Abstract
GCK-MODY, dominantly inherited mild fasting hyperglycemia, has been associated with >600 different mutations in the glucokinase (GK)-encoding gene (GCK). When expressed as recombinant pancreatic proteins, some mutations result in enzymes with normal/near-normal catalytic properties. The molecular mechanism(s) of GCK-MODY due to these mutations has remained elusive. Here, we aimed to explore the molecular mechanisms for two such catalytically ‘normal’ GCK mutations (S263P and G264S) in the F260-L270 loop of GK. When stably overexpressed in HEK293 cells and MIN6 β-cells, the S263P- and G264S-encoded mutations generated misfolded proteins with an increased rate of degradation (S263P>G264S) by the protein quality control machinery, and a propensity to self-associate (G264S>S263P) and form dimers (SDS resistant) and aggregates (partly Triton X-100 insoluble), as determined by pulse-chase experiments and subcellular fractionation. Thus, the GCK-MODY mutations S263P and G264S lead to protein misfolding causing destabilization, cellular dimerization/aggregation and enhanced rate of degradation. In silico predicted conformational changes of the F260-L270 loop structure is considered to mediate the dimerization of both mutant proteins by a domain swapping mechanism. Thus, similar properties may represent the molecular mechanisms for additional unexplained GCK-MODY mutations, and may also contribute to the disease mechanism in other previously characterized GCK-MODY inactivating mutations.
Keywords: GCK-MODY, Catalytic activity, Protein misfolding, Self-association, Dimerization, Aggregation, Degradation
1. Introduction
The enzyme glucokinase (GK), also denoted hexokinase IV (EC 2.7.1.1), is an important factor in the regulation of blood glucose, by acting as the ‘glucose sensor’ of the pancreatic β-cells. GK catalyzes the phosphorylation of α-D-glucose (Glc) to form glucose-6-phosphate, the entry point into glycolysis. The enzyme is expressed in hepatocytes [1], pancreatic β-cells [2,3], as well as in the brain and endocrine cells in the gut [4]. GK is activated by glucose binding, which in the hepatocytes (GK isoform 2) results in stimulation of glucose uptake, glycolysis and glycogen synthesis, and in the β-cells (GK isoform 1) mediates glucose-stimulated insulin secretion (GSIS) [5,6].
A complex network of protein-protein interactions has been reported for the posttranslational regulation of GK in the pancreatic β-cell and liver hepatocytes [7–10]. In hepatocytes, GK catalytic activity and cytoplasm↔nucleus transport is controlled by the GK regulatory protein (GKRP) [7,8]. The bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFK-2/FBPase-2), expressed in both pancreatic β-cells and liver, plays a role in GK activation [9,10]. Moreover, posttranslational modifications like S-nitrosylation [11] and ubiquitination [12] have also been associated with GK regulation.
The key role of GK in glucose homeostasis is illustrated by its three associated forms of ‘glucokinase disease’ caused by different mutations in its encoding gene (GCK). Heterozygous mutations that decrease GK enzyme activity result in one of the most common forms of maturity-onset diabetes of the young (MODY), termed GCK-MODY or MODY2 (MODY2; MIM#125851), and is characterized by mild fasting hyperglycemia [13, 14]. Homozygous or compound heterozygous inactivating mutations result in the more severe permanent neonatal diabetes mellitus [15]. The opposite phenotype exists in persistent hyperinsulinemic hypoglycemia of infancy (PHHI) due to mutations that increase GK enzyme activity [16]. Of the >600 GCK mutations identified to date, around 80 have been functionally characterized [17], most of them as recombinant pancreatic GST-fusion proteins. The majority of these mutations are associated with altered enzyme kinetic parameters and overall phosphorylating capacity [17], mainly by affecting the affinity for its substrates Glc and ATP [18,19]. In some cases, the GCK-MODY mutations have near normal enzyme kinetics, or even a mild increase in affinity for Glc, and show normal regulation by allosteric inhibitor/activator molecules (GKRP and GKAs) [20–23]. Thus, the molecular mechanism leading to their diabetes remains unexplained. For some mutations, a cellular instability or defect in S-nitrosylation has been indicated [24–26].
In the present work, we have studied two GCK-MODY mutations causing amino acid changes S263P and G264S in the F260-L270 loop structure of pancreatic hGK, and reported to result in GK enzymes with normal or near normal kinetic parameters, at least as GST fusion proteins [21,27]. We have reinvestigated their steady-state kinetic properties using both GST-tagged and tag-free proteins at two temperatures (30 and 37 °C). Also, we have studied their conformational, folding and stability properties by (i) in vitro susceptibility to limited proteolysis, (ii) stability in human embryonic kidney (HEK293) cells and mouse insulinoma (MIN6) β-cells in pulse-chase experiments, and (iii) in silico protein conformational changes in the loop region.
2. Materials and methods
2.1. Materials
We purchased factor Xa from Protein Engineering Technology ApS (Aarhus, Denmark), and Trypsin and Soybean trypsin inhibitor from Sigma-Aldrich (St. Louis, MO, USA). We obtained TnT® T7 Quick Coupled Transcription/Translation System, MagneHis Protein Purification System and FastBreak lysis buffer from Promega (Madison, WI, USA). Proteasomal inhibitor (MG132) and Ub aldehyde were from Biomol (Cambridge, MA, USA). Protease inhibitor cocktail was from F. Hoffmann-La Roche (Mannheim, Germany) and the bicinchoninic acid (BCA) protein assay kit from Pierce (Rockford, IL, USA). We bought the lysosomal protease inhibitor leupeptin from Sigma-Aldrich (St. Louis, MO, USA). SDS- and native-PAGE gels and buffers, and Hank’s balanced salt solution were from Invitrogen (Grand Island, NY, USA). Glutathione Sepharose 4B was obtained from GE Healthcare (Buckinghamshire, UK). Antibody (Ab) anti-GK (h-88) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and anti-V5 from Invitrogen. We obtained anti β-actin from Abcam (Cambridge, UK).
2.2. Expression and purification of recombinant hGK
We expressed and purified WT and mutant pancreatic hGK in pGEX-3X vector as glutathione-S-transferase (GST) fusion proteins [28]. The recovery of purified protein was 1.6 mg of soluble protein per liter of culture for WT hGK and 1.0–1.5 mg for the mutant proteins. Purified proteins were high-speed centrifuged (267000 g, 15 min, 4 °C) to remove any aggregates, then concentrated, aliquoted, and stored in liquid nitrogen in the absence of glucose. We determined protein concentrations using A280 (1 mg·ml−1·cm−1) of 1.05 (GST fusion protein) [28]. The recombinant proteins were isolated to a purity of >95% (SDS/PAGE) with an expected molecular mass of 76 kDa, as previously described for GST-tagged WT hGK [12]. To remove the GST partner the proteins were cleaved for 3 h at 4 °C by factor Xa protease, using a protease to substrate ratio of 1:25 (by mass).
2.3. Steady-state kinetics
We determined the steady-state kinetic properties of both GST- and non-tagged (cleaved) WT and mutant hGK proteins with glucose (1–60 mM; ATP 5mM) or ATP (0.025–5 mM; saturating amount of glucose) as the variable substrate. GK activity was measured, after preequlibration with glucose, by a glucose 6-phosphate dehydrogenase (G6PDH)-coupled assay as described previously [18,28]. Kinetic parameters were calculated by non-linear regression analysis using the Hill equation. The catalytic activity (kcat) was determined for the total enzyme pool (monomer and dimer; see Fig. 6H and I). We tested a minimum of three different protein preparations.
Fig. 6.
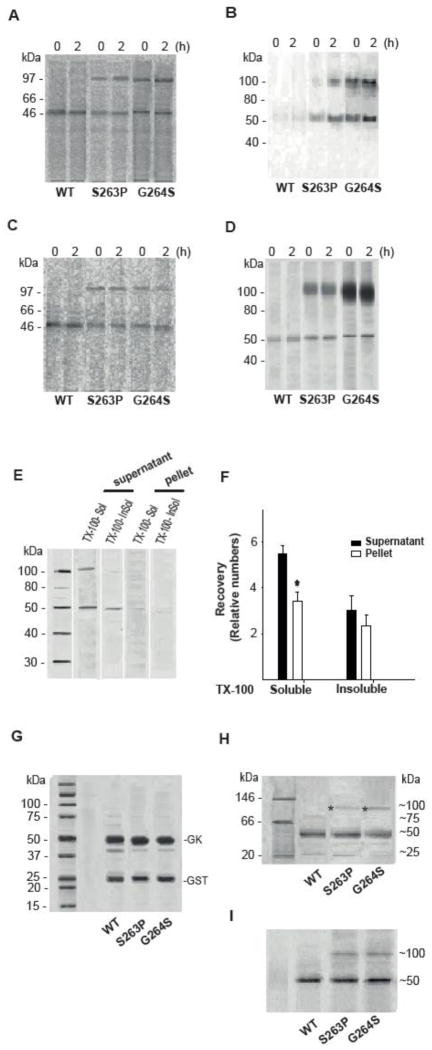
PAGE analysis of the native and denatured state of WT and mutant hGK forms over-expressed in cells and as recombinant proteins. Stably transfected HEK293 cells (A and B) and MIN6 cells (C and D) were metabolically labeled with [35S]Met/[35S]Cys for 30 min and chased for 2 h. WT, S263P and G264S mutant proteins were affinity isolated. After high-speed centrifugation of the eluate, the cytosolic fractions from HEK293 (A) and MIN6 cells (C) were analyzed by native-PAGE electrophoresis, and the pellets from HEK293 (B) and MIN6 cells (D) analyzed by SDS/PAGE (4–12 %) and immunoblotting (anti-V5 Ab) after denaturation (56 °C, 15 min). E: Analysis of Triton X-100 soluble and Triton X-100 insoluble (guanidine chloride soluble) forms of the G264S mutant protein. The post-nuclear supernatant fraction of MIN6 cells, stably expressing the G264S mutant form, was centrifuged and the recovered supernatant and pellet fractions treated with 1 % (v/v) Triton X-100. Following high-speed centrifugation, the supernatants were referred to as Triton X-100 soluble hGK protein. The resulting pellets were solubilized by sonication in 5 M guanidine chloride and high speed centrifuged (referred to as Triton X-100-insoluble (guanidine chloride-soluble) GK protein). The pellet samples were diluted 10-fold to reduce the concentration of denaturant, prior to denaturation and analysis by SDS/PAGE (4–12 %) and immunoblotting (anti-V5 Ab). (F): Densitometric quantification of immunoreactive proteins in the supernatant (black columns) and the pellet fractions (white columns) in E demonstrated more Triton X-100 soluble (supernatant > pellet) compared to Triton X-100 insoluble (guanidine chloride soluble) G264S protein (n = 3). For comparison, 10 μg of cleaved recombinant WT hGK and mutant proteins were denatured and subjected to SDS/PAGE (4–12 %) analyses and Coomassie blue staining (G), and to native-PAGE (Novex 3–12 % Bis-Tris Gel) analyses by running at 150 V for 2 h in dark blue cathode buffers followed by Coomassie blue staining (H) and by immunoblotting (anti-V5 Ab) (I). Asterix indicates dimeric forms. Monomeric and dimeric forms were quantitated by densitometric analysis (for numbers see main text) and represent the average of 3 (A–D) and 4 (H and I) experiments.
2.4. Limited proteolysis by trypsin
We subjected non-tagged WT and mutant pancreatic hGK proteins to limited proteolysis by trypsin at a hGK to trypsin ratio of 500:1 (by mass). Proteolysis of 30 μg hGK was performed at 25 °C in a 100 μl reaction mixture containing 20 mM Hepes (pH 7.0), 50 mM NaCl, 2 mM DTT, and in the absence/presence of 40 mM glucose. Over time (0–30 min), the reactions were stopped by adding soybean trypsin inhibitor, using a protease to inhibitor ratio of 1:1.5 (by mass). We analyzed samples (4.5 μg) by SDS/PAGE after denaturation at 56 °C for 15 min. Full-length hGK was quantified by densitometric analysis using the Quantity One 1-D analysis software (Systat Software, San Jose, CA, USA), and the data were plotted using the Sigma Plot Software version 11.0 (Systat Software).
2.5. Degradation of hGK in an in vitro reticulocyte lysate system
We assessed the degradation of newly synthesized and ubiquitinated [35S]Met labeled WT and mutant pancreatic hGK proteins in the in vitro TnT® T7 Quick Coupled Transcription/Translation System as described [12]. Thus, co-translational hGK degradation was measured after 30 min at 30 °C in the absence/presence of 100 μmol/l proteasomal inhibitor (MG132) and 2 μmol/l Ub aldehyde. Samples were denatured (56 °C, 15 min) and analyzed by SDS/PAGE. We quantified total hGK by densitometric analysis using the Image Gauge v4.0 software (Fuji Film, Tokyo, Japan).
2.6. Assay of GK activity in transfected HEK293 cell lysate
We measured the activity of hGK in cytosolic fractions from early passages (<F4) of stably transfected HEK293 cells (Fig. S1 – Supplementary data) (MIN6 cells express GK and thus unsuitable for studying the activity of transfected mutants). Cells (1 × 107) were washed, harvested and pelleted by centrifugation (1372 g, 5 min, 4 °C). We resuspended cell pellets in ice-cold buffer containing 25 mM Hepes, 2.5 mM MgCl2, 25 mM KCl (pH 7.4), 1× protease inhibitor cocktail, and homogenized the cells by 35 passages through a ball-bearing cell cracker (EMBL, Heidelberg, Germany) with a clearance of 0.01 mm. Collected lysates were cleared by centrifugation (267000 g, 15 min, 4 °C) and GK-specific activity was measured at 30 °C in 100 μg of total protein as described under steady-state kinetics (section 2.3). The background high-affinity hexokinase activity was measured at 0.5 mM glucose and subtracted from the GK activity measured at 1–60 mM glucose.
2.7. Metabolic labeling and pulse-chase of hGK in HEK293 cells and MIN6 β-cells
3.5 × 105 HEK293 and MIN6 cells stably expressing V5-His tagged pancreatic hGK, were rinsed (PBS), followed by 1 h incubation in Met/Cys and serum free DMEM medium. Cells were then supplemented with 5 μCi/ml of [35S]Met/[35S]Cys for 30 min (pulse), rinsed, and further incubated in DMEM medium containing an excess (50×) of cold Met and Cys (chase). We collected the cells at various time points. To determine the effect of proteasomal inhibitor (MG132) and the lysosomal protease inhibitor (leupeptin) on the rate of hGK degradation, cells were treated with 10 μmol/l MG132 (DMSO in the control) and 100 μg/ml leupeptin (ddH2O in the control).
2.8. Native-PAGE electrophoresis
Stably transfected HEK293 and MIN6 cells were metabolically labeled with [35S]Met/[35S]Cys for 30 min and chased for 2 h as described in section 2.7. We washed and lysed cells in 1× FastBreak lysis buffer (MagneHis kit) containing additional 40 mM imidazole, 500 mM NaCl and protease inhibitors. His-tagged hGK soluble forms were isolated using the MagneHis Protein Purification System according to the manufacturer’s protocol. We eluted bound proteins by 500 mM imidazole/100 mM Hepes buffer (pH 7.5) and cleared samples by high-speed centrifugation (417200 g, 1 h, 4 °C). The supernatants were analyzed by native-PAGE (Novex 3–12 % Bis-Tris Gel) by running at 150 V for 1 h in dark blue, then at 200 V for 1 h in light blue cathode buffers. The gels were fixed, dried and labeled bands detected by autoradiography. Pellets containing membrane-associated hGK were resuspended in a SDS-containing buffer, denatured (56 °C, 15 min) and analyzed by SDS/PAGE and immunoblot analysis. Recombinant purified proteins were similarly analyzed by native-PAGE. Gels were Coomassie stained or immunoblotted and bands corresponding to monomeric/dimeric forms were quantitated by densitometric analysis.
2.9. Determination of Triton X-100 soluble and Triton X-100 insoluble mutant hGK
We washed and harvested 4 × 107 MIN6 cells, stably expressing the G264S mutant, in Hank’s balanced salt solution, pelleted cells by centrifugation (300 g, 5 min), and resuspended in washing solution (30 mM KCl, 140 mM NaCl, 10 mM EDTA, 25 mM Tris-HCl (pH 7.4)). After repeating centrifugation (300 g, 5 min), pelleted cells were resuspended in ice-cold homogenization medium (HS) (130 mM KCl, 25 mM NaCl, 1 mM EGTA, 25 mM Tris-HCl (pH 7.4)), supplemented with protease inhibitors, and lysed/homogenized by 35 passages through a cell cracker (see section 2.6). Cell breakage was monitored by phase-contrast optics [29]. After centrifugation (600 g, 10 min, 4 °C), we removed the post nuclear supernatant (PNS) fraction and recentrifuged (3000 g, 10 min, 4 °C) the sample to pellet heavy mitochondria and lysosomes [30]. The supernatant and the pellet (resuspended in HS) fractions were treated with 1 % (w/v) Triton X–100 for 30 min and centrifuged (100 000 g, 1 h, 4 °C). The supernatant was referred to as Triton X-100 soluble protein. We solubilized the pellet by sonication in 5 M guanidine chloride, 50 mM Tris (pH 8.0), including protease inhibitors, incubated overnight at room temperature, and centrifuged (13000 g, 20 min). Samples were diluted 10-fold to reduce the concentration of denaturant. Both supernatant and pellet fractions were denatured (56 °C, 15 min) before analyzed by SDS/PAGE and immunoblotting.
2.10. CD spectroscopy
WT and G264S hGK proteins were isolated from the GST-tag as previously described [28]. The isolated proteins were further diluted in a sodium phosphate buffer (pH 7.2) with 5 mM DTT to a final concentration of 10 μM. The circular dichroism (CD) was recorded on a Jasco J-810 spectropolarimeter. Thermal unfolding (15–90 °C) was determined by following the change in ellipticity at 222 nm (light path 1mm) at a constant heating rate of 40 °C · hr−1. Using the first derivative of the smoothed curve, the midpoint of the transition (Tm) was determined. The spectra data were plotted using Sigma Plot Software version 11.0.
2.11
Plasmid constructs, cell lines and stable transfection, immunoprecipitation and mass spectrometry analyses; see Supplementary materials and methods.
2.12. Statistical analysis
Data obtained from independent experiments (n ≥ 3) are presented as mean ± SD and student’s t-test was conducted for statistical analysis of quantitative data (p<0.05 was considered significant).
2.13. Structural analyses
We computed the effect of the mutations on protein thermodynamic stability and protein conformation using the Medusa Force Field and Modeling Suite [31,32]. The algorithm (http://eris.dokhlab.org) features an all-atom force field, a fast side-chain packing algorithm and a backbone relaxation method. It models the backbone flexibility, thereby allowing for determination of the mutation-induced backbone conformational changes. We generated structural images using PyMol version 1.1 [33]. The static solvent accessibility of the individual residues in the two main conformational states was calculated using the CUPSAT algorithm [34].
3. Results
3.1. Catalytic properties of WT hGK and GCK-MODY mutant enzymes
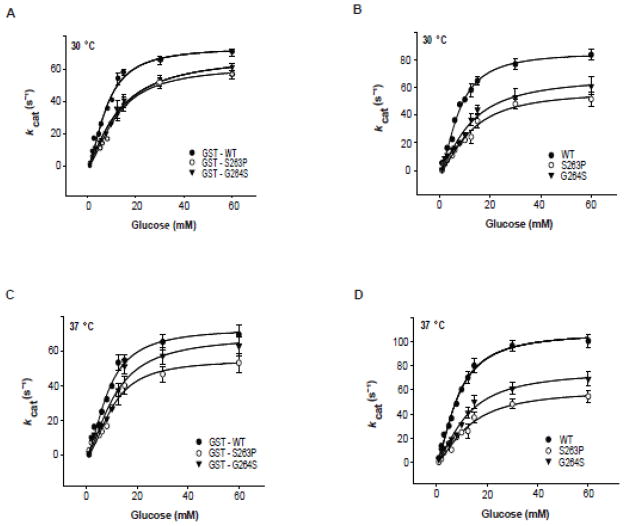
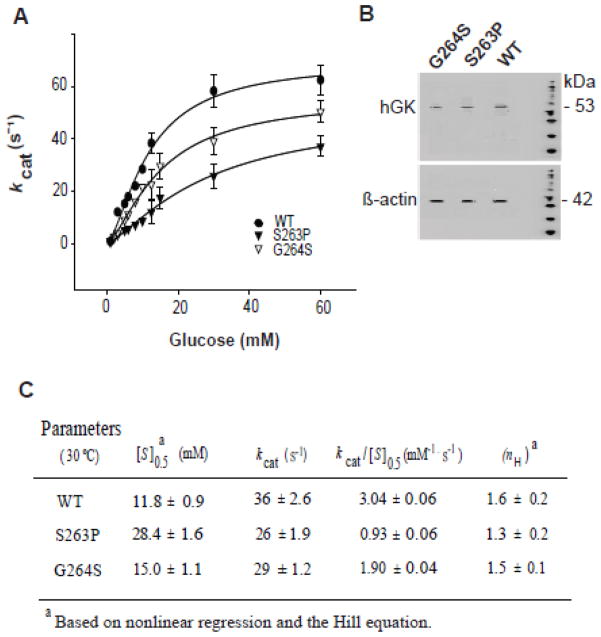
We subjected the recombinant WT and mutant pancreatic enzymes to steady-state kinetic analyses at 30 and 37 °C as GST-tagged and tag-free (cleaved) proteins (Fig. 1, Table 1). At both temperatures, the S263P and G264S mutants demonstrated slightly reduced overall turnover rates (kcat) and increased [S]0.5 values for Glc compared to WT hGK, consistent with previous reports [21,27]. Moreover, the Km value for ATP was reduced for S263P (Table S1 – Supplementary data). For this mutant, the reduced catalytic efficiency was most clearly observed with the tag-free enzyme at 37 °C (Table 1).
Fig. 1.
Steady-state kinetic properties of recombinant WT and mutant pancreatic hGK with Glc as the variable substrate. (A–D): The activity of 0.5 μg recombinant GST-tagged (A and C) and tag-free enzyme (B and D) as measured spectrophotometrically at 30 °C (A and B) and 37 °C (C and D). The data were analyzed by nonlinear regression analysis using the Hill equation. The steady-state kinetic constants are summarized in Table 1.
Table 1.
Steady-state kinetic parameters of WT and mutant hGK forms.
| kcat (s−1) | [S]0.5 (mM) | kcat/[S]0.5 (mM−1· s−1) | (n H) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| + GST | − GST | + GST | − GST | + GST | − GST | + GST | − GST | ||
| Parameters (30 °C) | WT | 55 ± 3 | 64 ± 2 | 8.0 ± 0.8 | 8.0 ± 0.5 | 6.9 ± 0.28 | 8.1 ± 0.24 | 1.7 ± 0.2 | 1.7 ± 0.1 |
| S263P | 46 ± 3 | 42 ± 4 | 12 ± 1.2 | 12 ± 1.8 | 3.9 ± 0.23 | 3.6 ± 0.08 | 1.6 ± 0.2 | 1.6 ± 0.3 | |
| G264S | 48 ± 5 | 49 ± 4 | 11 ± 1.9 | 11 ± 1.5 | 4.4 ± 0.09 | 4.4 ± 0.22 | 1.6 ± 0.3 | 1.6 ± 0.2 | |
|
| |||||||||
| Parameters (37 °C) | WT | 59 ± 5 | 79 ± 4 | 8.0 ± 1.2 | 8.0 ± 0.7 | 7.4 ± 0.39 | 9.9 ± 0.10 | 1.7 ± 0.3 | 1.7 ± 0.2 |
| S263P | 49 ± 5 | 44 ± 4 | 12 ± 1.5 | 12 ± 1.9 | 4.0 ± 0.16 | 3.6 ± 0.18 | 1.6 ± 0.2 | 1.6 ± 0.2 | |
| G264S | 52 ± 5 | 55 ± 5 | 11 ± 1.9 | 11 ± 1.7 | 4.7 ± 0.04 | 5.1 ± 0.33 | 1.6 ± 0.5 | 1.6 ± 0.3 | |
The catalytic activity was measured spectrophotometrically at 30 and 37 °C for GST-tagged and tag-free (factor Xa cleaved) enzymes. Analyses were performed using non-linear regression analysis and the Hill equation. The values were obtained from measurements at various glucose concentrations (1–60 mmol/l) and various ATP concentrations (0.025–5 mmol/l) and are representative of 3 different protein preparations.
3.2. Probing protein conformations by limited proteolysis with trypsin
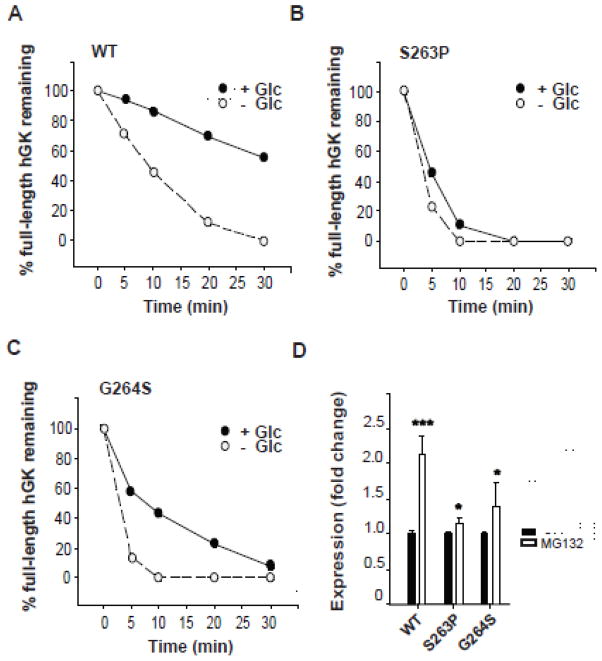
Limited proteolysis is a commonly used tool for probing the folding state of a protein and characterization of mutational effects on protein conformation and stability [35,36]. To evaluate the effect of the GK residue changes S263P and G264S on the native folding of the pancreatic protein, we compared the susceptibility of the WT and mutant enzymes to limited proteolysis by trypsin. As seen from Fig. 2 the S263P mutant protein demonstrated a higher susceptibility to limited proteolysis shown by a more rapid cleavage than the WT enzyme; the full-length protein was barely detectable after 10 min both in the absence and presence of glucose (Fig. 2B). The G264S mutant demonstrated a similar time course for its proteolysis as S263P in the absence of glucose (Fig. 2C), but was partly protected by glucose.
Fig. 2.
Probing protein conformations by limited proteolysis with trypsin and stability. (A–C): Time course for the limited proteolysis of WT and mutant forms by trypsin at 25 °C in the absence (white circles) and presence (black circles) of 40 mmol/l glucose. 4.5 μg of protein was removed at various time points, denatured and analyzed by SDS/PAGE (10 %), followed by Coomassie blue staining. Full-length protein was quantified by densitometric analysis and plotted as a function of time (A–C). Each time point represents the average of two individual experiments (n = 2). (D): In vitro stability of newly synthesized and partly ubiquitinated hGK forms, and the effect of the proteasome inhibitor MG132. His6-WT and -mutant forms were expressed ([35S]Met-labeled) in an in vitro coupled transcription-translation RRL system and their stability measured after 30 min at 30 °C, in the absence (black columns) and presence (white columns) of proteasome inhibitor (100 μmol/l MG132). Samples were denatured and analyzed by SDS/PAGE (10 %), autoradiography and densitometric quantification of total GK expression. Each column represents the mean ± SD of a minimum of five independent experiments (n = 5).
3.3. Proteasomal degradation of recombinant hGK
We further assessed the stability of WT and mutant pancreatic hGK enzymes by their susceptibility to proteasomal degradation [12]. [35S]Met-labeled His6-hGK proteins were expressed in an in vitro coupled transcription-translation system, and their degradation was followed in the same system (Fig. 2D). SDS/PAGE analysis and densitometric quantification of the protein samples revealed a ~2.1-fold increase in the recovered WT enzyme in the presence of the proteasomal inhibitor MG132. We observed a smaller effect of the inhibitor for the recovery of the S263P (~1.2-fold increase) and G264S (~1.4-fold increase) mutant proteins (Fig. 2D).
3.4. Degradation of hGK in stably transfected HEK293 cells and MIN6 β-cells
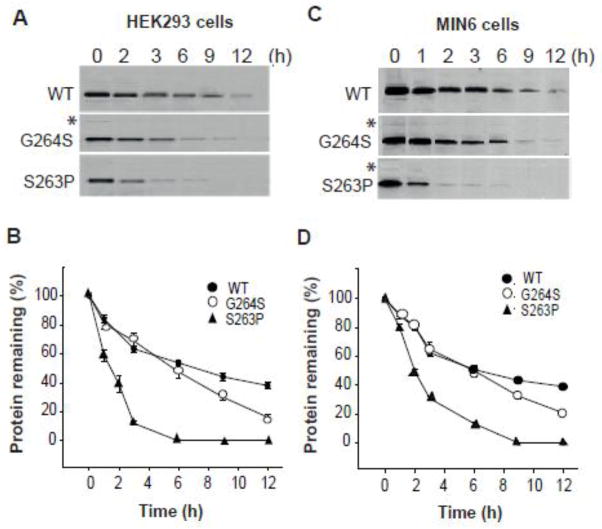
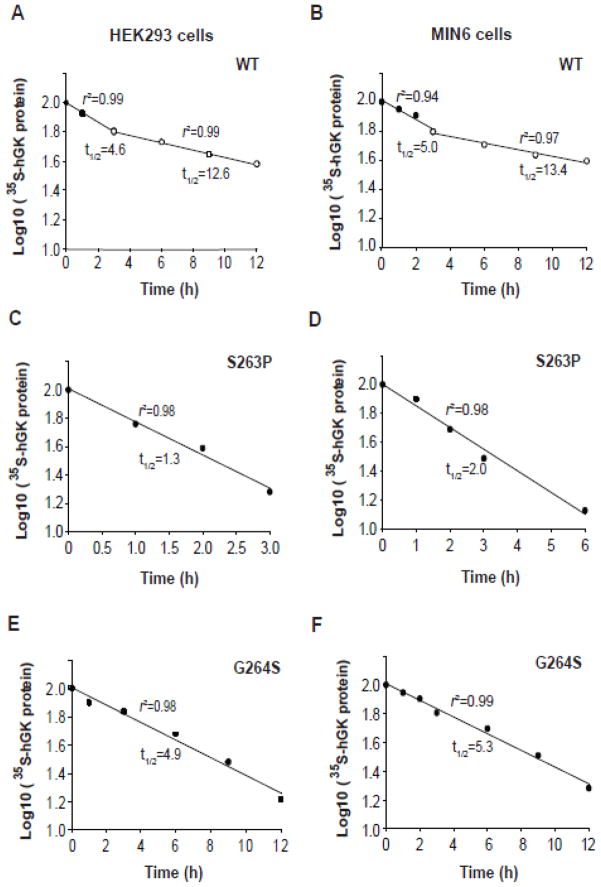
We then studied the stability/turnover of the S263P and G264S mutant pancreatic proteins in stably transfected HEK293 (Fig. 3A and B) and MIN6 cells (Fig. 3C and D). HEK293 cells were chosen as supplementary cell line due to its high efficiency of transfection and protein production. Cells were metabolically labelled for 30 min and chased for a period of 0–12 h, followed by SDS/PAGE analysis and quantification of the immunoisolated proteins (Fig. 3A and C). The recovery of full-length V5-tagged human GK (hGK) (~53 kDa) decreased as a function of time, and the apparent protein half-life (t1/2) was determined from semi-logarithmic plots (Fig. 4) of the data (Fig. 3). WT hGK revealed a biphasic time course for its degradation in both cell lines, with apparent t1/2 values of 5.0 and ~13 h (Fig. 4A and B). By contrast, we observed a monophasic time course for the recovery of the S263P and G264S mutant proteins (~53 kDa), with apparent half-lives of 1.3 and 2.0 h for S263P (Fig. 4C and D) and 4.9 and 5.3 h for G264S (Fig. 4E and F), in HEK293 and MIN6 cells, respectively.
Fig. 3.
Time course of the cellular degradation of WT and mutant hGK forms. Stably transfected HEK293 cells (A and B) and MIN6 β-cells (C and D) were labeled with [35S]Met/[35S]Cys for 30 min and chased for 12 h. The recovery of WT, S263P and G264S mutant proteins was monitored by immunoprecipitation (anti-V5 Ab). Samples were denatured at 56 °C and analyzed by SDS/PAGE (4–12 %) and autoradiography (A and C). The bands of full-length V5-His tagged monomeric protein (~53 kDa) were quantified by densitometric analysis and the data plotted as a function of chase time (B and D). The mean value of triplicates at time zero was normalized to 100 % and used as a reference value. Data for each time point represents the mean ± SD of triplicates examined on three independent days (n = 9). Asterix indicates trace amount of possible dimeric forms (see Fig. 6).
Fig. 4.
Determination of the apparent cellular half-life of WT and mutant hGK forms. (A–F): Semi-logarithmic plot of the time course of the degradation of WT, S263P and G264S mutant forms (data from Fig. 3). Linear regression analysis revealed a biphasic time course for WT hGK in HEK293 (A) and MIN6 cells (B), whereas a monophasic time course was observed for the S263P mutant in HEK293 (C) and MIN6 (D) cells, and the G264S mutant in HEK293 (E) and MIN6 (F) cells.
3.5. Effect of proteasomal and lysosomal inhibitors on the degradation of hGK in stably transfected HEK293 cells and MIN6 β-cells
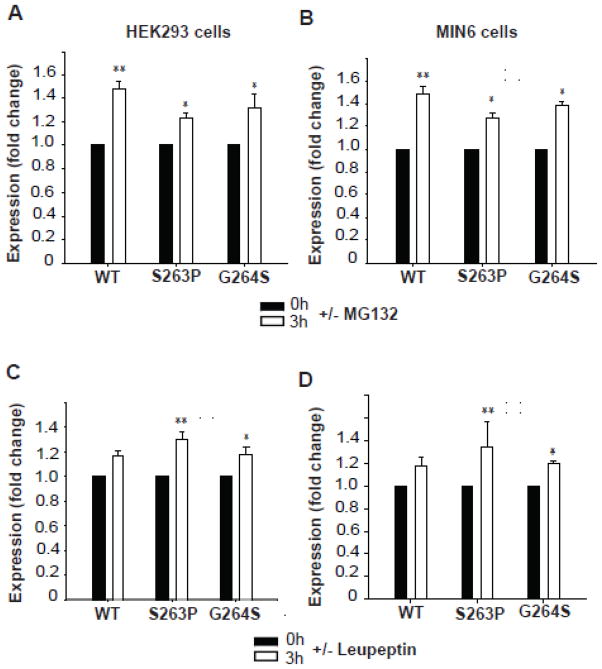
Considering the increased rate of degradation of the S263P and G264S mutant proteins in HEK293 and MIN6 cells (Figs. 3 and 4), we next investigated how the cellular proteolytic systems are involved in their quality control and turnover. The degradation was studied by pulse-chase experiments in stably transfected HEK293 and MIN6 cells in the presence and absence (during the chase) of the proteasomal inhibitor MG132 (10 μmol/l) (Fig. 5A and B). After a 3 h chase, the recovery of WT protein increased ~1.5-fold in HEK293 cells in the presence of inhibitor, while the S263P and G264S mutants increased ~1.2- and 1.3-fold, respectively (Fig. 5A). We observed a similar increase in recovery of WT and mutant proteins in stably transfected MIN6 cells (Fig. 5B) that also correlated with our observations in the in vitro RRL system (Fig. 2D).
Fig. 5.
Effect of proteasomal and lysosomal inhibitors on the apparent cellular stability of WT and mutant hGK forms. The recovery of the ~53 kDa band of WT hGK and the S263P and G264S mutant forms in stably transfected HEK293 cells (A and C) and MIN6 cells (B and D) was followed in pulse-chase experiments. The cells were labeled with [35S]Met/[35S]Cys for 30 min and chased at 0 h (black columns) and 3 h (white columns), in the presence and absence of proteasomal inhibitor (A and B) and lysosomal inhibitor (C and D). Cell lysis was followed by immunoprecipitation (anti-V5 Ab) of total detergent soluble GK. Samples were denatured and analyzed by SDS/PAGE (4–12 %), autoradiography and densitometric analysis of the radioactive bands. Each column represents the mean ± SD of triplicates examined on three independent days (n = 9). The fold change after 3 h refers to the difference in the average recovery at t = 3 h relative to the average recovery at t = 0 h (=1.0).
Due to the relatively long half-life of WT hGK in HEK293 and MIN6 cells (Fig. 4), we also investigated the contribution of the lysosomal degradation system in the turnover of hGK in both cell lines. Here, the chase period (0–3 h) was conducted in the presence or absence of leupeptin, an inhibitor of lysosomal protein degradation (Fig. 5C and D). The fold increase in the recovery of cellular mutant proteins was significantly (p<0.004) higher (S263P>G264S) than for WT hGK in both HEK293 and MIN6 cells (Fig. 5C and D).
3.6. Intracellular dimerization and aggregation of the S263P and G264S mutant proteins
We further compared the recovery of [35S]Met/[35S]Cys-labeled WT and mutant enzymes, isolated from cell extracts (HEK293 and MIN6 cells) (Fig. 6A and C), by native-PAGE analyses. Moreover, the E. coli expressed and purified recombinant proteins were also analyzed and compared by native-PAGE electrophoresis (Fig. 6H and I). In the cytosolic fractions from both cell lines, the immunoisolated mutant proteins revealed radioactive bands with a relative mobility corresponding to dimeric forms (~100 kDa), in addition to their monomeric forms of ~53 kDa (proteins with V5- and His-tag), which were present at zero time and during chase time (Fig. 6A and C). The relative content of the dimeric form represented 50–60 % (both mutants in HEK293), and 45–55 % (S263P) and 30–35 % (G264S) in MIN6 cells (n = 3). A similar high molecular-mass (dimeric) band (~100 kDa) was also detected by native-PAGE for both mutant recombinant enzymes (Fig. 6H and I), and estimated to represent ~5 % and ~10 % of S263P and G264S total protein, respectively (n = 4). These dimeric species were confirmed to represent 55 % (S263P) and 68 % (G264S) GK peptide sequence by mass spectrophotometric (LC-MS/MS) analyses (Orbitrap Velos Pro) (data not shown). Moreover, dimeric and aggregated forms were also recovered for the mutants from the pellets of the PNS fractions, enriched in lysosomes and mitochondria (Fig. 6B and D). These aggregated forms were most pronounced in MIN6 cells (Fig. 6D), representing 70–80 % (G264S>S263P) of total immunoreactive protein. Only the monomeric form was detected for the WT enzyme (Fig. 6A–D). Since the G264S mutant revealed the highest propensity to form SDS-resistant dimers (and aggregates), we further investigated the subcellular distribution of this mutant. The majority of the monomeric and dimeric species was recovered in the PNS fraction (Fig. 6E and F) and was mostly Triton X-100 soluble and partly Triton X-100 insoluble, but soluble in guanidine chloride (Fig. 6E).
3.7. Catalytic activity of WT, S263P and G264S mutant enzymes stably expressed in HEK 293 cells
To assess how the mutations affect the cellular GK catalytic activity, we measured the activity of WT and mutant pancreatic enzymes at 30 °C in cytosolic fractions from stably transfected HEK293 cells (Fig. 7) with glucose as the variable substrate (1–60 mM). Using equal amounts of cellular monomeric GK protein, as determined by immunoblotting (Fig. 7B), the homospecific activity of both mutants was lower than of the WT enzyme (Fig. 7C), resembling to that determined for the purified recombinant enzymes measured at 30 °C (Fig. 1, Table 1), however with the S263P cellular enzyme demonstrating a much lower affinity for Glc.
Fig. 7.
Catalytic activity of WT, S263P and G264S enzymes in HEK293 cell extracts. (A): Glucose-dependent GK activity in cytosolic fractions of stably transfected HEK293 cells, prepared by mechanical shear forces. The activity was measured spectrophotometrically at 30 °C at equal amounts of cellular GK (~100 μg of total protein), as determined by SDS/PAGE (10 %) and immunoblotting (anti-GK and anti-actin Ab) (B). (C): Kinetic parameters calculated from data in (A) by nonlinear regression analysis using the Hill equation. Data represents the mean ± SD of three independent experiments (n = 3).
3.8. Structural analysis
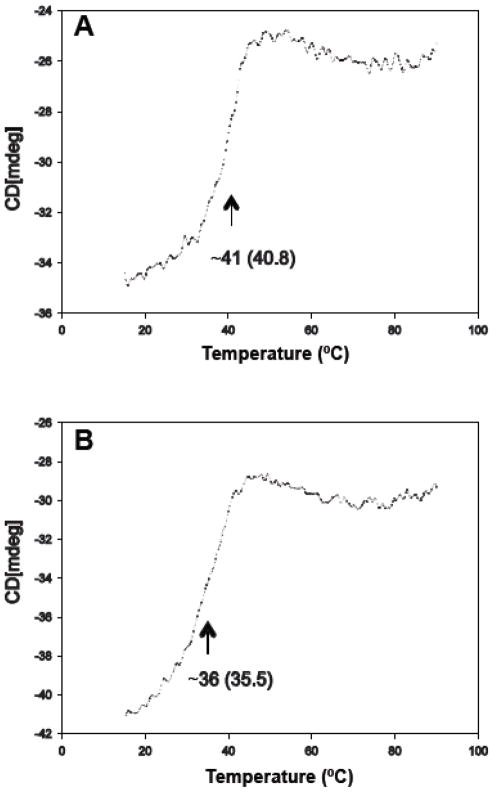
Recombinant hGK without a fusion partner is marginally stable under physiological conditions. It has a low intrinsic thermal stability with an apparent Tm of ~ 41 °C (Fig. 8A) and a propensity to aggregate at Tp>40 °C [37,38]. Here we obtained a Tm of 36 °C for the G264S mutant enzyme (Fig. 8B), ~5 °C below the WT, compatible with a misfolded protein. Moreover, using an ANS fluorescence binding assay, a reduction in solvent exposed hydrophobic clusters in the mutant vs WT enzyme was demonstrated (Fig. S2).
Fig. 8.
Thermal unfolding of WT and G264S hGK. The thermal unfolding profile of WT (A) and G264S (B) at a concentration of 10 μM in the absence of Glc was obtained by following the change in ellipticity at 222 nm at a constant heating rate of 40 °C · h−1. From the first derivative of the smoothed denaturation curve, apparent transition temperatures of ~40.8 °C (A) and ~35.5 °C (B) were determined for the WT and G264S hGK, respectively.
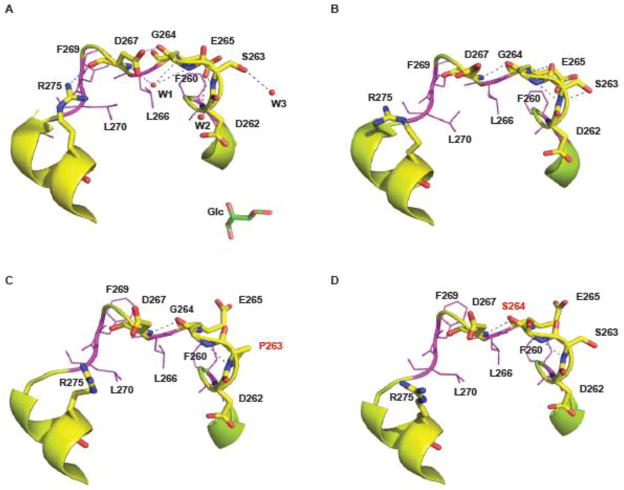
In the closed, glucose-bound conformation of pancreatic hGK (PDB i.d. 1v4s), the residues S263 and G264 are positioned in an eleven-residue (F260 – L270) loop structure (Fig. 9). In this loop, the polypeptide chain folds back on itself, and its conformation is stabilized by a network of hydrogen bonds (possibly involving three water molecules) and hydrophobic interactions (residues F260, L266 and L270), which project into the center of the loop, forming a cluster of packed apolar side-chains with very low static solvent accessibility. A very similar backbone conformation is observed for the loop structure in the ligand-free open conformation (PDB i.d. 1v4t), with only minor differences in the phi and psi angles and in the hydrogen-bonding network (Fig. 9B). Using the flexible-backbone method the Eris algorithm predicts local changes in the loop structure including side-chain motions (E265, D267 and F269) and changes in the hydrogen-bonding network of the loop (Fig. 9C and D). Moreover, a shortening of the L271-S280 α-helix is indicated. Direct simulation approaches have not been attempted to further predict the structural changes associated with the mutations, including the interphase of the homodimers observed for both mutant proteins.
Fig. 9.
The position of S263 and G264 in the 3D structure of (A) the Glc-bound closed conformation (PDB i.d. 1v4s) and (B) the ligand-free conformation (PDB i.d. 1v4t) of pancreatic hGK. The Cα-atom trace of the eleven residue loop structure (F260-L270) is shown in ribbon, with key residues in stick model (D262, S263, G264 and E265) and line model (F260, L266, F269 and L270). The polypeptide chain folds back on itself and the 3D structure is stabilized by hydrogen bonds and hydrophobic interactions formed by a cluster of packed apolar sidechains. (C) and (D): In silico predicted conformational changes of the loop structure in the S263P (C) and G264S (D) mutant proteins using the Eris algorithm. The figure was created using PyMol, version 1.1.
4. Discussion
The current spectrum of GCK-MODY mutations consists of >600 mutations [17], and various mechanisms have been demonstrated/proposed to explain how the mutations cause diabetes. Functional characterization of recombinant enzymes in vitro have identified some mutant forms (V62M, E70K, G72R, L165F, V203A, S263P, E265K, M298K, E300K and K414E) with an apparent reduced thermal stability, as measured by heat-inactivation of the GST-GK fusion proteins at 30 – 52.5 °C [20,21,23,39–44]. This in vitro instability has been suggested to represent the molecular mechanism for the hyperglycemic phenotype either alone or in combination with a reduced catalytic efficiency and/or a ‘dysregulation’ by the liver specific inhibitory protein GKRP or the activator protein PFK-2/FBPase-2 [7,17,21,22, 25,45,46]. In the present study, we have focused on two mutations (amino acid substitutions S263P and G264S) previously described in one patient with GCK-MODY and one with neonatal diabetes, respectively [21,27]. The G264S mutation has later also been found to coseggregate with MODY diabetes in three families (Christine Bellanné-Chantelot, personal communication). Both mutations have been reported to result in recombinant enzymes with normal/near normal steady-state kinetics [20,21,27,42], and normal regulation by GKRP [21,27], however lower cellular GK specific activity [25]. The S263P mutation is also reported to cause a thermal instability of the recombinant GST-GK fusion protein [21,44], as well as a cellular instability [25].
4.1. Comparison of GST-GK and tag-free GK at 30 and 37 °C
Up to date multiple studies [18,22,28,47] comparing the functional properties of GST-GK and the tag-free pancreatic enzyme, have revealed similar enzyme kinetic parameters for the two forms. By contrast, in terms of thermal stability, the GST fusion partner may have a stabilizing effect on GK, which is indicated by our comparison of WT and S263P mutant protein with regard to the apparent overall turnover rate (kcat) assayed at 30 and 37 °C, in the presence and absence of GST (Fig. 1, Table 1). A stabilizing effect of GST on the mutant protein is not surprising since WT hGK without the fusion partner has a marginal thermal stability (apparent Tm ~41 °C in Fig. 8A) and a propensity to aggregate at Tp ≥ 40 °C as measured by CD spectroscopy [18]. Our data are consistent with GST being a transiently stabilizing protein for hGK, and notably for its mutant proteins with a lower Tm value than for the WT (Fig. 8B), as previously observed for the GST fusion protein with the catalytic domain of the nuclear inclusion protease from tobacco etch virus (TEV) [48], an effect which may be explained by dimerization of the GST fusion protein [49].
4.2. Cellular degradation and self-association of the S263P and G264S mutant proteins
While it is established that a complex network of mechanisms is involved in the regulation of GK catalytic activity in pancreatic β–cells and hepatocytes, relatively little is known regarding the cellular protein quality control (PQC) machinery involved in the recognition and targeted degradation of GK and its many diabetes-associated mutant forms [17]. When stably expressed in HEK293 cells and insulin-secreting MIN6 β-cells, both mutant proteins (~53 kDa) revealed an increased rate of degradation (Fig. 3) with monophasic decay curves and reduced apparent half-lives, most pronounced for S263P (Fig. 4C and D). Whether the increased rate of degradation of the mutants is compensated by an increased level of their cellular mRNA expression was not investigated here, but has not been found to be the case in other GCK instability mutations [24,50]. By contrast, WT hGK revealed a biphasic decay curve (Fig. 4A and B) in which the second and longest apparent half-life (~13 h) corresponds well with the value (12.7 h) previously estimated for rat GK (rGK) in rat liver hepatocytes [51], where the enzyme was considered to be non-selectively sequestered and degraded by the autophagosomal-lysosomal pathway. In our study, when the lysosomal proteolytic activity was inhibited by leupeptin, the recovery of cellular GK proteins (~53 kDa) increased (Fig. 5C and D). Our data indicate that pancreatic hGK is also partly cleared by autophagy in both HEK293 and MIN6 cells, possibly due to a preference for targeting and degradation of oligomeric/aggregated mutant forms [52]. For both mutant proteins, aggregates were observed (G264S>S263P; G264S aggregates were Triton X-100-insoluble (guanidine chloride soluble)) in HEK293 and MIN6 cells, in both the cytosolic fraction and a subcellular fraction enriched in lysosomes and mitochondria (Fig. 6). Previously, we have demonstrated that WT pancreatic and liver hGK are covalently modified by ubiquitination, serving as a signal for the degradation by the ubiquitin-proteasome system (UPS) [12]. Here, when the proteasomal proteolytic activity was blocked by the inhibitor MG132, the recovery of the two misfolded mutant proteins slightly increased, both in the in vitro coupled transcription/translation (RRL) system (Fig. 2D) and when stably expressed in HEK293 or MIN6 cells (Fig. 5A and B).
4.3. In cellulo dimerization of S263P and G264S mutant proteins
Whereas the tag-free recombinant forms of E. coli expressed S263P and G264S revealed a single monomeric (~ 50 kDa) band on SDS/PAGE (Fig. 6G), both mutant proteins demonstrated an additional band with a relative mobility corresponding to homodimeric GK species on native-PAGE (Fig. 6H and I), and confirmed by MS analyses (data not shown). Similar dimeric species were seen when stably expressed as V5-His tagged proteins in cells (Fig. 6A–D). This observation requires a further comment. In general, 3D domain swapping represents a common oligomerization mechanism in which two or more polypeptide chains exchange identical units, and when two monomers are involved, homodimers are formed [53, 54]. In search for local signal that may cause swapping, attention has focused on loop regions which adopt a different conformation in monomeric and domain swapped forms [55]. Proline residues are frequently found in loop regions involved in domain swapped dimer formation, representing the thermodynamically most stable state [56]. The F260-L270 loop of hGK may similarly promote domain swapping when S263 and G264 are mutated to Pro and Ser, respectively. Our in silico analyses predict conformational changes in the loop of both mutant forms (Fig. 9C and D), forming a conformation that promotes the formation of homodimers. A similar mechanism has previously been reported for the dimerization of monomeric immunoglobulin-binding domain B1 of streptococcal protein G (GB1) by a single-point mutation [57]. The resistance of the hGK mutant dimers to denaturation by SDS at 56 °C (Fig. 6B and D) suggests the formation of a metastable compact, globular dimeric structure, as previously demonstrated for dimeric diphtheria toxin [53] and dimeric cyanivirin-N [56]. Moreover, using an ANS fluorescence binding assay, it is demonstrated (Fig. S2) a reduction in solvent exposed hydrophobic clusters in the mutant vs the WT enzyme, possibly related to a reduced accessibility of the hydrophobic residues in the F260-L270 loop structure of the mutant and dimerization involving these residues. Considering the stabilizing effect of E265 on the WT loop conformation (Fig. 9A and B), it is not surprising that the GCK-MODY mutant E265K has been reported to be very thermolabile, even as GST-GK fusion protein, while the mutation barely affected the protein catalytic efficiency at 30 °C [41]. That formation of dimers/oligomers of S263P and G264S is observed most pronounced when the mutants are expressed in cells (Fig. 6), may be related to the stabilization of a conformationally more “open” monomer by molecular chaperones and the frequently observed effect of high protein concentration on protein self-association in the cellular environment. Thus, macromolecular crowding influences the structure and function of proteins under physiological conditions and promotes protein-protein interactions [55,58–61]. It remains to be seen how protein self-association influences GK catalytic properties, as well as its association with previously reported subcellular localizations, including mitochondria, insulin granula, microtubuli and nucleus [62–65].
4.4. Relevance for the study of GK aggregation and degradation
In summary, our study has revealed an unexpected structural role of the F260-L270 loop of hGK and demonstrated that the GCK-MODY mutations S263P and G264S generate misfolded and conformationally unstable monomeric proteins which result in (i) an increased sensitivity to limited proteolysis in vitro, (ii) an enhanced rate of degradation when expressed in HEK293 cells and MIN6 cells (S263P>G264S), and (iii) an increased propensity to self-associate and form homodimers/higher oligomers and aggregates (G264S>S263P) in these cells. Our data highlight the variety of molecular mechanisms leading to GCK-MODY diabetes and underline the importance of a cell-biological approach to fully characterize the mutant enzymes, since protein destabilization/aggregation and degradation may contribute to the molecular cause behind other GCK-MODY mutations. The study also suggests a possible new therapeutic approach, different from GK allosteric activators, by prevention of self-association (conformational rescue) of some mutant proteins using bioactive small molecules (chemical/pharmaceutical chaperones).
Supplementary Material
Highlights.
GCK-MODY mutations S263P and G264S cause reduced GK catalytic activity (S263P<G264S)
The mutations cause misfolded proteins (S263P>G264S)
The mutant proteins self-associate (G264S>S263P) and form homo-dimers/aggregates
Prevention of self-association may represent new therapeutic approach
Acknowledgments
This work was supported by funds from the Research Council of Norway, Helse Vest, the Novo Nordisk Foundation, the Nils Norman Foundation, the Bergen Medical Research Foundation, the Meltzer Foundation, and the University of Bergen. We thank João Leandro, Department of Biomedicine, University of Bergen, for his supporting information and Ali Sepulveda Munõz, Department of Biomedicine, University of Bergen, for expert technical assistance with the French press. The mass spectrometry analyses were conducted by the Proteomics Unit at the University of Bergen (PROBE), supported by the National Program for Research in Functional Genomics (FUGE) funded by the Norwegian Research Council.
Abbreviations
- Ab
antibody
- GK
glucokinase
- GKRP
glucokinase regulatory protein
- Glc
α-D-glucose
- HEK293
Human Embryonic Kidney 293 cells
- hGK
human glucokinase
- MIN6
mouse insulinoma cell
- MODY
maturity-onset diabetes of the young
- PDB
protein data bank
- PNS
post nuclear supernatant
- RRL
rabbit reticulocyte lysate
- TEV protease
tobacco etch virus protease
- UPS
ubiquitin-proteasome system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Katzen HM, Schimke RT. Multiple forms of hexokinase in the rat: tissue distribution, age dependency, and properties. Proc Natl Acad Sci U S A. 1965;54:1218–1225. doi: 10.1073/pnas.54.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meglasson MD, Burch PT, Berner DK, Najafi H, Vogin AP, Matschinsky FM. Chromatographic resolution and kinetic characterization of glucokinase from islets of Langerhans. Proc Natl Acad Sci U S A. 1983;80:85–89. doi: 10.1073/pnas.80.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magnuson MA, Shelton KD. An alternate promoter in the glucokinase gene is active in the pancreatic beta cell. J Biol Chem. 1989;264:15936–15942. [PubMed] [Google Scholar]
- 4.Jetton TL, Liang Y, Pettepher CC, Zimmerman EC, Cox FG, Horvath K, Matschinsky FM, Magnuson MA. Analysis of upstream glucokinase promoter activity in transgenic mice and identification of glucokinase in rare neuroendocrine cells in the brain and gut. J Biol Chem. 1994;269:3641–3654. [PubMed] [Google Scholar]
- 5.Matschinsky FM, Ellerman JE. Metabolism of glucose in the islets of Langerhans. J Biol Chem. 1968;243:2730–2736. [PubMed] [Google Scholar]
- 6.Matschinsky FM. Glucokinase as glucose sensor and metabolic signal generator in pancreatic beta-cells and hepatocytes. Diabetes. 1990;39:647–652. doi: 10.2337/diab.39.6.647. [DOI] [PubMed] [Google Scholar]
- 7.Van Schaftingen E, Detheux M, Veiga da Cunha M. Short-term control of glucokinase activity: role of a regulatory protein. FASEB J. 1994;8:414–419. doi: 10.1096/fasebj.8.6.8168691. [DOI] [PubMed] [Google Scholar]
- 8.Shiota C, Coffey J, Grimsby J, Grippo JF, Magnuson MA. Nuclear import of hepatic glucokinase depends upon glucokinase regulatory protein, whereas export is due to a nuclear export signal sequence in glucokinase. J Biol Chem. 1999;274:37125–37130. doi: 10.1074/jbc.274.52.37125. [DOI] [PubMed] [Google Scholar]
- 9.Baltrusch S, Lenzen S, Okar DA, Lange AJ, Tiedge M. Characterization of glucokinase-binding protein epitopes by a phage-displayed peptide library. Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel interaction partner. J Biol Chem. 2001;276:43915–43923. doi: 10.1074/jbc.M105470200. [DOI] [PubMed] [Google Scholar]
- 10.Massa L, Baltrusch S, Okar DA, Lange AJ, Lenzen S, Tiedge M. Interaction of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFK-2/FBPase-2) with glucokinase activates glucose phosphorylation and glucose metabolism in insulin-producing cells. Diabetes. 2004;53:1020–1029. doi: 10.2337/diabetes.53.4.1020. [DOI] [PubMed] [Google Scholar]
- 11.Rizzo MA, Piston DW. Regulation of beta cell glucokinase by S-nitrosylation and association with nitric oxide synthase. J Cell Biol. 2003;161:243–248. doi: 10.1083/jcb.200301063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjorkhaug L, Molnes J, Sovik O, Njolstad PR, Flatmark T. Allosteric activation of human glucokinase by free polyubiquitin chains and its ubiquitin-dependent cotranslational proteasomal degradation. J Biol Chem. 2007;282:22757–22764. doi: 10.1074/jbc.M700517200. [DOI] [PubMed] [Google Scholar]
- 13.Froguel P, Zouali H, Vionnet N, Velho G, Vaxillaire M, Sun F, Lesage S, Stoffel M, Takeda J, Passa P, et al. Familial hyperglycemia due to mutations in glucokinase. Definition of a subtype of diabetes mellitus. New Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 14.Timsit J, Bellanne-Chantelot C, Dubois-Laforgue D, Velho G. Diagnosis and management of maturity-onset diabetes of the young. Treat Endocrinol. 2005;4:9–18. doi: 10.2165/00024677-200504010-00002. [DOI] [PubMed] [Google Scholar]
- 15.Njolstad PR, Sovik O, Cuesta-Munoz A, Bjorkhaug L, Massa O, Barbetti F, Undlien DE, Shiota C, Magnuson MA, Molven A, Matschinsky FM, Bell GI. Neonatal diabetes mellitus due to complete glucokinase deficiency. New Engl J Med. 2001;344:1588–1592. doi: 10.1056/NEJM200105243442104. [DOI] [PubMed] [Google Scholar]
- 16.Glaser B, Kesavan P, Heyman M, Davis E, Cuesta A, Buchs A, Stanley CA, Thornton PS, Permutt MA, Matschinsky FM, Herold KC. Familial hyperinsulinism caused by an activating glucokinase mutation. New Engl J Med. 1998;338:226–230. doi: 10.1056/NEJM199801223380404. [DOI] [PubMed] [Google Scholar]
- 17.Osbak KK, Colclough K, Saint-Martin C, Beer NL, Bellanne-Chantelot C, Ellard S, Gloyn AL. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum mutat. 2009;30:1512–1526. doi: 10.1002/humu.21110. [DOI] [PubMed] [Google Scholar]
- 18.Molnes J, Teigen K, Aukrust I, Bjorkhaug L, Sovik O, Flatmark T, Njolstad PR. Binding of ATP at the active site of human pancreatic glucokinase--nucleotide-induced conformational changes with possible implications for its kinetic cooperativity. FEBS J. 2011;278:2372–2386. doi: 10.1111/j.1742-4658.2011.08160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.OS, Gloyn AL, Buettger C, Njølstad PR, Shiota C, Magnuson MA, Matschinsky FM. Glucokinase and the regulation of blood sugar. a mathematical model predicts the threshold for glucose stimulated insulin release for GCK gene mutations that cause hyper- and hypoglycemia. In: Matschinsky FM, Magnuson MA, editors. Glucokinase and Glycemic Disease: From Basics to Novel Therapeutics. Karger, Basel: 2004. pp. 92–109. [Google Scholar]
- 20.Davis EA, Cuesta-Munoz A, Raoul M, Buettger C, Sweet I, Moates M, Magnuson MA, Matschinsky FM. Mutants of glucokinase cause hypoglycaemia- and hyperglycaemia syndromes and their analysis illuminates fundamental quantitative concepts of glucose homeostasis. Diabetologia. 1999;42:1175–1186. doi: 10.1007/s001250051289. [DOI] [PubMed] [Google Scholar]
- 21.Sagen JV, Odili S, Bjorkhaug L, Zelent D, Buettger C, Kwagh J, Stanley C, Dahl-Jorgensen K, de Beaufort C, Bell GI, Han Y, Grimsby J, Taub R, Molven A, Sovik O, Njolstad PR, Matschinsky FM. From clinicogenetic studies of maturity-onset diabetes of the young to unraveling complex mechanisms of glucokinase regulation. Diabetes. 2006;55:1713–1722. doi: 10.2337/db05-1513. [DOI] [PubMed] [Google Scholar]
- 22.Zelent B, Odili S, Buettger C, Zelent DK, Chen P, Fenner D, Bass J, Stanley CA, Laberge M, Vanderkooi JM, Sarabu R, Grimsby J, Matschinsky FM. Mutational analysis of allosteric activation and inhibition of glucokinase. Biochem J. 2011 doi: 10.1042/BJ20110440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gloyn AL. Glucokinase (GCK) mutations in hyper- and hypoglycemia: maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemia of infancy. Hum mutat. 2003;22:353–362. doi: 10.1002/humu.10277. [DOI] [PubMed] [Google Scholar]
- 24.Arden C, Trainer A, de la Iglesia N, Scougall KT, Gloyn AL, Lange AJ, Shaw JA, Matschinsky FM, Agius L. Cell biology assessment of glucokinase mutations V62M and G72R in pancreatic beta-cells: evidence for cellular instability of catalytic activity. Diabetes. 2007;56:1773–1782. doi: 10.2337/db06-1151. [DOI] [PubMed] [Google Scholar]
- 25.Cullen KS, Matschinsky FM, Agius L, Arden C. Susceptibility of Glucokinase-MODY Mutants to Inactivation by Oxidative Stress in Pancreatic beta-Cells, Diabetes. 2011 doi: 10.2337/db11-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding SY, Tribble ND, Kraft CA, Markwardt M, Gloyn AL, Rizzo MA. Naturally occurring glucokinase mutations are associated with defects in posttranslational S-nitrosylation. J Mol Endocrinol. 2010;24:171–177. doi: 10.1210/me.2009-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Njolstad PR, Sagen JV, Bjorkhaug L, Odili S, Shehadeh N, Bakry D, Sarici SU, Alpay F, Molnes J, Molven A, Sovik O, Matschinsky FM. Permanent neonatal diabetes caused by glucokinase deficiency: inborn error of the glucose-insulin signaling pathway. Diabetes. 2003;52:2854–2860. doi: 10.2337/diabetes.52.11.2854. [DOI] [PubMed] [Google Scholar]
- 28.Molnes J, Bjorkhaug L, Sovik O, Njolstad PR, Flatmark T. Catalytic activation of human glucokinase by substrate binding: residue contacts involved in the binding of D-glucose to the super-open form and conformational transitions. FEBS J. 2008;275:2467–2481. doi: 10.1111/j.1742-4658.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- 29.Sannerud R, Marie M, Hansen BB, Saraste J. Use of polarized PC12 cells to monitor protein localization in the early biosynthetic pathway. Methods Mol Biol. 2008;457:253–265. doi: 10.1007/978-1-59745-261-8_19. [DOI] [PubMed] [Google Scholar]
- 30.Johansson BB, Torsvik J, Bjorkhaug L, Vesterhus M, Ragvin A, Tjora E, Fjeld K, Hoem D, Johansson S, Raeder H, Lindquist S, Hernell O, Cnop M, Saraste J, Flatmark T, Molven A, Njolstad PR. Diabetes and Pancreatic Exocrine Dysfunction Due to Mutations in the Carboxyl Ester Lipase Gene-Maturity Onset Diabetes of the Young (CEL-MODY): A PROTEIN MISFOLDING DISEASE. J Biol Chem. 2011;286:34593–34605. doi: 10.1074/jbc.M111.222679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 32.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nature Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 33.DS LLC. The PyMOL Molecular Graphics System. South San Francisco, CA, USA: 2008. [Google Scholar]
- 34.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontana A, Polverino de Laureto P, De Filippis V, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold Des. 1997;2:R17–26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 36.Villanueva J, Villegas V, Querol E, Aviles FX, Serrano L. Protein secondary structure and stability determined by combining exoproteolysis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 2002;37:974–984. doi: 10.1002/jms.356. [DOI] [PubMed] [Google Scholar]
- 37.Molnes J, Teigen K, Aukrust I, Bjorkhaug L, Sovik O, Flatmark T, Njolstad PR. Binding of ATP at the active site of human pancreatic glucokinase--nucleotide-induced conformational changes with possible implications for its kinetic cooperativity. FEBS J. 2011;278:2372–2386. doi: 10.1111/j.1742-4658.2011.08160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larion M, Salinas RK, Bruschweiler-Li L, Bruschweiler R, Miller BG. Direct evidence of conformational heterogeneity in human pancreatic glucokinase from high-resolution nuclear magnetic resonance. Biochemistry. 2010;49:7969–7971. doi: 10.1021/bi101098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, Kesavan P, Wang LQ, Niswender K, Tanizawa Y, Permutt MA, Magnuson MA, Matschinsky FM. Variable effects of maturity-onset-diabetes-of-youth (MODY)-associated glucokinase mutations on substrate interactions and stability of the enzyme. Biochem J. 1995;309(Pt 1):167–173. doi: 10.1042/bj3090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kesavan P, Wang L, Davis E, Cuesta A, Sweet I, Niswender K, Magnuson MA, Matschinsky FM. Structural instability of mutant beta-cell glucokinase: implications for the molecular pathogenesis of maturity-onset diabetes of the young (type-2) Biochem J. 1997;322(Pt 1):57–63. doi: 10.1042/bj3220057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galan M, Vincent O, Roncero I, Azriel S, Boix-Pallares P, Delgado-Alvarez E, Diaz-Cadorniga F, Blazquez E, Navas MA. Effects of novel maturity-onset diabetes of the young (MODY)-associated mutations on glucokinase activity and protein stability. Biochem J. 2006;393:389–396. doi: 10.1042/BJ20051137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller SP, Anand GR, Karschnia EJ, Bell GI, LaPorte DC, Lange AJ. Characterization of glucokinase mutations associated with maturity-onset diabetes of the young type 2 (MODY-2): different glucokinase defects lead to a common phenotype. Diabetes. 1999;48:1645–1651. doi: 10.2337/diabetes.48.8.1645. [DOI] [PubMed] [Google Scholar]
- 43.Pino MF, Kim KA, Shelton KD, Lindner J, Odili S, Li C, Collins HW, Shiota M, Matschinsky FM, Magnuson MA. Glucokinase thermolability and hepatic regulatory protein binding are essential factors for predicting the blood glucose phenotype of missense mutations. J Biol Chem. 2007;282:13906–13916. doi: 10.1074/jbc.M610094200. [DOI] [PubMed] [Google Scholar]
- 44.Fenner D, Odili S, Hong HK, Kobayashi Y, Kohsaka A, Siepka SM, Vitaterna MH, Chen P, Zelent B, Grimsby J, Takahashi JS, Matschinsky FM, Bass J. Generation of N-ethyl-N-nitrosourea (ENU) diabetes models in mice demonstrates genotype-specific action of glucokinase activators. J Biol Chem. 2011;286:39560–39572. doi: 10.1074/jbc.M111.269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gloyn AL, Odili S, Zelent D, Buettger C, Castleden HA, Steele AM, Stride A, Shiota C, Magnuson MA, Lorini R, d’Annunzio G, Stanley CA, Kwagh J, van Schaftingen E, Veiga-da-Cunha M, Barbetti F, Dunten P, Han Y, Grimsby J, Taub R, Ellard S, Hattersley AT, Matschinsky FM. Insights into the structure and regulation of glucokinase from a novel mutation (V62M), which causes maturity-onset diabetes of the young. J Biol Chem. 2005;280:14105–14113. doi: 10.1074/jbc.M413146200. [DOI] [PubMed] [Google Scholar]
- 46.Ralph EC, Sun S. Biochemical characterization of MODY2 glucokinase variants V62M and G72R reveals reduced enzymatic activities relative to wild type. Biochemistry. 2009;48:2514–2521. doi: 10.1021/bi900020n. [DOI] [PubMed] [Google Scholar]
- 47.Zelent B, Odili S, Buettger C, Shiota C, Grimsby J, Taub R, Magnuson MA, Vanderkooi JM, Matschinsky FM. Sugar binding to recombinant wild-type and mutant glucokinase monitored by kinetic measurement and tryptophan fluorescence. Biochem J. 2008;413:269–280. doi: 10.1042/BJ20071718. [DOI] [PubMed] [Google Scholar]
- 48.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim K, Ho JX, Keeling K, Gilliland GL, Ji X, Ruker F, Carter DC. Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV. Protein Sci. 1994;3:2233–2244. doi: 10.1002/pro.5560031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke CV, Buettger CW, Davis EA, McClane SJ, Matschinsky FM, Raper SE. Cell-biological assessment of human glucokinase mutants causing maturity-onset diabetes of the young type 2 (MODY-2) or glucokinase-linked hyperinsulinaemia (GK-HI) Biochem J. 1999;342(Pt 2):345–352. [PMC free article] [PubMed] [Google Scholar]
- 51.Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990;111:941–953. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim KL. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett MJ, Eisenberg D. Refined structure of monomeric diphtheria toxin at 2.3 A resolution. Protein Sci. 1994;3:1464–1475. doi: 10.1002/pro.5560030912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gronenborn AM. Protein acrobatics in pairs--dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrientos LG, Louis JM, Botos I, Mori T, Han Z, O’Keefe BR, Boyd MR, Wlodawer A, Gronenborn AM. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures. Structure. 2002;10:673–686. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- 57.Byeon IJ, Louis JM, Gronenborn AM. A protein contortionist: core mutations of GB1 that induce dimerization and domain swapping. J Mol Biol. 2003;333:141–152. doi: 10.1016/s0022-2836(03)00928-8. [DOI] [PubMed] [Google Scholar]
- 58.Ellis RJ. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr Opin Struct Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 59.Minton AP. Influence of macromolecular crowding upon the stability and state of association of proteins: predictions and observations. J Pharm Sci. 2005;94:1668–1675. doi: 10.1002/jps.20417. [DOI] [PubMed] [Google Scholar]
- 60.Munishkina LA, Ahmad A, Fink AL, Uversky VN. Guiding protein aggregation with macromolecular crowding. Biochemistry. 2008;47:8993–9006. doi: 10.1021/bi8008399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellis RJ, Minton AP. Protein aggregation in crowded environments. Biol Chem. 2006;387:485–497. doi: 10.1515/BC.2006.064. [DOI] [PubMed] [Google Scholar]
- 62.Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 63.Toyoda Y, Miwa I, Satake S, Anai M, Oka Y. Nuclear location of the regulatory protein of glucokinase in rat liver and translocation of the regulator to the cytoplasm in response to high glucose. Biochem Biophys Res Commun. 1995;215:467–473. doi: 10.1006/bbrc.1995.2488. [DOI] [PubMed] [Google Scholar]
- 64.Arden C, Harbottle A, Baltrusch S, Tiedge M, Agius L. Glucokinase is an integral component of the insulin granules in glucose-responsive insulin secretory cells and does not translocate during glucose stimulation. Diabetes. 2004;53:2346–2352. doi: 10.2337/diabetes.53.9.2346. [DOI] [PubMed] [Google Scholar]
- 65.Murata T, Katagiri H, Ishihara H, Shibasaki Y, Asano T, Toyoda Y, Pekiner B, Pekiner C, Miwa I, Oka Y. Co-localization of glucokinase with actin filaments. FEBS Lett. 1997;406:109–113. doi: 10.1016/s0014-5793(97)00253-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.