Abstract
Viral double-stranded RNA (dsRNA) activates protein kinase R (PKR), which phosphorylates eIF2α and inhibits translation. In response, viruses have evolved various strategies to evade the antiviral impact of PKR. We investigated whether guinea pig cytomegalovirus (GPCMV), a useful model of congenital CMV infection, encodes a gene that interferes with the PKR pathway. Using a proteomic screen, we identified several GPCMV dsRNA-binding proteins, among which only gp145 rescued replication of a vaccinia virus mutant that lacks E3L. gp145 also reversed the inhibitory effects of PKR on expression of a cotransfected reporter gene. Mapping studies demonstrated that the gp145 dsRNA-binding domain has homology to the PKR antagonists of other CMVs. However, dsRNA-binding by gp145 is not sufficient for it to block PKR. gp145 differs from the PKR antagonists of murine CMV in that is functions alone and from those encoded by human CMV in functioning in cells from both primates and rodents.
Keywords: Cytomegalovirus, Guinea Pig, Protein Kinase R, eIF2α, gp145, TRS1, double-stranded RNA, US22 gene family
INTRODUCTION
Human cytomegalovirus (HCMV) is the most common viral infection in newborns, infecting between 0.5 and 2% of infants in utero (Demmler, 1996). Each year in the United States there are over 5500 congenital HCMV infections that result in permanent neurologic disability or death (Bate, Dollard, and Cannon, 2010). HCMV is thought to have coevolved with humans (McGeoch, Rixon, and Davison, 2006) and replicates well only in human cells. This species-specificity precludes the study of HCMV in animal models and has led to the development of several primate and rodent CMVs as models for studying pathogenesis of and immunity to HCMV (Barry et al., 2006; Powers and Fruh, 2008; Schleiss, 2006). Among these, guinea pig cytomegalovirus (GPCMV) is a particularly valuable model to study congenital infection since, like HCMV, GPCMV crosses the placenta and infects the developing fetus in utero (Schleiss, 2006). Application of the GPCMV model requires an understanding of the similarities and differences between GPCMV and HCMV genes and mechanisms, particularly those involved in evasion of rapidly evolving host defense systems.
Double-stranded RNA (dsRNA), which accumulates during the replication of HCMV and many other viruses (Marshall et al., 2009; Weber et al., 2006), activates several cellular antiviral pathways, including one mediated by protein kinase R (PKR). Upon binding to dsRNA, PKR dimerizes and autophosphorylates to form active PKR, which then phosphorylates the α-subunit of eukaryotic initiation factor 2 (eIF2α). Phosphorylated eIF2α inhibits the activity of the guanine nucleotide exchange factor eIF2B and thereby limits formation of the eIF2α-tRNAiMet-GTP ternary complex, inhibiting translation initiation and viral replication (Garcia et al., 2006).
To counteract the effects of PKR, many viruses have evolved factors that block the pathway at one or more steps (Langland et al., 2006; Mohr, Pe’ery, and Mathews, 2007). In the case of HCMV, two genes, IRS1 and TRS1, encode proteins that bind both to dsRNA and to PKR, preventing PKR activation in human cells (Child et al., 2004; Hakki and Geballe, 2005; Hakki et al., 2006). TRS1 and IRS1 are members of the β-herpesvirus US22 gene family, which also includes PKR antagonists of MCMV (m142 and m143) and RhCMV (rhTRS1). The proteins encoded by these genes all bind to dsRNA and interact with PKR with varying species-specificities (Budt et al., 2009; Child et al., 2012; Child et al., 2006; Valchanova et al., 2006). Deletion of both the PKR antagonists of HCMV or either one from MCMV eliminates viral replication (Marshall et al., 2009; Valchanova et al., 2006). Productive infection can be restored to these viruses by providing an active PKR antagonist in cis- or trans- (Marshall et al., 2009; Valchanova et al., 2006) or, in the case of MCMV, by abolishing PKR activity in infected cells (Budt et al., 2009).
To better understand the function and evolution of PKR inhibition by CMVs, we sought to identify a PKR antagonist encoded by GPCMV. We found that gp145, a member of the US22 protein family, binds to dsRNA and inhibits the PKR pathway. gp145 shares a noncanonical dsRNA-binding domain and an ability to multimerize with TRS1 and m142/m143. Also, like TRS1, dsRNA-binding and self-association by gp145 are insufficient to antagonize PKR. Our results reveal that while dsRNA-binding is a conserved feature of the CMV PKR antagonists, mechanistic differences between GPCMV gp145 and the PKR antagonists of other rodent and primate CMVs may reflect adaptations to evolutionary changes in the PKR genes of their host species.
RESULTS
Identification of GPCMV dsRNA-binding proteins
To test whether GPCMV encodes a PKR antagonist, we initially investigated whether GPCMV infection could rescue a vaccinia virus mutant (VVΔE3L) that lacks its PKR antagonist E3L and thus replicates poorly in many cell types (Beattie et al., 1996). VVΔE3L replication can be rescued by coinfection with a second virus that blocks PKR (Jacobs, Langland, and Brandt, 1998), as was observed previously by coinfection with HCMV and VVΔE3L (Child et al., 2002). However, VVΔE3L replication was much less restricted in guinea pig lung fibroblasts (GPL) and other GPCMV-permissive cell lines than it is in other cell types such as human fibroblasts or HeLa cells. Prior infection with GPCMV did not further increase VVΔE3L replication in GPL cells (data not shown). We also tested whether a second vaccinia protein, K3L (Beattie, Tartaglia, and Paoletti, 1991), might be acting as the primary PKR antagonist in GPL cells but found that VVΔK3L also displayed only a modest growth defect in GPL cells and prior infection with GPCMV resulted in no substantial rescue of VVΔK3L (data not shown). Thus we were unable to utilize rescue of VVΔE3L in GPCMV-permissive cell lines to determine whether GPCMV encodes a PKR antagonist.
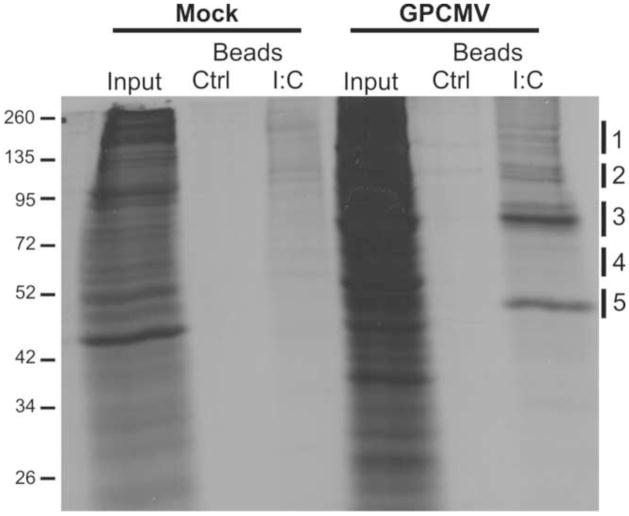
The unusually weak phenotypes of VVΔE3L and VVΔK3L viruses in GPCMV-permissive cell lines led us to explore alternative means to screen for a GPCMV PKR antagonist. Because the PKR antagonists of many viruses are dsRNA-binding proteins (Child et al., 2012; Child et al., 2006; Hakki and Geballe, 2005; Langland et al., 2006; Mohr, Pe’ery, and Mathews, 2007), we hypothesized that a GPCMV PKR antagonist might also bind dsRNA. To identify GPCMV dsRNA-binding proteins, we incubated lysates from mock or GPCMV-infected GPL cells with poly I:C agarose beads (Fig. 1) as described in Materials and Methods. Several bands unique to GPCMV infection were detected, and these likely represented either viral or virally-induced host dsRNA-binding proteins. To identify these proteins, the prominent bands were excised, digested with trypsin, and subjected to liquid chromatography–tandem mass spectrometry (Table 1). Several cellular dsRNA-binding proteins were detected in this experiment, including mammalian Staufen and ILF3 (Buaas et al., 1999; Ramos et al., 2000), supporting the specificity of the poly I:C pull-down assay. The most intense bands (~72 and ~47 kDa) corresponded to the GPCMV proteins gp145 and GP44. gp145 is a member of the US22 gene family and has been annotated as a possible homologue of HCMV TRS1, while GP44 is homologous to UL44, a HCMV DNA polymerase accessory protein that has been previously shown to interact with TRS1 (Schleiss et al., 2008; Strang, Geballe, and Coen, 2010). gp145 was detected in multiple samples that migrated above and below the predicted molecular weight of the protein, suggesting that multiple or modified forms of the gp145 might be produced during infection. Alternatively, the smaller fragments could be degradation products generated during sample preparation. Other abundant GPCMV proteins identified in this screen included gp3, a US22 family protein with no obvious homologue in HCMV, and GP122, the GPCMV homologue of the HCMV IE2 transcriptional transactivator. A third US22 family protein identified in this experiment, gp139 (Schleiss et al., 2008), was not detected when this experiment was repeated.
Figure 1.
dsRNA-binding proteins produced during GPCMV infection. GPL cells infected with GPCMV were radiolabeled with 35S at 4 days post infection, lysed, and proteins bound to control or poly I:C conjugated agarose beads were visualized by autoradiography following SDS-PAGE. Regions indicated on the right (1–5) correspond to samples isolated from a second experiment that were analyzed by mass spectrometry (see Table 1).
Table 1.
dsRNA-binding proteins in GPCMV infected cells.
| Samplea | Gene | # Pep.b | % Cov.c | MW (kDa) | Gene function |
|---|---|---|---|---|---|
| 1 | ILF3 | 22 | 21.9 | 95.3 | interleukin enhancer binding factor 3 |
| gp3 | 13 | 14.5 | 87.7 | homology to THV T5b; US22 superfamily | |
| gp145 | 10 | 12.0 | 68.9 | homology to HCMV IRS1/TRS1; US22 superfamily | |
| GP69 | 7 | 7.0 | 116.3 | UL69 homolog | |
| GP122 | 6 | 13.7 | 35.4 | UL122 homolog; HCMV IE2 | |
| 2 | ILF3 | 32 | 21.7 | 95.3 | interleukin enhancer binding factor 3 |
| gp145 | 12 | 16.1 | 68.9 | homology to HCMV IRS1/TRS1; US22 superfamily | |
| gp139 | 5 | 6.5 | 79.5 | homology to THV T5b; US22 superfamily | |
| 3 | gp145 | 38 | 24.4 | 68.9 | homology to HCMV IRS1/TRS1; US22 superfamily |
| 4 | STAU1 | 22 | 23.7 | 62.2 | staufen, RNA binding protein, homolog 1 |
| GP55 | 12 | 12.7 | 102.2 | UL55 homolog; glycoprotein B | |
| gp145 | 12 | 13.0 | 68.9 | homology to HCMV IRS1/TRS1; US22 superfamily | |
| 5 | GP44 | 87 | 44.7 | 44.8 | UL44 homolog, polymerase accessory protein |
See Figure 1 for approximate regions of the gel represented in each sample.
Number of peptides detected. Percent coverage
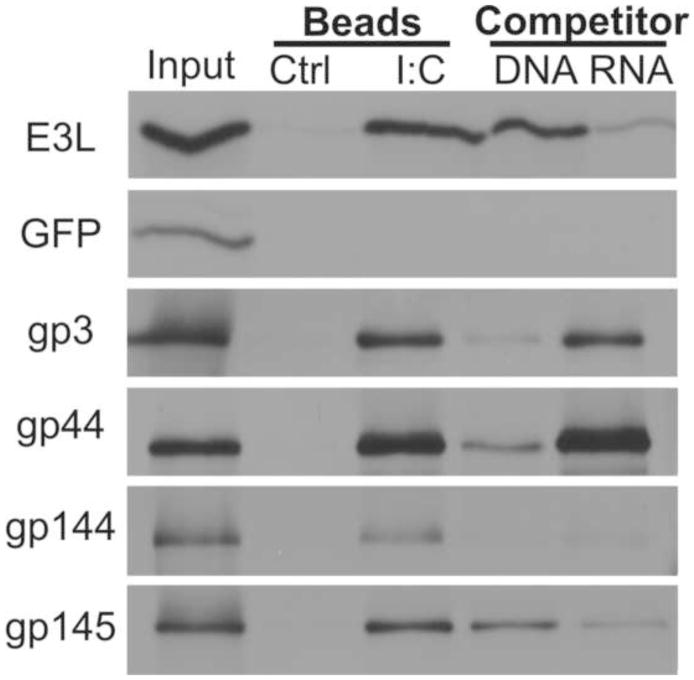
We next sought to differentiate direct dsRNA-binding by GPCMV proteins from indirect binding as part of a multi-protein complex. In addition to testing gp3, gp44, and gp145, we also tested the dsRNA-binding of three additional US22 family proteins that have some sequence similarity to TRS1 (gp141, gp144, and gp146) but that were not detected in the proteomic analysis. The GPCMV proteins were expressed by in vitro translation and analyzed by the dsRNA-binding assay. With the exception of gp141, which failed to express either by in vitro translation or transient transfection, all of the GPCMV proteins tested bound to the poly I:C beads (Fig. 2 and data not shown). To test the specificity of the observed dsRNA-binding, the in vitro translation products were preincubated with either DNA or poly I:C prior to incubation with poly I:C beads. As expected, E3L binding to the poly I:C beads was competed by dsRNA but not by DNA, and gp145 binding was also more strongly competed by preincubation with dsRNA. The opposite pattern was observed from gp3 and GP44, suggesting that these proteins preferentially bind to DNA. These experiments demonstrated GPCMV encodes several dsRNA-binding proteins, although only gp145 appears to preferentially bind dsRNA over DNA.
Figure 2.
dsRNA-binding by in vitro translated GPCMV proteins. In vitro translated, radiolabeled GPCMV and control (E3L and EGFP) proteins were bound to either control or poly I:C conjugated agarose beads. To test the specificity of binding, samples were preincubated with or without DNA or poly I:C competitor prior to binding to poly I:C beads. 10% of the protein used in each binding reaction was run as an input control. The samples were analyzed by SDS-PAGE and autoradiography.
gp145 rescues VVΔE3L
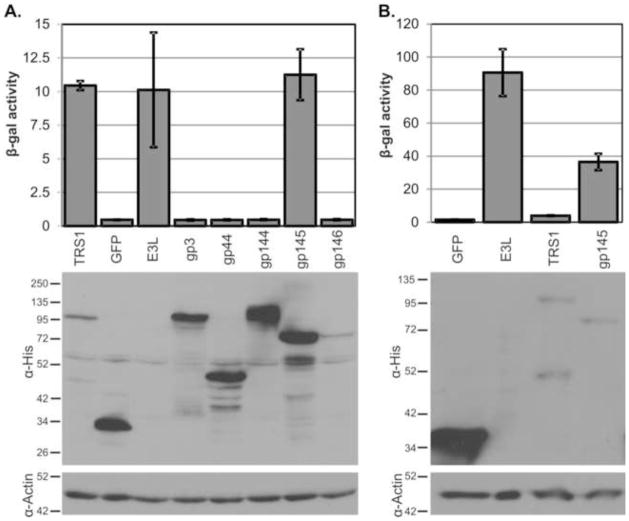
Having identified GPCMV proteins capable of dsRNA-binding, we next tested their ability to inhibit the PKR pathway by measuring the impact of the individual proteins on VVΔE3L replication (Jacobs, Langland, and Brandt, 1998). We transfected HeLa cells with plasmids expressing the GPCMV proteins, infected the cells with VVΔE3L, and measured viral replication at 24 hours post infection. Of the GPCMV proteins tested, only gp145 rescued the replication of VVΔE3L (Fig. 3A). Similar results were also observed in African green monkey cells (COS-7, data not shown). Unlike the MCMV proteins m142 and m143, which function as a heteromeric complex (Child et al., 2006), gp145 was both stably expressed and sufficient to rescue VVΔE3L when expressed by itself. We tested whether VVΔE3L rescue by gp145 might be enhanced by cotransfection of HeLa cells with both gp145 and either gp3, GP44, gp144, gp146, or GFP, but observed no further increase in VVΔE3L replication by mixed transfection of GPCMV proteins over the gp145 plus GFP control (data not shown).
Figure 3.
gp145 rescues VVΔE3L replication. (A) HeLa or (B) GPC-16 cells were transfected with the indicated plasmids and infected with VVΔE3L 24 hours later. Viral replication was measured at 24 hours post infection and the expression of the transfected proteins was measured by α-His immunoblot. E3L was not detectable in this blot because it is not 6X-His-tagged.
Because GPCMV evolved in guinea pigs and its PKR antagonist is likely to be most active against guinea pig PKR, we sought to identify a guinea pig cell line with which we could assess rescue of VVΔE3L replication by transfection. We found that GPC-16 cells (ATCC CCL-242), a transformed line of guinea pig epithelial cells, are highly restrictive to VVΔE3L replication. This cell line is not permissive to GPCMV replication (Nozawa et al., 2008) and so we could not use it to test whether GPCMV infection could rescue VVΔE3L replication. However, transient transfection of either gp145 or E3L did rescue VVΔE3L replication in GPC-16 cells (Fig. 3B). TRS1 failed to rescue VVΔE3L in GPC-16 cells, suggesting that TRS1 may be inactive against guinea pig PKR. These results demonstrated that gp145 can rescue VVΔE3L replication in human, African green monkey, and guinea pig cells and suggest that gp145 activity against PKR is less species-specific than the PKR antagonists of primate CMVs (Child et al., 2012). Additionally, gp145 is more similar to the HCMV and RhCMV PKR antagonists than to MCMV m142/m143 in that a single protein is sufficient to block the PKR pathway.
gp145 antagonizes PKR
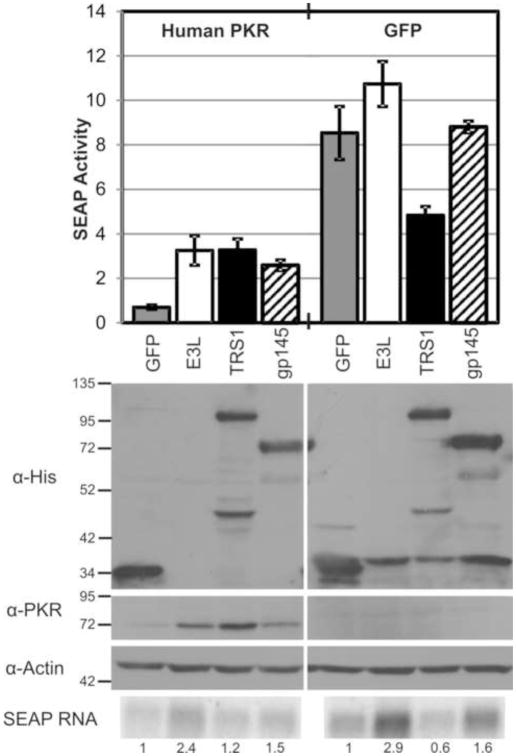
Because E3L interferes with a number of cellular antiviral pathways in addition to PKR (Perdiguero and Esteban, 2009), the ability of a protein to rescue VVΔE3L is only an indirect measure of PKR antagonism. We sought additional evidence demonstrating specific activity of gp145 against PKR, but the lack of available guinea pig reactive reagents prevented us from assaying whether gp145 can prevent phosphorylation of guinea pig PKR. Therefore, we tested whether gp145 can block PKR by measuring its ability to reverse the inhibitory effects of PKR on expression of cotransfected reporter genes (Jacobs, Langland, and Brandt, 1998; Rothenburg et al., 2009). We transfected plasmids expressing secreted alkaline phosphatase (SEAP), human PKR or GFP, and either GFP, E3L, TRS1, or gp145 into HeLa cells in which endogenous PKR has been stably knocked down by an shRNA (Zhang and Samuel, 2007). Expression of PKR with SEAP resulted in a 12-fold reduction in protein expression relative to the GFP control (Fig. 4). E3L, TRS1, and gp145 all rescued SEAP expression when coexpressed with human PKR.
Figure 4.
gp145 inhibits the activity of PKR. HeLa PKR knockdown cells were transfected with plasmids expressing SEAP, either knockdown-resistant human PKR or GFP, and GFP, E3L, TRS1, or gp145. SEAP activity of the medium was measured at 24 hours post transfection. Expression of the transfected proteins was measured by immunoblot. SEAP mRNA was detected by northern blot, and values indicate the amount of transcript relative to the GFP control in each set of transfections.
We observed that PKR expression was higher in cells that also expressed a PKR antagonist. This suggests that active antagonists enable increased expression of all transfected genes, including PKR and the SEAP reporter, and has been observed by others (Rothenburg et al., 2009). We suspect that the increase in PKR expression ultimately overwhelms the antagonists and prevents the full recovery of SEAP levels. To exclude the possibility that increased SEAP expression is the result of an increased level of transcription, we assessed the abundance of SEAP mRNA by northern blot. While transfection of E3L and gp145 both increased the abundance of SEAP mRNA, this effect did not result in a significant increase in SEAP production in the absence of PKR. Surprisingly, although TRS1 has previously been described as a transcriptional transactivator (Romanowski and Shenk, 1997; Stasiak and Mocarski, 1992), expression of TRS1 inhibited SEAP mRNA and protein production in the absence of PKR. These results support to our conclusion from the previous VVΔE3L rescue experiments that gp145 antagonizes the PKR pathway.
dsRNA-binding activity maps to the gp145 US22 domain
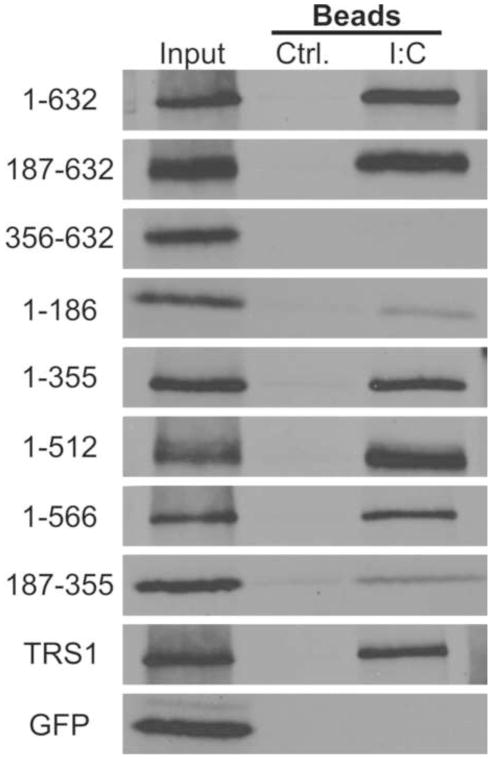
Having demonstrated the anti-PKR activity of gp145, we next sought to identify its functional domains. To delineate the dsRNA-binding domain of gp145, we engineered a series of N- and C-terminal gp145 truncations and analyzed their dsRNA-binding properties (Fig. 5). gp145[187–632], [1–355], and all longer C-terminal truncations bound to poly I:C beads at a level similar to that of full length gp145 while the deletion of residues 1–355 (gp145[356–632]) eliminated dsRNA-binding activity. Both gp145[1–186] and gp145[187–355] retained the ability to bind dsRNA, although the level of binding was substantially reduced. dsRNA-binding in TRS1 also maps to the N-terminus, and the TRS1 minimal dsRNA-binding fragment (TRS1[74–246]) and gp145[187–355] are among the most similar regions of the two proteins, having ~45% amino acid sequence similarity (Hakki and Geballe, 2005). These data suggest that the gp145[187–355] and TRS1[74–246] are a homologous dsRNA-binding domain and that the common ancestor of these proteins predates the divergence of human and guinea pig CMVs.
Figure 5.
The core dsRNA-binding domain maps to gp145 amino acids 187–355.. gp145 N- and C- terminal truncations were expressed by in vitro translation and pulled down with either control or poly I:C beads. The numbers (left side) indicate the amino acid range included in each gp145 plasmid (diagramed in Figure 8). 5% of the protein used in each binding reaction was run as an input control.
dsRNA-binding by gp145 is not sufficient to rescue VVΔE3L
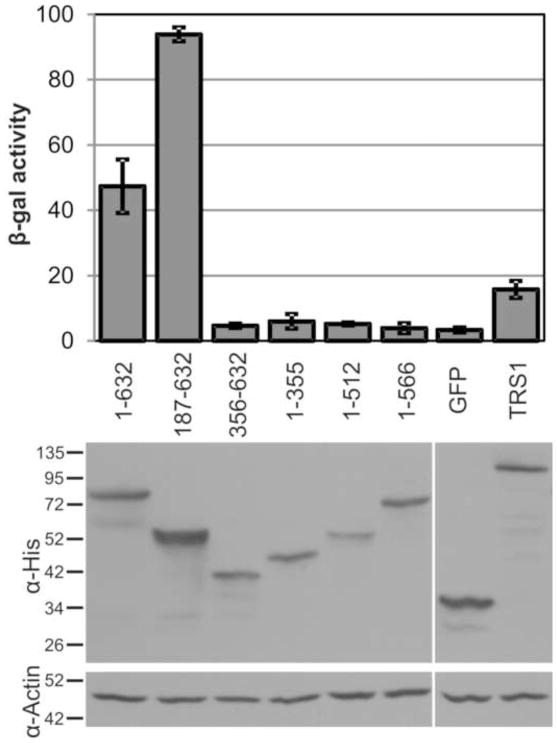
In addition to the N-terminal dsRNA-binding domain, TRS1 requires most of its C-terminal end to rescue VVΔE3L replication and to bind to PKR (Hakki and Geballe, 2005; Hakki et al., 2006). Outside of the shared US22 domain, gp145 and TRS1 are highly divergent and gp145 lacks similarity to the C-terminal end of TRS1 that is necessary for PKR interactions. We hypothesized that gp145 functions primarily by binding and sequestering dsRNA; such a mechanism is sufficient for E3L to limit PKR activation in cell culture (Shors et al., 1997). To evaluate the requirement of dsRNA-binding and/or other regions of gp145 to antagonize PKR, we tested a selection of gp145 truncations for their ability to rescue VVΔE3L replication following transfection of COS-7 cells. Only full length gp145 and gp145[187–632] rescued the replication of VVΔE3L (Fig. 6). Notably, gp145[1–355], gp145[1–512] and gp145[1–566] all failed to rescue VVΔE3L replication despite having dsRNA-binding activity comparable to full length gp145 (Fig. 5). Similar results were observed in VVΔE3L rescue experiments in both HeLa and GPC-16 cells (data not shown). These data led us to suspect that the C-terminus of gp145 provides some function other than dsRNA-binding that is required for VVΔE3L rescue.
Figure 6.
The gp145 dsRNA-binding domain is not sufficient to rescue VVΔE3L. COS-7 cells were transfected with gp145 truncations and infected with VVΔE3L 24 hours later. Viral replication was measured at 24 h.p.i. by β-gal expression and the expression of transfected proteins was monitored by α-His immunoblot.
Because direct interaction with PKR appears to be a necessary function of the PKR antagonists expressed by HCMV, MCMV, and RhCMV (Child et al., 2012; Child and Geballe, 2009; Hakki et al., 2006), we investigated whether gp145 could bind to PKR. In cotransfection experiments, we detected only weak and inconsistent binding of gp145 to either human or guinea pig PKR (data not shown). Thus, whether gp145 interacts with PKR remains unclear, but we suspect there is a weak protein-protein interaction between gp145 and PKR that might be facilitated by a dsRNA tether.
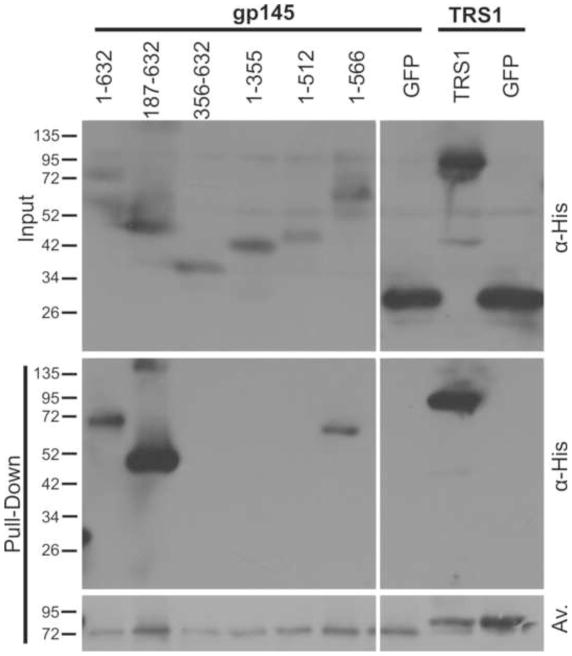
MCMV m142 and m143 cooperatively bind dsRNA and both are needed to block PKR (Child et al., 2006; Valchanova et al., 2006), and thus we hypothesized that gp145 may need to multimerize to antagonize PKR. To test for self-interaction, we transfected COS-7 cells with plasmids expressing biotin ligase and biotinylation signal-tagged gp145 along with full length or truncated 6X-His-tagged gp145 expression plasmids. Following avidin pull-down of the biotinylated gp145, we detected co-precipitated 6X-His-tagged proteins by immunoblot assay (Fig. 7). Three gp145 constructs—full length, gp145[187-632] and gp145[1–566]—co-precipitated with full length gp145, while the shorter C-terminal truncations did not. Because gp145[1–566] interacted with full length gp145 but did not rescue VVΔE3L replication (Fig. 6), gp145 self-interaction appears to be insufficient for PKR inhibition. Additionally, gp145 self-interaction cannot be explained as exclusively mediated by a dsRNA bridge as several truncations that bind dsRNA fail to self-interact, such as gp145[1–355] and gp145[1–512]. Using this approach, we found that TRS1 also self-interacted. As with gp145, C-terminal truncations of TRS1 that fail to rescue VVΔE3L retain the ability to self-associate and some truncations do not self-interact even though they retain the dsRNA-binding activity (data not shown). In summary, dsRNA-binding and self-interaction are common features shared between gp145, TRS1, and the other known CMV PKR antagonists. While these activities may be necessary for gp145 function they appear to be insufficient to rescue VVΔE3L. C-terminal residues of gp145 not required for dsRNA-binding may either participate in another currently unidentified protein interaction or enhance dsRNA-binding to facilitate gp145 rescue of VVΔE3L replication.
Figure 7.
gp145 and TRS1 self-interact. COS-7 cells were transfected with plasmids expressing biotin ligase, biotin signal-tagged gp145 or TRS1 as indicated at the top, and with 6X-His tagged gp145 truncations, TRS1, or GFP. Biotinylated proteins were pulled down by binding to avidin beads and detected with Avidex-AP (Av), while associated proteins were detected by α-His immunoblot.
DISCUSSION
PKR recognizes dsRNAs that accumulate during viral infection and can inhibit viral replication by shutting down protein synthesis (Garcia et al., 2006; Weber et al., 2006). Viral dependence on the host translation machinery to replicate has led to the evolution of numerous viral strategies to antagonize PKR function. In response, PKR has evolved rapidly during speciation, presumably driven by selection to evade viral antagonists (Elde et al., 2008; Rothenburg et al., 2009). The cytomegaloviruses are believed to have cospeciated with their respective hosts over the past 105 million years (McGeoch, Rixon, and Davison, 2006). As the diversity of available host and viral genome sequences increases, CMVs present a unique model to study the coevolution of host defenses and viral countermeasures. Prior to this study, the PKR antagonists of two primate viruses (HCMV and RhCMV) and one rodent virus (MCMV) had been characterized (Budt et al., 2009; Child et al., 2012; Marshall et al., 2009; Valchanova et al., 2006). In this study, we demonstrate that the GPCMV protein gp145 antagonizes the PKR pathway.
The ability to bind to dsRNA is a common feature of the PKR antagonists encoded by many RNA viruses, poxviruses, and herpesviruses (Chang, Watson, and Jacobs, 1992; Child et al., 2012; Child et al., 2006; Hakki and Geballe, 2005; Mohr and Gluzman, 1996; Poppers et al., 2003). Thus, we suspected that a dsRNA-binding protein may contribute to PKR inhibition in GPCMV, where no single protein could be identified as an obvious homologue to either TRS1 or m142/m143. We identified several viral dsRNA-binding proteins present in GPCMV-infected cell lysates by mass spectrometry and assessed their ability to rescue VVΔE3L replication, a surrogate measure of PKR antagonism (Jacobs, Langland, and Brandt, 1998). The GPCMV protein gp145 was detected in this proteomic screen and subsequently found to bind dsRNA when translated in vitro, to rescue VVΔE3L replication, and to reverse the inhibitory effects of PKR activation on the expression of a cotransfected reporter gene.
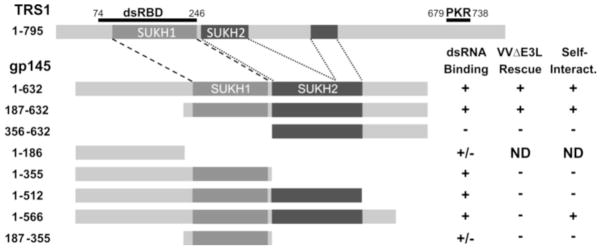
Like the PKR antagonists of HCMV, RhCMV, and MCMV, gp145 is a member of the US22 gene family, which is one of several gene families unique to beta herpesviruses (Chee et al., 1990; Hansen et al., 2003; Rawlinson, Farrell, and Barrell, 1996; Schleiss et al., 2008; Weston and Barrell, 1986). A recent study included the US22 family in the SUKH gene superfamily, which predominantly includes bacterial nuclease toxin-immunity systems (Zhang, Iyer, and Aravind, 2011). Most US22 family proteins are comprised of 2 SUKH domains in tandem, which are referred to here as SUKH1 and SUKH2 of TRS1 and gp145 (Fig. 8). Little is known about either the function of most US22 family proteins or the shared structural domain(s) of these proteins (Menard et al., 2003). However, we found dsRNA-binding maps to the gp145 US22 domain, specifically to SUKH1, as is the case for TRS1 (Fig. 8). As with MCMV, several GPCMV US22 proteins bind dsRNA but only a subset of these proteins can antagonize PKR (Child et al., 2006). It remains to be seen whether binding to dsRNA or other nucleic acids plays a central role in the presently uncharacterized functions of the other US22 proteins.
Figure 8.
Summary of gp145 domain mapping experiments. Shown are diagrams of the approximate boundaries of the TRS1 and gp145 SUKH domains, along with two regions of TRS1—the minimal dsRNA-binding domain and C-terminal residues necessary for direct PKR-binding—that are essential for rescue of VVΔE3L. The gp145 truncations used in Figs. 5–7 and summaries from the dsRNA-binding, VVΔE3L rescue, and self-interaction experiments are also shown, unless not determined (ND).
In addition to binding dsRNA TRS1, rhTRS1, and m142/m143 all directly interact with PKR, in some cases in a species-specific manner (Budt et al., 2009; Child et al., 2012; Child and Geballe, 2009; Hakki et al., 2006). gp145 lacks residues homologous to the C-terminal end of TRS1 that are essential for interaction with human PKR and for VVΔE3L rescue. Our failure to identify robust gp145-PKR interactions led us to speculate that the activity of gp145 against PKR utilizes a mechanism that relied exclusively on dsRNA-binding and sequestration. However, we observed that the C-terminal half of gp145, which is dispensable for dsRNA-binding, is required for VVΔE3L rescue (Fig. 8). We speculated that, similar to m142 and m143, gp145 may need to multimerize to function. While gp145 does self-interact, the region of gp145 that is sufficient for self-interaction is incapable of rescuing VVΔE3L. It remains unclear whether the function of the gp145 C-terminus in intact cells is to potentiate binding to dsRNA or PKR, self-interaction, or for some other interaction.
PKR has been found to be evolving under strong positive selection in several mammalian lineages (Elde et al., 2008; Rothenburg et al., 2009). Complex patterns of species-specificity have been observed in the sensitivities of the vaccinia virus eIF2α mimic K3L (Elde et al., 2008) and more recently in the anti-PKR response of the TRS1 genes of HCMV and RhCMV (Child et al., 2012). While human and rhesus CMV TRS1s are only active against PKRs originating from close relatives of each virus’s natural host (Child et al., 2012), gp145 appears to have activity against human, African green monkey, and guinea pig PKRs. As previous studies have found that MCMV m142 and m143 are active against human and mouse PKR (Child et al., 2006; Valchanova et al., 2006), rodent CMVs may have maintained a broader range of activity against PKR genes than have the primate CMVs (Child et al., 2012). Nonetheless, GPCMV and MCMV PKR differ in their requirement for one versus two proteins to antagonize PKR (Child et al., 2006; Valchanova et al., 2006). Such differences are not entirely surprising since mice and guinea pigs last shared a common ancestor approximately 65 million years ago (Meredith et al., 2011) and the GPCMV genome appears to be more similar to primate CMVs than to MCMV (Schleiss et al., 2008).
Deletion of the PKR antagonists of either HCMV or MCMV results in a replication incompetent virus. While this study does not address whether gp145 is essential for GPCMV replication, a previous study found that GPCMVΔgp138–149 had an approximately three-log replication defect compared to wild type virus while both Δgp123–143 and Δgp147–149 viruses replicated to normal titers (Cui et al., 2009). These results are consistent with the possibility that gp145 plays a critical role in efficient GPCMV replication. It is surprising that GPCMVΔgp138–149 can replicate at all in normal cells, since HCMVΔIRS1/ΔTRS1 and MCMVΔm142 or Δm143 cannot. However, the Δgp138–149 virus was propagated in GPL cells, which we found also support the replication of VVΔE3L and VVΔK3, and it is possible that GPL cells are less sensitive to dsRNA accumulation or PKR activation than many other cell types. Alternatively, as is the case for herpes simplex virus and vaccinia virus (Davies et al., 1993; Mohr and Gluzman, 1996), GPCMV may encode a second PKR antagonist that has yet to be identified. Future analyses of a gp145 knockout virus will help clarify these issues.
Viruses that lack a normal PKR response, such as VVΔE3L, have been explored as potential live-attenuated or disabled infectious single cycle vaccines (Jentarra et al., 2008). The urgent need for an effective vaccine to prevent congenital CMV infection and the known replication defect of HCMVΔIRS1/ΔTRS1 (Marshall et al., 2009) make GPCMVΔgp145 an attractive model for vaccine development. GPCMV has an advantage over MCMV in that the virus better models congenital infection and may require only a single protein to antagonize PKR. That said, many viral proteins are multifunctional and TRS1 is no exception. Previous studies have demonstrated possible roles for TRS1 as a coactivator with IE1 and IE2 transcriptional transactivators (Romanowski and Shenk, 1997; Stasiak and Mocarski, 1992), in virion assembly (Adamo, Schroer, and Shenk, 2004), in viral DNA replication (Pari, Kacica, and Anders, 1993; Strang, Geballe, and Coen, 2010), and in viral manipulation of autophagy (Chaumorcel et al., 2011). Determining whether gp145 shares any of these additional activities will be of great interest for understanding the conserved roles of TRS1 and related US22 family proteins in viral replication and pathogenesis.
MATERIALS AND METHODS
Cells and viruses
HeLa PKR knockdown cells (HeLa PKRKD) cells were provided by Charles Samuel (University of California, Santa Barbara) (Zhang and Samuel, 2007). GPL (ATCC CCL-158), HeLa, GPC-16 (ATCC CCL-242), HeLa PKRKD, and COS-7 (ATCC CRL-1651) cells were propagated on Dulbecco’s modified Eagle’s medium supplemented with 10% NuSerum (BD Biosciences) as previously described (Child et al., 2004). VVΔE3L, obtained from Bertram Jacobs (Arizona State University), was propagated and titered in BHK cells (Beattie et al., 1995; Child et al., 2004). GPCMV strain 22122 (ATTC VR682) was propagated and titered on GPL cells.
Plasmids
GPCMV genes were PCR amplified using the following oligonucleotide pairs and cloned into pcDNA3.1/V5-His-TOPO (Invitrogen): pKTS777 (gp141, 5′-ACCATGTGGCGACTGAACAG-3′ and 5′-TATCAGGTAGCTAGAAAAGT-3′), pKTS778 (gp44, 5′-ACCATGGAACGTAAGACGTC-3′ and 5′-GGCGGAACATTTCTGCTTCTT-3′), pKTS780 (gp3, 5′-ACCATGAAGCGATCGGAAGC-3′ and 5′-ATCGAGATCCAGTTCGAACG-3′), pKTS784 (gp144, 5′-ACCATGCCCAAGCTCGTCTC-3′ and 5′-GTTCTGATGTTTTCTCGGCG-3′), pKTS786 (gp145, 5′-ACCATGACCGAGGCGTTTC-3′ and 5′-GGAACACTCCTCGTCGCTGG-3′), and pEQ1396 (gp146, #960 5′-ACCATGTCGCGTTCTGTATCTCG-3′ and #961 5′-CGCGAGTCCCGTCGGG-3′).
Two E3L-expressing plasmids were used in this study: pMTE3L (Chang, Watson, and Jacobs, 1992), generously provided by Bert Jacobs, was used for transient transfections while pEQ1119 (Child et al., 2012) was used for in vitro translation and dsRNA-binding assays. TRS1 was expressed using pEQ1180 (formerly pEQ981) (Hakki and Geballe, 2005). An EGFP construct tagged with 6X-His only, pEQ1159, was cloned from pEQ1100 (Child et al., 2006), which expresses GFP tagged with both a biotinylation signal and 6X-His (Bio-His), by digestion with EcoRV and SacII, blunted with Klenow, and ligated to excise the biotinylation signal. Knockdown-resistant human PKR (SR#329) was generously provided by Stefan Rothenburg (Kansas State University) (Rothenburg et al., 2009). The SEAP expression vector (pEQ886) has been previously described (Janzen and Geballe, 2004). pEQ1338 was cloned by removing the CMV and T7 promoters of the EGFP expressing pEQ1100 by digestion with SspI and Asp718 and inserting a PvuII - Asp718 fragment from SR#329 containing the SV40 promoter and β-Globin intron.
gp145 truncations were amplified from pKTS786 using either the PCR Extender System (5 Prime) or Phusion (New England Biolabs) polymerases with the following oligonucleotide pairs before cloning into pcDNA3.1/V5-His-TOPO (Invitrogen): pEQ1336 (gp145[187–632], #899 5′-ACCATGGCCAGGGAAAGCTACG-3′ and #900 5′-GGAACACTCCTCGTCGCTGG-3′); pEQ1337 (gp145[356–632], #901 5′-ACCATGGCCTATCTGCCCGCCAGC-3′and #900); pEQ1412 (gp145[1–186], #902 5′-ACCATGCACCGAGGCGTTTC-3′ and #988 5′ CATCTTGACCATCTTCAGCTGCA 3′ );pEQ1353 (gp145[1–355], #902 and #903 5′-GTCCAGGTACTCGGCGCT-3′); pEQ1347 (gp145[1–512], #902 and #904 5′-GAGATACGTGTTTCCCGGCAG-3′); pEQ1352 (gp145[1–566], #902 and #905 5′-GTCGTAGCGCTCGTCGGTG-3′); pEQ1355 (gp145[187–355], #899 and #903).
pCS2+BirA expressing the Escherichia coli biotin ligase was obtained from Bruce Clurman (FHCRC). Biotinylation signal (Bio) tagged TRS1[1–738], pEQ1133, was engineered by digesting the Bio-His tagged pEQ1068 (Child and Geballe, 2009) with PmeI and BstBI, blunting with the Klenow fragment, and re-ligating. This plasmid was then digested with Asp718 and NotI and TRS1 was replaced with gp145 excised from pKTS786 using the same enzymes to make pEQ1376, Bio-tagged gp145.
GPCMV Infection
Confluent GPL cells were infected with GPCMV at a multiplicity of infection (MOI) of 3. Virus was allowed to adsorb to the cells for 2 hours at 37°C before the inoculum was aspirated and replaced with fresh media. At 4 days post infection, cells were either harvested directly for dsRNA-binding assays or radiolabeled with EasyTag Express35S Protein Labeling Mix (50 μCi/ml; Perkin-Elmer) for 2 hours, washed once with PBS, harvested by scraping, and lysed with Buffer A (100 mM KCl, 20 mM HEPES [pH 7.5], 10% glycerol, 5 mM MgOAc, 1 mM dithiothreitol, 1 mM benzamidine (Sigma), and1% NP-40) (Hakki and Geballe, 2005).
dsRNA-binding assay
dsRNA [poly(rI · rC)] agarose beads were prepared as described previously (Langland, Pettiford, and Jacobs, 1995). Binding assays using GPCMV-infected cells were performed by harvesting the cells and lysing in Buffer A by incubating for 20 minutes on ice and pelleting the nuclei by centrifuging (16,000 × g) for 15 minutes at 4°C. The lysate was split between samples containing a mixture of 10 or 20% dsRNA agarose beads in carrier Sepharose CL-6B (Sigma-Aldrich) or with control Sepharose CL-6B alone and incubated for 2 hours at 4°C on a rotating mixer. After binding, the beads were pelleted (845 × g, 3 min) and washed in Buffer A 3–4 times after which bound proteins were denatured, separated on 10% SDS-PAGE gels and visualized either by silver-staining or autoradiography. To test the dsRNA-binding of an individual protein, the indicated plasmid was in vitro translated using the TNT Quick Coupled Transcription/Translation System (Promega) and radiolabeled with [35S] (Easy Tag Express35S Protein Labeling Mix, Perkin-Elmer) per reaction. The in vitro translated protein was then diluted with Buffer A and subjected to the above binding assay. For competition assays, samples were incubated with either 200 μg of free poly(rI · rC) (Sigma) or salmon sperm DNA (Sigma) competitor for 1 hour at 4 °C on a rotating mixer before addition of the dsRNA bead mixture. Following separation on 10% SDS-PAGE gels and fluorographic enhancement (EN3HANCE [PerkinElmer]), samples were visualized by autoradiography.
LC-MS/MS analysis
Samples from a GPCMV infection that had been subjected to a dsRNA-binding assay were run on a 10% SDS-PAGE gel before being stained and destained using the SilverQuest kit (Invitrogen). The bands were excised, dehydrated using acetonitrile, and digested overnight with 5 ng/μL trypsin (Promega) in 50 mM ammonium bicarbonate at 37 °C. The peptides were first extracted using 5% v/v formic acid in water after 30 min incubation, then with acetonitrile. The pooled extracts were dried in a speed vac and cleaned using ZipTip™ C18 (Millipore) before mass spectrometry (MS).
Liquid chromatography (LC)–tandem mass spectrometry (MS/MS) analysis was performed using a LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific). The LC system was configured in a vented format consisted of a fused-silica nanospray needle packed in-house with Magic C18 AQ 100A reverse-phase media (Michrom Bioresources Inc.) and a trap containing Magic C18 AQ 200A reverse-phase media (Licklider et al., 2002). The peptide samples were loaded onto the column and chromatographic separation was performed using a two-mobile-phase solvent system consisting of 0.1% formic acid in water (A) and 0.1% acetic acid in acetonitrile (B). The mass spectrometer operated in a data-dependent MS/MS mode over the m/z range of 400–1800. For each cycle, the five most abundant ions from each MS scan were selected for MS/MS analysis using 35% normalized collision energy. Selected ions were dynamically excluded for 45 seconds.
Raw MS/MS data were submitted to the Computational Proteomics Analysis System (CPAS), a web-based system built on the LabKey Server and searched using the X! Tandem search engine against a protein database derived from the GPCMV genome sequence (GenBank ID: NC_011587.1) and the guinea pig protein database (Broad Institute: cavPor3), which also included additional common contaminants such as human keratin (Craig and Beavis, 2004; Rauch et al., 2006). The search output files were analyzed and validated by ProteinProphet (Nesvizhskii et al., 2003). Proteins and peptides with a probability score of 1.0 were accepted.
VVΔE3L rescue by transient transfection
Triplicate wells of subconfluent HeLa, GPC-16, or COS-7 cells were transfected with the indicated plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. 24 hours post transfection, cells were infected with VVΔE3L at a MOI of 1 and viral replication was quantified by 4-methylumbelliferyl β-D-galactopyranoside (MUG) assay at 24 hours post infection as described previously (Hakki and Geballe, 2005).
HeLa PKRKD reporter gene rescue assay
Subconfluent HeLa PKRKD cells in 24 well plates were transfected with 2 μl Lippofectamine 2000 (Invitrogen) per well and the following amounts of plasmid DNA: 0.05 μg of SEAP reporter (pEQ886), 0.1 μg of knockdown resistant PKR or EGFP (SR#329 or pEQ1338), and 0.65 μg of EGFP or PKR antagonist (pEQ1159, pMTE3L, pEQ1180, or pKTS786) and (Rothenburg et al., 2009). The transfection medium was replaced with fresh medium after 24 hours and at 48 hours post transfection, the medium was harvested and SEAP activity was measured as described previously (Janzen and Geballe, 2004).
Northern Blot
RNA was harvested from transfected HeLa PKRKD cells using Trizol reagent (Invitrogen) and resolved on a 1% formaldehyde agarose gel prior to capillary transfer to a supported nitrocellulose membrane (Optitran, GE Healthcare). Northern hybridization was performed as described previously (Child et al., 2004) using a probe specific to SEAP, which was generated from a fragment of pEQ886 gel isolated following digestion with XhoI and HindIII. The blot was visualized by phosphorimager analysis (Typhoon Trio [GE Healthcare]) and bands quantified using the ImageQuant software.
Avidin agarose pull-down
Subconfluent COS-7 cells in 6 well plates were transfected using Lipofectamine 2000 with a mixture of 1.3 μg of each of 3 plasmids: 1) pCS2+BirA, 2) Bio-tagged TRS1 (pEQ1133) or gp145 (pEQ1376), and 3) a plasmid expressing 6X-His-tagged protein. 24 hours post transfection, the transfection medium was replaced with medium supplemented with 35 μM Biotin (Sigma). At 48 hours post transfection, cells were harvested by trypsanization and lysed in 110 μl of radioimmunoprecipitation assay buffer ([RIPA] 50 mM Tris-HCl, 150 mM NaCl, 0.1% sodium deoxycholate, 0.05% SDS, and 1% Triton X-100) by incubation for 20 minutes on ice followed by centrifugation (16,000 × g) for 15 minutes at 4°C to pellet the nuclei. After reserving a portion, the lysate was added to avidin agarose (Pierce) and incubated on a rotating mixer at 4°C for 2 hours. After binding, the beads were pelleted (845 × g, 3 min) and washed 3X with RIPA buffer before denaturing and separation by 10% SDS-PAGE and visualization by immunoblot as described below.
Immunoblot analyses
Unless otherwise noted, cells were lysed with 2% SDS, DNA sheared with a Bransonic bath sonicator, and protein concentrations were determined using fluoraldehyde o-phthalaldehyde (Pierce) assay (Geballe and Mocarski, 1988). Equivalent amounts of protein were separated by SDS-PAGE prior to transfer to polyvinylidene difluoride membranes (GE Life Sciences) by electroblotting. Proteins were detected using the Western-Star™ chemiluminescent detection system (Applied Biosystems) according to the manufacturer’s instructions using the following commercial primary antibodies diluted 1:1000: Actin (Sigma A-2066), Penta-His (Qiagen, 34660), and PKR (Santa Cruz Biotechnology, Inc. sc-6282). Biotinylated proteins were detected using avidin-alkaline phosphatase conjugate (Avidx-AP, Applied Biosystems).
HIGHLIGHTS.
The guinea pig cytomegalovirus (CMV) protein gp145 antagonizes the protein kinase R pathway.
gp145 binds dsRNA and its minimal dsRNA-binding domain is conserved with HCMV TRS1.
Unlike mouse CMV m142/m143, gp145 alone is sufficient to block PKR.
All known CMV PKR antagonists are members of the US22 gene family.
Acknowledgments
We thank Michael McVoy (Virginia Commonwealth School of Medicine) for reagents, helpful discussions, and critical review of the manuscript, Pau Mezquita (Fred Hutchinson Cancer Research Center) and Michael Leviton (University of Minnesota) for technical help, and Erik Mattsen (Fred Hutchinson Cancer Research Center) for helpful discussions. Reagents were kindly provided by Bruce Clurman (Fred Hutchinson Cancer Research Center), Bertram Jacobs (Arizona State University), Stefan Rothenburg (Kansas State University), and Charles Samuel (University of California, Santa Barbara). We thank Phil Gafken, Deepa Hegde, and Yuko Ogata of the Fred Hutchinson Cancer Research Center Proteomics Core and Elizabeth Jensen of the Genomic Core for technical assistance.
This work was supported by NIH grants AI26672 (to A.P.G) and HD044864 (to M.R.S and A.P.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamo JE, Schroer J, Shenk T. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J Virol. 2004;78(19):10221–9. doi: 10.1128/JVI.78.19.10221-10229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry PA, Lockridge KM, Salamat S, Tinling SP, Yue Y, Zhou SS, Gospe SM, Jr, Britt WJ, Tarantal AF. Nonhuman primate models of intrauterine cytomegalovirus infection. ILAR J. 2006;47(1):49–64. doi: 10.1093/ilar.47.1.49. [DOI] [PubMed] [Google Scholar]
- Bate SL, Dollard SC, Cannon MJ. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin Infect Dis. 2010;50(11):1439–47. doi: 10.1086/652438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69(1):499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E, Kauffman EB, Martinez H, Perkus ME, Jacobs BL, Paoletti E, Tartaglia J. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes. 1996;12(1):89–94. doi: 10.1007/BF00370005. [DOI] [PubMed] [Google Scholar]
- Beattie E, Tartaglia J, Paoletti E. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology. 1991;183(1):419–22. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Lee K, Edelhoff S, Disteche C, Braun RE. Cloning and characterization of the mouse interleukin enhancer binding factor 3 (Ilf3) homolog in a screen for RNA binding proteins. Mamm Genome. 1999;10(5):451–6. doi: 10.1007/s003359901022. [DOI] [PubMed] [Google Scholar]
- Budt M, Niederstadt L, Valchanova RS, Jonjic S, Brune W. Specific inhibition of the PKR-mediated antiviral response by the murine cytomegalovirus proteins m142 and m143. J Virol. 2009;83(3):1260–70. doi: 10.1128/JVI.01558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 1992;89(11):4825–9. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumorcel M, Lussignol M, Mouna L, Cavignac Y, Fahie K, Cotte-Laffitte J, Geballe A, Brune W, Beau I, Codogno P, Esclatine A. The Human Cytomegalovirus Protein TRS1 Inhibits Autophagy via Its Interaction with Beclin 1. J Virol. 2011 doi: 10.1128/JVI.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA, 3rd, Kouzarides T, Martignetti JA, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–69. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Child SJ, Brennan G, Braggin JE, Geballe AP. Species specificity of protein kinase R antagonism by cytomegalovirus TRS1 genes. J Virol. 2012;86(7):3880–9. doi: 10.1128/JVI.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ, Geballe AP. Binding and relocalization of protein kinase R by murine cytomegalovirus. J Virol. 2009;83(4):1790–9. doi: 10.1128/JVI.01484-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ, Hakki M, De Niro KL, Geballe AP. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J Virol. 2004;78(1):197–205. doi: 10.1128/JVI.78.1.197-205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ, Hanson LK, Brown CE, Janzen DM, Geballe AP. Double-stranded RNA binding by a heterodimeric complex of murine cytomegalovirus m142 and m143 proteins. J Virol. 2006;80(20):10173–80. doi: 10.1128/JVI.00905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ, Jarrahian S, Harper VM, Geballe AP. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J Virol. 2002;76(10):4912–8. doi: 10.1128/JVI.76.10.4912-4918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–7. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- Cui X, McGregor A, Schleiss MR, McVoy MA. The impact of genome length on replication and genome stability of the herpesvirus guinea pig cytomegalovirus. Virology. 2009;386(1):132–8. doi: 10.1016/j.virol.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MV, Chang HW, Jacobs BL, Kaufman RJ. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67(3):1688–92. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmler GJ. Congenital cytomegalovirus infection and disease. Adv Pediatr Infect Dis. 1996;11:135–62. [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2008 doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70(4):1032–60. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe AP, Mocarski ES. Translational control of cytomegalovirus gene expression is mediated by upstream AUG codons. J Virol. 1988;62(9):3334–40. doi: 10.1128/jvi.62.9.3334-3340.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M, Geballe AP. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol. 2005;79(12):7311–8. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M, Marshall EE, De Niro KL, Geballe AP. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J Virol. 2006;80(23):11817–26. doi: 10.1128/JVI.00957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol. 2003;77(12):6620–36. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs BL, Langland JO, Brandt T. Characterization of viral double-stranded RNA-binding proteins. Methods. 1998;15(3):225–32. doi: 10.1006/meth.1998.0626. [DOI] [PubMed] [Google Scholar]
- Janzen DM, Geballe AP. The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Res. 2004;32(15):4491–502. doi: 10.1093/nar/gkh791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentarra GM, Heck MC, Youn JW, Kibler K, Langland JO, Baskin CR, Ananieva O, Chang Y, Jacobs BL. Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: scarification vaccination. Vaccine. 2008;26 (23):2860–72. doi: 10.1016/j.vaccine.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119(1):100–10. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Langland JO, Pettiford SM, Jacobs BL. Nucleic acid affinity chromatography: preparation and characterization of double-stranded RNA agarose. Protein Expr Purif. 1995;6(1):25–32. doi: 10.1006/prep.1995.1004. [DOI] [PubMed] [Google Scholar]
- Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem. 2002;74(13):3076–83. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- Marshall EE, Bierle CJ, Brune W, Geballe AP. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol. 2009;83(9):4112–20. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117(1):90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Menard C, Wagner M, Ruzsics Z, Holak K, Brune W, Campbell AE, Koszinowski UH. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J Virol. 2003;77(10):5557–70. doi: 10.1128/JVI.77.10.5557-5570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, Murphy WJ. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–4. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Mohr I, Gluzman Y. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 1996;15(17):4759–66. [PMC free article] [PubMed] [Google Scholar]
- Mohr I, Pe’ery T, Mathews MB. Protein Synthesis and Translational Control during Viral Infection. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 545–599. [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nozawa N, Yamamoto Y, Fukui Y, Katano H, Tsutsui Y, Sato Y, Yamada S, Inami Y, Nakamura K, Yokoi M, Kurane I, Inoue N. Identification of a 1.6 kb genome locus of guinea pig cytomegalovirus required for efficient viral growth in animals but not in cell culture. Virology. 2008;379(1):45–54. doi: 10.1016/j.virol.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Pari GS, Kacica MA, Anders DG. Open reading frames UL44, IRS1/TRS1, and UL36–38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993;67(5):2575–82. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero B, Esteban M. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res. 2009;29(9):581–98. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- Poppers J, Mulvey M, Perez C, Khoo D, Mohr I. Identification of a lytic-cycle Epstein-Barr virus gene product that can regulate PKR activation. J Virol. 2003;77(1):228–36. doi: 10.1128/JVI.77.1.228-236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers C, Fruh K. Rhesus CMV: an emerging animal model for human CMV. Med Microbiol Immunol. 2008;197(2):109–15. doi: 10.1007/s00430-007-0073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19(5):997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Bellew M, Eng J, Fitzgibbon M, Holzman T, Hussey P, Igra M, Maclean B, Lin CW, Detter A, Fang R, Faca V, Gafken P, Zhang H, Whiteaker J, States D, Hanash S, Paulovich A, McIntosh MW. Computational Proteomics Analysis System (CPAS): an extensible, open-source analytic system for evaluating and publishing proteomic data and high throughput biological experiments. J Proteome Res. 2006;5(1):112–21. doi: 10.1021/pr0503533. [DOI] [PubMed] [Google Scholar]
- Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70(12):8833–49. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski MJ, Shenk T. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J Virol. 1997;71(2):1485–96. doi: 10.1128/jvi.71.2.1485-1496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol. 2009;16(1):63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiss MR. Nonprimate models of congenital cytomegalovirus (CMV) infection: gaining insight into pathogenesis and prevention of disease in newborns. ILAR J. 2006;47(1):65–72. doi: 10.1093/ilar.47.1.65. [DOI] [PubMed] [Google Scholar]
- Schleiss MR, McGregor A, Choi KY, Date SV, Cui X, McVoy MA. Analysis of the nucleotide sequence of the guinea pig cytomegalovirus (GPCMV) genome. Virol J. 2008;5:139. doi: 10.1186/1743-422X-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shors T, Kibler KV, Perkins KB, Seidler-Wulff R, Banaszak MP, Jacobs BL. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology. 1997;239(2):269–76. doi: 10.1006/viro.1997.8881. [DOI] [PubMed] [Google Scholar]
- Stasiak PC, Mocarski ES. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66(2):1050–8. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang BL, Geballe AP, Coen DM. Association of human cytomegalovirus proteins IRS1 and TRS1 with the viral DNA polymerase accessory subunit UL44. J Gen Virol. 2010;91(Pt 9):2167–75. doi: 10.1099/vir.0.022640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valchanova RS, Picard-Maureau M, Budt M, Brune W. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J Virol. 2006;80(20):10181–90. doi: 10.1128/JVI.00908-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80(10):5059–64. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K, Barrell BG. Sequence of the short unique region, short repeats, and part of the long repeats of human cytomegalovirus. J Mol Biol. 1986;192(2):177–208. doi: 10.1016/0022-2836(86)90359-1. [DOI] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Samuel CE. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J Virol. 2007;81(15):8192–200. doi: 10.1128/JVI.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]