-RFP-LC3 and treated with A) chloroquine (50μM) or B) trehalose (10mM) as assay controls. Cells were fixed and harvested at 4 hours post-treatment for fluorescent imaging. Chloroquine treated cells display continued GFP signal but trehalose activation of autophagy results in a shift to RFP signal due to autophagosome acidification. GFP-RFP-LC3 transfected cells were inoculated with C) mock or D) WNV (MOI 3) and cells were harvested at 24 hours post-infection. Mock infected cells display persistent GFP signal (green and yellow) while WNV infected cells exhibit a shift to RFP (red) signal. Images shown at 600x original magnification with boxes representing digitally magnified cells in the same row (1000x). Images representative of 3 replicate experiments and all nuclei were stained blue with DAPI.
WNV growth is independent of autophagy activation
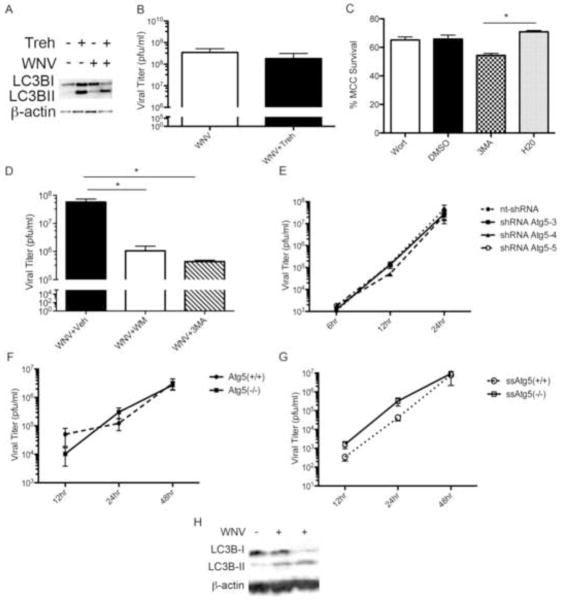
Our data show that WNV-induces autophagy and lysosomal associated acidification of the autophagosome. Despite virus induced autophagy activation, WNV-induced autophagy has little effect on basal p62/SQSTM1 degradation. We next determined whether autophagy activation functions to clear WNV infection or support viral infection. First, we determined the effect of stimulating autophagy on WNV growth using the mTOR-independent activator of autophagy, trehalose. Primary MCCs were inoculated with WNV (MOI 3 for Western Blot analysis) followed by treatment with vehicle control or trehalose (10mM) at time 0 and daily until MCCs were harvested 72 hours post-infection. Primary MCCs treated with trehalose, exhibit robust activation of autophagy indicated by increased LC3B-II on western blot of whole cell lysates (Fig. 6A). There was no significant difference in autophagy induction between trehalose treated WNV-inoculated and trehalose treated mock-inoculated MCCs during replicate experiments. Using standard plaque assay of the cell pellet at 72 hours post-infection, treatment of WNV-inoculated (MOI 0.1) MCCs with trehalose has no significant effect on WNV growth (n=5, 1.8×108 pfu/ml ± 2.9×108) compared to vehicle-treated, WNV-inoculated MCCs (n=5, p=0.5, 3.5×108 pfu/ml ± 3.8×108, Fig. 6B).
Figure 6.
WNV growth is independent of WNV-induced autophagy. A) MCCs were inoculated with mock or WNV (MOI 3) followed by treatment with trehalose (10mM) or vehicle control at time 0. Cells were harvested at 24 hours post-infection and analyzed for LC3B-II with western blot. B) MCCs were inoculated with WNV (MOI 0.1) followed by treatment with vehicle control or trehalose (10mM) at time 0 and every 24 hours with media change until cells were harvested at 72 hours post-infection in 5 replicate experiments. C) MCCs were treated daily with wortmannin (Wort, 0.2mM), DMSO control, 3MA (10mM), or water control for 48 hours and percent neuron survival calculated using trypan exclusion (*p=0.03). D) BSCs were infected with WNV (104 pfu/well), treated with vehicle controls, 3MA (10mM) or wortmannin (0.2mM) every 24 hours, and harvested at 72 hours post-infection for WNV titer using standard viral plaque assay (*p=0.003, n=6). E) BHK cells were transduced with lentivirus expressing one of three constructs of shRNA to Atg5 (shRNA Atg5-3, shRNA Atg5-4, shRNA Atg5-5) or a nontargeting shRNA (nt-shRNA). At 48 hours post-transduction, cells were inoculated with WNV (MOI 3) and viral titer of the supernatant determined at indicated time points (n=3). F) Serum fed Atg5(−/−) and Atg5(+/+) MEFs were infected with WNV (MOI 0.1). WNV titer assay of media at the specified time points post-infection show WNV growth of Atg5(−/−) MEFs compared to Atg5(+/+) MEFs (n=7). G) Atg5(−/−) and Atg5(+/+) MEFs were inoculated with WNV (MOI 0.1) and then fed media without serum at time 0 post-infection followed by viral titer of media at specified time points. Serum-starved (ss)Atg5 (−/−) and ssAtg5 (+/+) MEFs exhibit no significant difference in viral growth (n=9). H) Atg5 (+/+) MEF cells were infected with WNV (MOI 3) and cells harvested at 24 hours post-infection for western blot analysis of LC3B-II expression.
We next determined the effect of autophagy inhibition on viral growth in models of neuronal WNV infection. We used wortmannin and 3MA as targeted pharmacologic inhibitors of autophagy activation. Wormannin and 3MA are PI3 kinase inhibitors that were previously shown to inhibit autophagy (Blommaart et al., 1997; Klionsky et al., 2008). Wortmannin treatment shows no evidence of significant toxicity in MCCs (48 hours post-treatment, Fig. 6C) or BSCs using MTT assay (72 hours post-treatment, Supp. Fig. E). 3MA treatment results in increased toxicity to MCCs (48 hours post-treatment, Fig. 6C) but no evidence toxicity to BSCs (72 hours post-treatment, Supp. Fig. E). BSCs were inoculated with WNV (NY99, 104 pfu/well) followed by treatment with vehicle control, 3MA (10mM), or wortmannin (0.2mM) at time 0 hours and every 24 hours with media changes until BSCs were harvested at 72 hours post-infection. WNV-inoculated BSCs treated with 3MA exhibit decreased viral growth (4.4×105 pfu/ml ±1×105) by 129-fold compared to vehicle treated, WNV-inoculated BSCs (5.7×107 pfu/ml ±4×107, p=0.0034, Fig. 6D). Similarly, treatment of WNV-inoculated BSCs with wortmannin decreased viral growth by 57-fold (1×106 pfu/ml ± 9×105) compared with vehicle-treated, WNV-infected BSCs (p=0.02, Fig. 6D).
These data suggest that WNV growth is dependent on an early step in autophagy activation. Given the possible off-target effects of chemical inhibitors and the broad-range of effects of the PI3 kinase targets, we verified these findings with gene-specific knockdown of autophagy and evaluated viral growth by standard plaque assay. Our previous data show that shRNA-mediated knockdown of Atg5 inhibits WNV-induced autophagy (Fig. 2C). We next evaluated the effect of shRNA Atg5-mediated inhibition on WNV growth. BHK cells were tranduced with one of three shRNA Atg5 constructs or a nontargeting shRNA control using a lentivirus vector. At 48 hours, cells were inoculated with WNV (MOI 3) and supernatant harvested at indicated time points post-infection (Fig. 6E). There was statistical difference in WNV growth between treatments groups using a two-way ANOVA analysis with repeated measures.
Given the possible off target effects of shRNA approaches, we used a gene knockout model for further analysis of the effects of Atg5 on WNV growth and autophagy inhibition. We obtained Atg5 (+/+) and Atg5 (−/−) mouse embryonic fibroblasts (MEFs, gift from Dr. Mizushima). Wild-type Atg5 (+/+) MEF cells and Atg5 (−/−) MEF cells were infected with WNV (MOI 0.1) and supernatants were evaluated for WNV growth at indicated timepoints post-infection (Fig. 6F). Following analysis of 6 replicate experiments, WNV was able to grow to equivalent viral titers in Atg5 (+/+) cells and in Atg5 (−/−) cells (two-way ANOVA, Fig. 6F) in a serum fed state. Given that autophagy is an important pathway to maintain cell survival during periods of cellular stress and is activated by serum starvation, we determined whether Atg5 was required to support viral replication during stress induced by serum starvation. In nine replicate experiments, Atg5 (+/+) and Atg5 (−/−) cells were infected with WNV (MOI 1) and fed media without serum followed by sample collection of the supernatant for viral titer at the indicated time points. Again, WNV was able to grow to statistically equivalent titers in both Atg5 (+/+) and Atg5 (−/−) MEF cells (Fig. 6G). Analysis of growth curves using a Two-Way ANOVA with repeated measures shows no statistical difference in viral titers. Comparison of WNV-infected Atg5 (+/+) and Atg5 (−/−) MEF cells at 12 and 24 hours post-infection using an unpaired t-test with Welch’s correction demonstrate no statistically significant difference between groups (p=0.1, n=7). To ensure that WNV-induced autophagy occurs in MEF cells, we mock or WNV-inoculated (MOI 3) wild-type MEF cells and evaluated whole cell lysates for evidence of LC3B-II using western blot analysis (Fig. 6H). We found that WNV induces autophagy in Atg5 (+/+) MEF cells and found no evidence of autophagy induction in Atg5 (−/−) MEF cells. These data show that WNV growth is not dependent on the host gene Atg5 and that WNV exhibits no change in growth kinetics despite gene-mediated inhibition of autophagy.
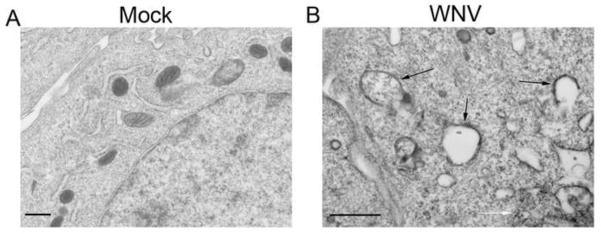
Based on our immunofluorescence data, WNV-induced autophagosomes localize in the perinuclear cytoplasm of cells. Using electron microscopy (EM), we determined the relationship between autophagosome structures and WNV replication centers and virions at the endoplasmic reticulum (ER). Following mock or WNV inoculation (MOI 5), BHK cells were fixed with gluteraldehyde solution and prepared for EM imaging using freeze substitution. EM images revealed structures consistent with autophagosomes in the cytoplasm that had no relationship with virions, viral replication complexes, or the ER (Fig. 7).
Figure 7.
Electron Microscope images of WNV-induced Autophagosome-like structures. Cells were inoculated with A) mock or B) WNV (MOI 5) and harvested at 24 hours post-infection. Black arrows indicated autophagosome like structures and white arrows indicate virions. Scale bar=500nm.
Discussion
Microorganisms and eukaryotic autophagy responses have co-evolved over billions of years and understanding the interactions between infection and evolutionarily conserved cellular responses is vital to decipher the pathogenesis of intracellular pathogens (Levine et al., 2011). To our knowledge, this is the first description of WNV interactions with the autophagy pathway in cells and neurons. We have shown that like other flaviviruses, WNV activates autophagy. Distinct from other flaviviruses, WNV is not dependent on Atg5 or autophagy for viral growth and unlike other viruses such as Sindbis virus, is not restricted by autophagy induction. We also show that basal p62/SQSTM1 levels are unchanged despite activation of autophagy.
Infection with several viruses in cell culture and in the nervous system is associated with activation of autophagy (Dreux et al., 2009; Heaton and Randall, 2010; Jackson et al., 2005; Lee et al., 2008; McLean et al., 2011; Orvedahl et al., 2007; Orvedahl et al., 2010; Yoon et al., 2008). Given this background, we first determined whether WNV infection activates autophagy in a variety of cell lines and in non-replicating, terminally differentiated neurons. Our data show that WNV infection increases the lipidated product of LC3B (LC3B-II) in Vero cells and in a neuronal model of viral encephalitis using BSCs and MCCs infected with WNV. Recently, LC3 lipidation was associated with the formation of single membrane structures including phagosomes, macroendosomes, and entotic vacuoles (Florey et al., 2011; Sanjuan et al., 2007). Therefore, we determined whether the presence of LC3B lipidated products were secondary to activated autophagy signaling by inhibiting autophagy with 3MA and shRNA to Atg5. In both cases, we found that WNV-induced LC3B-II production was reduced when autophagy was inhibited. These data show that LC3B-II production following WNV infection was indeed related to autophagy. This data supports prior finding in other flaviviruses including Dengue and Japanese encephalitis virus.(Heaton and Randall, 2010; Lee et al., 2008; Li et al., 2011)
Next, we determined whether the increase in LC3B-II was secondary to increased production due to an increase in autophagy activity or due to a block in downstream LC3B-II turnover. We employed several strategies to evaluate this question including assaying levels of WNV-induced LC3B-II following inhibition of autophagosome acidification with chloroquine and bafilomycin, evaluating the degree of p62 turnover, evaluating co-localization of GFP-labeled LC3+ puncta with LAMP1+ lysosomes, and by evaluating signal changes in the autophagosome using a GFP-RFP-LC3 expressing plasmid. With the exception of p62 turnover, all approaches indicate that WNV infection results in increased autophagosome turnover and acidification of the autophagosome by lysosomes.
In general, p62/SQSTM1 turnover during autophagy induction is another indicator of flux through the autophagy system (Johansen and Lamark, 2011). When autophagy is activated, p62 levels typically decrease within the cell. We found that p62/SQSTM1 levels were unchanged following WNV-infection despite robust activation of autophagy. Treatment of cells with acidification inhibitors (chloroquine and bafilomycin) or an autophagy inhibitor (3MA) resulted in an increase in p62/SQSTM1 levels indicating that p62/SQSTM1 turnover was autophagy dependent, but p62 levels were not altered following WNV-infection. Next, we determined whether WNV prevented p62 from being delivered from the cytosol to the membrane compartment or autophagosomes. We found that after infection with a clone-derived neuroinvasive strain of WNV or a clone-derived strain of WNV isolated from Kenya with decreased virulence, p62/SQSTM1 is still delivered to the membrane following infection and again p62 levels did not significantly change compared to mock infection. These findings indicate that basal levels of p62/SQSTM1 are maintained despite virus-induced activation of autophagy and turnover of p62 at the autophagosome. We are currently working to understand the role of cellular maintenance of p62 levels in the cells despite autophagy induction and define the role of p62 in WNV infection.
Previous studies have shown that activation of autophagy can result in either antiviral responses following Sindbis and HSV infection, or autophagy can be subverted to support viral replication in the cases of picornoviruses, Dengue virus, and HCV (Jackson et al., 2005; Ke and Chen, 2011; Kirkegaard, 2009; Lee et al., 2008; Orvedahl et al., 2010; Talloczy et al., 2006). Our data show that during serum fed and serum starvation conditions, following inhibition of autophagy with shRNA to Atg5 or with Atg5 KO MEF cells, autophagy is not required to support WNV growth. Activation of autophagy with trehalose also had no effect on viral growth. Other flaviviruses including Dengue and Japanese encephalitis virus were previously shown to be dependent on autophagy for viral replication but the mechanism of flavivirus-dependent replication remains unclear (Dreux et al., 2009; Heaton and Randall, 2010; Lee et al., 2008; Li et al., 2011). The mechanism for these differences is not clear but may be exploited to understand the difference in host range of viruses or neuroinvasive phenotypes. Further studies are ongoing in the laboratory to specifically understand the mechanisms of autophagy activation and viral dependent replication in Dengue virus and WNV. Prior studies have suggested that the ER is the site of autophagy initiation and flavivirus replication complexes, however, our EM data show no evidence of interactions between autophagosome-like structures and viral replication centers or virions. However, our EM studies were completed to just evaluate structure and further studies will need to be completed with labeled proteins to correlate specific structural interactions with specific cellular and viral proteins.
In neuronal and Vero cell models of WNV infection, we found a significant decrease in WNV replication following inhibition of PI3 kinase with chemical inhibitors 3MA and wortmannin in serum fed cells. Inhibition of autophagy with 3MA resulted in a 129-fold decrease in WNV growth and treatment with wortmannin caused a 57-fold decrease in viral replication. Given that we found no difference in WNV growth following Atg5 specific knockdown and gene deletion of Atg5 in serum fed cells, we conclude that the effect of these pharmacologic PI3 kinase inhibitors on viral growth are due to other cellular mechanisms not related to autophagy but dependent on PI3 kinase activation. We are currently working to understand the role of PI3 kinase activation following WNV infection in Vero cells and in primary neuronal culture systems.
Materials and Methods
Cell culture and Virus Stocks
Vero and BHK cells (ATCC) were cultured in MEM (+Earle’s salts and L-glutamine, Gibco) supplemented with 10% heat inactivated fetal bovine serum (FBS) at 37°C and 5% CO2. Primary mouse cortical cells (MCC) (Swiss Webster; Harlan) were harvested and plated using methods previously described (Richardson-Burns et al., 2002). Briefly, E15 NIH Swiss Webster pups were harvested and cortical slices cultured in Neurobasal (Gibco) supplemented with B27 (Gibco) at 37°C and 5% CO2. All MCCs were fed every 2–3 days and were used in experiments at day 6 ex vivo. MEF cells were maintained in MEM with 20% FBS. In starvation studies, MEF cells were fed MEM without FBS following WNV infection.
West Nile virus stocks were obtained from clone derived 382–99 (NY99) strain and a Kenyan strain of WNV as previously described (Brault et al., 2007). After growth in Vero cells, virus was passaged once in C6/36 cells (ATCC). In general, MOI of 3 was used for western blot analysis and immunocytochemistry studies and MOI of 0.1 was used for viral growth assays unless otherwise stated.
Brain Slice Culture
Organotypic brain slice cultures (BSC) were prepared from C57 Black 6 (Harlan) mice according to methods previously described (Dionne et al., 2011b). Briefly, 48–72 hr neonates were sacrificed and their cerebrum and cerebellums removed and 400μM sections obtained using a Leica VT 1000S vibrating microtome in ice cold slicing media (MEM, 10mM Tris, 28mM D-glucose, pH 7.2) and cultured in neurobasal supplemented with 10mM HEPES, 1× B27, 600μM Glutamax, 10% FBS. Slices were infected with 1×105 pfu of WNV 24 hrs after plating by diluting virus stock in 40μl sterile PBS and adding drop wise to slices. Mock infections were performed in a similar manner using PBS alone. Slices were maintained in humidified incubator at 37°C, 5% CO2.
Pharmacologic Inhibitors and treatments
Primary mouse cortical cultures, Vero cells, and BSCs were treated with pharmacologic agents at the indicated concentrations and times post-infection as defined in the text. Pharmacologic treatments used in these studies included trehalose (Sigma-Aldrich, St. Louis, MO), 3-methyladenine (3MA, Sigma-Aldrich), Wortmannin (Sigma-Aldrich), Chloroquine (Sigma-Aldrich) and Bafilomycin (Tocris Bioscience). Appropriate vehicle controls were used for each experiment.
Western blot Analysis
Cells were harvested at various times post-infection in lysis buffer consisting of 1% Triton-X, 10mM triethanolamine-HCL, 150mM NaCl, 5mM EDTA, 1mM MSF, 1x Halt protease and phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL) and disrupted using an ultrasonic processor (Sonics & Materials) VCX130. Lysates were cleared by centrifugation and whole cell extracts were run on standard SDS-PAGE gels (Criterion system; Biorad, USA). Membrane-cytosol extractions were completed using the Pierce Mem-Per Eukaryotic Membrane protein extraction per the manufacturer’s instructions (Thermo/Fisher). The separated proteins were electrically transferred to PVDF membranes (Millipore, USA). For all WB analysis, membranes were activated for 10–15 seconds in methanol and blocked with 5% NFDM (Blocking Grade Buffer, Biorad) and probed with primary antibodies: LC3B and beta-actin (Cell Signaling, USA), p62 C-terminus/STSQM1 (American Research Products, Inc.), mouse monoclonal antibody to WNV envelope (ATCC), and LAMP1 (Abcam). After washing, membranes were probed with appropriate HRP-conjugated secondary antibodies (Jackson ImunoResearch, West Grove, PA) and images obtained after incubation with Western Lightning ECL Pro (Perkin Elmer, MA) and visualized with ChemiDoc XRS+ system (Bio-Rad, Hercules, CA). Band density was measured using AUC measurements with ImageJ software (NIH) and corrected for βactin band density.
Plaque assay
BHK cells were plated into 6 well plates (8 ×105 cells/well) and cultured as above. After 1 hour of adsorption, the inoculum was removed and cells were overlayed with agar and culture media. Cells were incubated for 5 days and plaques fixed with 4% PFA, rinsed with PBS and visualized with crystal violet solution (1% crystal violet in 95% EtOH) followed by gentle washing with deionized H2O. Viral plaques were counted and titer was calculated based on the initial inoculum.
Transfections and shRNA Studies
Vero cells were transfected with GFP-LC3 expressing plasmid (CBA401, Cell Biolabs, Inc) or GFP-RFP-LC3 expressing plasmid (ptfLC3, Addgene) using Mirus transIT-2020 transfection reagent according to manufacturer instructions. Empty vector controls were used for each transfection. After 24–48 hrs of transfection cells were infected with WNV at specified MOIs, labeled with primary and secondary immunofluorescent antibodies, and imaged using the Zeiss LSM 510 Meta Confocal microscope. Puncta counts per cell were completed in observer-blinded fashion and were also completed specifically in antigen positive and negative cells. No primary controls were completed with each IF experiment.
For shRNA experiments, 3 construct lentivirus vectors with shRNA to Atg5 and one non-targeting control vector were obtained from the Functional Genomics Facility at University of Colorado Boulder: TRCN0000150645 (shRNA Atg5-5), TRCN0000151474 (shRNA Atg5-4), TRCN0000151963 (shRNA Atg5-3), SHC002 non-targeting contol. Lentiviral stocks were tittered at 106 pfu/ml. BHK cells were plated in 12 well plates, transduced at MOI 3 and media changed at 24 hours post-transduction. After 48 hours, cells were inoculated with mock or WNV infection (MOI 3) for one hour at 36C. After inoculation, the inoculum was removed, cells were washed with PBS and fed with fresh media. Cells and supernatant were collected at specified time points post-infection for standard plaque assay of supernatant or whole cell lysate of cell pellets.
Electron Microscopy
BHK cells were maintained in MEM, WNV infected (MOI 5) for 24 hours and processed for EM by standard procedures. Briefly, cells were pelleted, fixed in 2% gluteraldehyde and resuspended in 0.1M cacodylate buffer, loaded into 0.25mm aluminum hats and HPF in a high pressure freezer (HPM 010; Balzers Union AG) with an automatic freeze substitution unit (AFS; Leica) for freeze substitution with 0.1% uranyl acetate and 0.25% glutaraldehyde in anhydrous acetone embedded in embedding media and polymerized at −60°C(West et al., 2011). An unltramicrotome (Leica) was used to cut 80nm serial thin sections and collected onto 1% Formvar films adhered to rhodium plated copper grids (Electron Microscopy Sciences). Images obtained using SerialEM at 300kV (Tecnai 30; FEI).
Statistics and Analysis
All values were analyzed using Graphpad Prism version 5.0c. Paired samples were analyzed using unpaired T-test with Welch’s correction and multiple samples in an experiment were analyzed using 1 way ANOVA analysis with Dunn’s multiple comparisons test or 2 way ANOVA for repeated observations.
Supplementary Material
A) BSCs were snap frozen and embedded in OCT followed by sectioning on cryostat. IHC was completed at day 4 ex vivo and neurons labeled with antibody to NeuN (green). B) BSCs were infected with WNV (104 pfu/well with 4 slices/well) and viral titer of BSC tissue was determined at indicated time points post-infection using plaque assay. C) MCCs were fixed with 4% PFA and labeled with anti-neuronal antibodies to neuN (green) and map2 (red). D) MCCs were infected with WNV (MOI 0.1) and viral titer of cell pellet determined. E) BSCs were treated with 3MA (10mM), or wortmannin (0.2mM) every 24 hours until harvested at 48 hours ex-vivo. BSCs were subjected to MTT assay and exhibited normal uptake of MTT compared to vehicle controls.
Acknowledgments
We would like to thank Dr. Mizushima for providing Atg5 knockout and wildtype MEF cells. We appreciate the assistance of the University of Colorado Boulder Electron Microscopy Core and Dr. Thomas Perkins in the preparation and imaging of samples using electron microscopy. We also appreciate the use of the University of Colorado AMC Advanced Light Microscopy Core for confocal imaging studies. We would like to thank Dohun Pyeon and Laura Griffin for assistance with reagents. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) grant K08AI076518 and the NIAID supported Rocky Mountain Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research grant U54AI065357. KLT is supported by a VA Merit award and NIH NINDS R01NS076512. We would also like to thank Micayla Medek for her work on our campus and dedicate this work to her memory and the memory of the other 11 victims from 7/20/11.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Brault AC, Huang CY, Langevin SA, Kinney RM, Bowen RA, Ramey WN, Panella NA, Holmes EC, Powers AM, Miller BR. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne KR, Leser JS, Lorenzen KA, Beckham JD, Tyler KL. A brain slice culture model of viral encephalitis reveals an innate CNS cytokine response profile and the therapeutic potential of caspase inhibition. Exp Neurol. 2011;228(2):222–231. doi: 10.1016/j.expneurol.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K. Subversion of the cellular autophagy pathway by viruses. Curr Top Microbiol Immunol. 2009;335:323–333. doi: 10.1007/978-3-642-00302-8_16. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, Lin YS, Yeh TM, Liu CC, Liu HS. Autophagic machinery activated by Dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JK, Liang JJ, Liao CL, Lin YL. Autophagy is involved in the early step of Japanese encephalitis virus infection. Microbes Infect. 2012;14:159–168. doi: 10.1016/j.micinf.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z. Flavivirus NS4A-induced Autophagy Protects Cells against Death and Enhances Virus Replication. J Biol Chem. 2011;286:22147–22159. doi: 10.1074/jbc.M110.192500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson-Burns SM, Kominsky DJ, Tyler KL. Reovirus-induced neuronal apoptosis is mediated by caspase 3 and is associated with the activation of death receptors. J Neurovirol. 2002;8:365–380. doi: 10.1080/13550280260422677. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro JA, Rodriguez L, Casarejos MJ, Solano RM, Gomez A, Perucho J, Cuervo AM, Garcia de Yebenes J, Mena MA. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010;39:423–438. doi: 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Talloczy Z, Virgin HWt, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4:286–289. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kweon HS, Lee JY, Kim DH. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol. 2008;82:11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) BSCs were snap frozen and embedded in OCT followed by sectioning on cryostat. IHC was completed at day 4 ex vivo and neurons labeled with antibody to NeuN (green). B) BSCs were infected with WNV (104 pfu/well with 4 slices/well) and viral titer of BSC tissue was determined at indicated time points post-infection using plaque assay. C) MCCs were fixed with 4% PFA and labeled with anti-neuronal antibodies to neuN (green) and map2 (red). D) MCCs were infected with WNV (MOI 0.1) and viral titer of cell pellet determined. E) BSCs were treated with 3MA (10mM), or wortmannin (0.2mM) every 24 hours until harvested at 48 hours ex-vivo. BSCs were subjected to MTT assay and exhibited normal uptake of MTT compared to vehicle controls.