SUMMARY
Obstacles in elucidating the role of oxidative stress in aging include difficulties in 1) tracking in vivo oxidants, in 2) identifying affected proteins, and in 3) correlating changes in oxidant levels with lifespan. Here, we used quantitative redox proteomics to determine the onset and the cellular targets of oxidative stress during Caenorhabditis elegans’ lifespan. In parallel, we used genetically encoded sensor proteins to determine peroxide levels in live animals in real time. We discovered that C. elegans encounters significant levels of oxidants as early as during larval development. Oxidant levels drop rapidly as animals mature and reducing conditions prevail throughout the reproductive age, after which age-accompanied protein oxidation sets in. Long-lived daf-2 mutants transition faster to reducing conditions, whereas short-lived daf-16 mutants retain higher oxidant levels throughout their mature life. These results suggest that animals with improved capacity to recover from early oxidative stress have significant advantages later in life.
INTRODUCTION
The oxidative stress theory of aging postulates that oxidative damage to cellular macromolecules, caused by the progressive accumulation of reactive oxygen species (ROS), contributes and possibly even leads to the decline in physiological functions observed in aging organisms (Finkel and Holbrook, 2000; Harman, 1956). Since its inception, extensive correlative evidence has been collected that corroborates this popular theory. For instance, it has been shown that oxidative damage to proteins, lipids, and DNA increases with age (Finkel and Holbrook, 2000) and that interventions that delay aging (e.g., reduced caloric intake) decrease the extent of oxidative damage and mediate the rapid removal of damaged macromolecules through proteolysis and autophagy (Cavallini et al., 2008). Moreover, one of the unifying features that distinguish many long-lived mutant invertebrates and rodents from their wild-type cohorts appears to be a significant increase in oxidative stress resistance (Salmon et al., 2005). However, the validity of the free radical theory of aging recently came under dispute when a series of genetic studies, particularly those in mice, failed to generate conclusive results about the role of ROS in aging (Perez et al., 2009). These findings suggested a more complex aging mechanism, potentially driven by more subtle changes in ROS levels or the cellular redox state.
One aspect of redox biology that has long been overlooked when testing the free radical theory of aging is the fact that ROS, like peroxide, also play important regulatory roles as intracellular signaling molecules. While high levels of peroxide are thought to be toxic, low levels of peroxide, which are continuously produced during mitochondrial respiration and other processes, set the pace of numerous metabolic and signaling pathways in the cell (D’Autreaux and Toledano, 2007). Proteins that are regulated by peroxide and potentially other reactive oxygen and nitrogen species commonly contain highly oxidation-sensitive cysteine residues whose thiol oxidation status controls the protein’s activity, and by extension, the pathway that the protein is part of (Cross and Templeton, 2006). Given this fact, it is thus very likely that depletion of pro-oxidants by either dietary antioxidants or genetic manipulation profoundly impacts development, differentiation, and stress responses and, when applied at the wrong stage in life, potentially outweighs the beneficial effects.
We decided to take a different approach in evaluating the role that ROS play in lifespan. We used recently developed quantitative probes to determine when ROS accumulate, which proteins and pathways they affect, and whether a correlation exists between the onset and extent of endogenous oxidative stress and the lifespan of the organism. We chose C. elegans as our model system because it is a well-established eukaryotic aging model and is well-suited for redox proteomic analyses and the application of fluorescent biosensors (Johnson, 2008; Kumsta et al., 2010). To monitor global changes in the cellular redox environment, we applied the quantitative redox proteomic technique OxICAT at distinct time points in C. elegans lifespan to determine the thiol redox status of almost 140 different proteins. In parallel, we employed the chromosomally encoded hydrogen peroxide (H2O2) sensor protein HyPer (Belousov et al., 2006) as a tissue-specific read-out for changes in endogenous peroxide levels. By using these quantitative redox-sensors, we discovered that C. elegans is exposed to high oxidant levels at two distinct stages in life: during development and during aging. Comparative analysis with long- and short-lived mutants of the insulin/IGF-1 signaling (ILS) pathway revealed that long-lived mutant worms recovered faster from increased developmental ROS levels and reached lower steady-state redox levels during their reproductive period than short-lived worms, which failed to completely restore redox homeostasis. These results suggest that changes in the cellular redox homeostasis, encountered at a very early stage in life, determine subsequent redox levels and potentially the lifespan of organisms.
RESULTS
The C. elegans Redoxome: Establishing the Redox Baseline
We recently developed a redox proteomic technique, termed OxICAT, which allows us, in a single experiment, to determine the in vivo oxidation status of hundreds of different protein thiols, many of which are redox sensitive (Kumsta et al., 2010; Leichert et al., 2008). We reasoned that by monitoring the in vivo redox state of protein thiols over the lifespan of C. elegans, we should obtain information about temporal and spatial changes in cellular redox homeostasis and identify processes and pathways that might be affected by the prevailing redox conditions. With OxICAT, we use the quantitative properties of the thiol-reactive isotope-coded affinity tag (ICAT) to differentially label in vivo reduced and in vivo oxidized protein thiols (Fig. S1). HPLC is used to separate the ICAT-labeled peptides, followed by mass spectrometry (MS) and tandem MS/MS, which allows us to identify the thiol-containing peptides and quantify their in vivo oxidation status (Leichert et al., 2008). Due to its ratio-based nature, OxICAT is independent of the protein amount present in cells and tissues, making it ideally suited to monitor changes in protein oxidation over the lifespan of the organism.
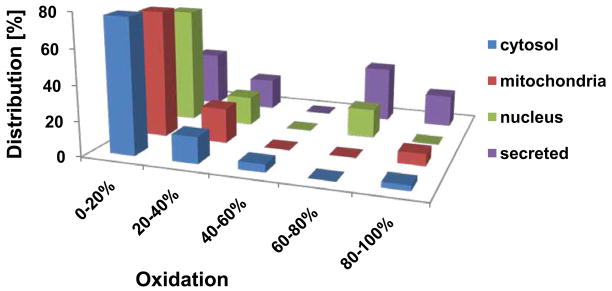
We applied OxICAT to analyze the redox status of proteins in synchronized wild-type C. elegans on day 2 of adulthood. This analysis in young adults was intended to serve as a reference point for all subsequent studies so that relative changes in the redox status of the proteins could be determined at different time points during C. elegans lifespan. As shown in Table S1, we reproducibly determined the redox status of 170 thiol-containing peptides, representing 137 different C. elegans proteins. Database analysis revealed that we identified the redox status of numerous ubiquitously expressed proteins (e.g., HSP-1, 17 ribosomal proteins) as well as proteins selectively expressed in body wall muscle (e.g., ANC-1, DIM-1, myosin-3), intestine (e.g., vitellogenin-6, neprilysin), nervous system (e.g., degenerin), and pharynx (e.g., myosin-2, myosin-4, annexin). Of the identified protein thiols, 65% exhibited oxidation states of less than 20%, with most of them being less than 10% oxidized at this point in life. These protein thiols were found predominantly in cytosolic and mitochondrial proteins (Fig. 1). About 25% of our identified peptides contained cysteines with oxidation levels between 20% and 60% (Fig. 1). This partially oxidized group of proteins included enzymes involved in glucose metabolism (e.g., alcohol dehydrogenase, fructose-bisphosphate aldolase) and ATP-homeostasis (e.g., nucleoside diphosphate kinase, vacuolar ATP synthases), as well as proteins involved in motility (e.g., UNC-87, paramyosin), signal transduction (e.g., phosphoinositide kinase AGE-1), and protein homeostasis (e.g., ATP-dependent chaperone CDC-48) (Table S1). Many of these proteins have been previously shown to contain redox-sensitive cysteines (Kumsta et al., 2010) (Table 1), suggesting that they are partially oxidized in response to either local or global ROS accumulation or due to changes in cellular redox homeostasis. Less than 10% of our identified protein thiols were oxidized greater than 80%. Most of these highly oxidized thiols belonged to secreted proteins with known disulfides (Fig. 1).
Fig. 1. In Vivo Thiol Oxidation Status of C. elegans Proteins.
Wild-type N2 worms were synchronized and cultivated at 15°C. At day 2 of adulthood, worms were lysed and the protein thiol oxidation status was determined using the differential thiol trapping technique OxICAT. Proteins are categorized by their oxidation status and sub-cellular localization, according to WormBase (http://www.wormbase.org/). The complete list of proteins and their respective oxidation states with standard deviations can be found in Table S1.
Table 1.
Oxidation status of select proteins during C. elegans lifespan
| Protein (affected cysteine) | Acc. Nr. | Tissuee | Average Oxidation Statusa | Oxidation sensitive | ||
|---|---|---|---|---|---|---|
| Devb | Youngc | Oldd | ||||
|
| ||||||
| Tubulin (alpha-2 chain) (C314) | CE09692 | N | 23 +/− 6 | 7 +/− 2 | n.d. | Yesh |
| GTP-binding protein (TAG-210) (C54) | CE14708 | I | 20 +/− 1* | 7 +/− 6 | n.d. | |
| Protein CE32871 (C374) | CE32871 | 26 +/− 3 | 12 +/− 3 | n.d. | ||
| Proteasome alpha subunit (PAS-3) (C74) | CE30307 | 26 +/− 6* | 13 +/− 1 | n.d. | ||
| Heat Shock Protein 70 HSP-1 (C243) | CE09682 | P, I, BW | 31 +/− 2* | 4 +/−2 | 9 +/− 4 | Yesg |
| Aspartyl-tRNA synthetase (C233) | CE00015 | 27 +/− 8 | 6 +/− 2 | 17 +/− 11 | ||
| Vacuolar ATP synthase (subunit A) (C218) | CE22210 | I, BW | 28 +/− 5 | 10 +/− 2 | 11 +/− 4 | Yesf |
| T-complex protein 1 (subunit zeta) (C517) | CE01234 | U | 30 +/− 4 | 12 +/− 2 | 26 +/− 14 | Yesg |
| 40S ribosomal protein S28 (C22) | CE21842 | U | 20 +/− 3 | 6 +/− 3 | 29 +/− 4 | Yesf |
| DIM1 (C234) | CE09308 | BW | 18 +/− 5 | 8 +/− 3 | 33 +/− 14 | Yesf |
| COPII coatomer (subunit SAR-1) (C174) | CE07622 | 23 +/− 5 | 10 +/− 2 | 25 +/− 7 | ||
| Methylcrotonyl-CoA carboxylase (C212) | CE00136 | P | 26 +/− 5 | 10 +/− 6 | 22 +/− 2 | |
| 40S ribosomal protein S12 (C114) | CE26896 | U | 30 +/− 5 | 13 +/− 4 | 34 +/− 6 | Yesf |
| Neprilysin (C41) | CE43217 | I | 66 +/− 4* | 22 +/− 6 | 62 +/− 6 | |
| Serine hydroxymethyltransferase MEL-32 (C391) | CE01130 | I, H | 63 +/− 3** | 31 +/− 4 | 44 +/− 3 | Yesf |
| UNC-87 (C472) | CE36924 | BW, M | 68 +/− 6 | 40 +/− 2 | 54 +/− 6 | |
| Vitellogenin-6 (C468) | CE28594 | I | 89 +/− 5 | 65 +/− 4 | 80 +/− 8 | |
| Chitinase (C738/747) | CE32592 | G | 94 +/− 5 | 68 +/− 1 | 91 +/− 2 | Yesh |
| Vitellogenin-3 (C626) | CE20900 | I | n.d. | 10 +/− 2 | 76 +/− 9 | |
| RNA helicase CGH-1 (C336) | CE00839 | G | n.d. | 22 +/− 4 | 45 +/− 10 | |
| 60S ribosomal protein L22 (C27) | CE04102 | U | 8 +/− 2 | 6 +/− 2 | 19 +/− 5 | Yesf |
| Protein K07C5.4 (C390) | CE06114 | 12 +/− 4* | 9 +/− 5 | 28 +/− 8 | ||
| Small molecule methylase (C28H8.7) (C224) | CE01829 | 2 +/− 2 | 12 +/− 1 | 26 +/− 6 | ||
| Actin-2 (C258) | CE13150 | BW, N | 16 +/− 10* | 16 +/− 3 | 30 +/− 4 | Yesg |
| Glutamyl-tRNA synthetase (C377) | CE06580 | 14 +/− 5* | 22 +/− 2 | 33 +/− 3 | ||
| Translation initiation factor 5A (C111) | CE37787 | G | 17 +/− 3* | 27 +/− 6 | 47 +/− 2 | Yesf |
| Nucleoside diphosphate kinase NDPK (C109) | CE09650 | U | 35 +/− 6 | 28 +/− 4 | 47 +/− 8 | Yesf |
| Nucleoside diphosphate kinase NDPK (C117) | CE09650 | U | 36 +/− 2 | 30 +/− 5 | 54 +/− 13 | Yesf |
| Phosphoinositide 3-kinase AGE-1 (C710) | CE43431 | I, N | 56 +/− 4 | 50 +/− 12 | 72 +/− 5 | |
Average oxidation of at least three individual replicates;
Thiol oxidation status of peptides in L4 stage; thiol oxidation status of peptide in L2 stage was used * if corresponding peptide was not identified in L4 larva, or ** if thiol oxidation in L2 was substantially higher than in L4;
Thiol oxidation status of peptides of young adults (day 2);
Thiol oxidation status of peptides in 15-days-old adults;
Tissue localization according to WormBase (www.wormbase.org); U, ubiquitous; BW, body wall muscle; M, muscle; N, neuron; I, intestine, P, pharynx; H, hypodermis; G, gonads; R, reproductive system;
Cysteine/
protein identified to be peroxide-sensitive in C. elegans (Kumsta et al., 2010);
protein identified as redox stress-sensitive in other systems (Joe et al., 2008) (Maeda et al., 2004); n.d., peptide not reproducibly identified at this stage.
Monitoring Changes in Protein Thiol Redox State during the Lifespan of C. elegans
To investigate the redox state of the identified thiol-containing proteins over the lifespan of C. elegans, we took aliquots of ~100,000 worms from a synchronized wild-type population during early (i.e., L2) and late (i.e., L4) development and at days 2, 8, and 15 of adulthood for subsequent OxICAT analysis. Each of the time points was selected based on the fact that they represent distinct physiological stages in C. elegans. The larval stages L2 and L4 represent early and late post-embryonic developmental stages of C. elegans. Worms transition to their reproductive phase, which peaks around day 2 to 3 of adulthood. At day 8, most animals have ceased reproduction, yet worms still reveal juvenile mobility and pharyngeal pumping. By day 15, significant mobility defects are observed in the majority of worms, however the mean survival of the population is still about 80%. We separated live worms from eggs and any dead worms by sucrose flotation, lysed the worms in trichloroacetic acid to maintain the in vivo redox status of the protein thiols, and conducted our differential OxICAT thiol trapping. We were able to reproducibly determine the redox status of many of our previously identified peptides at each time point, with the exception of proteins that are either not expressed during early development (e.g., vitellogenin) or whose expression decreases as the worms age (e.g., lysozyme).
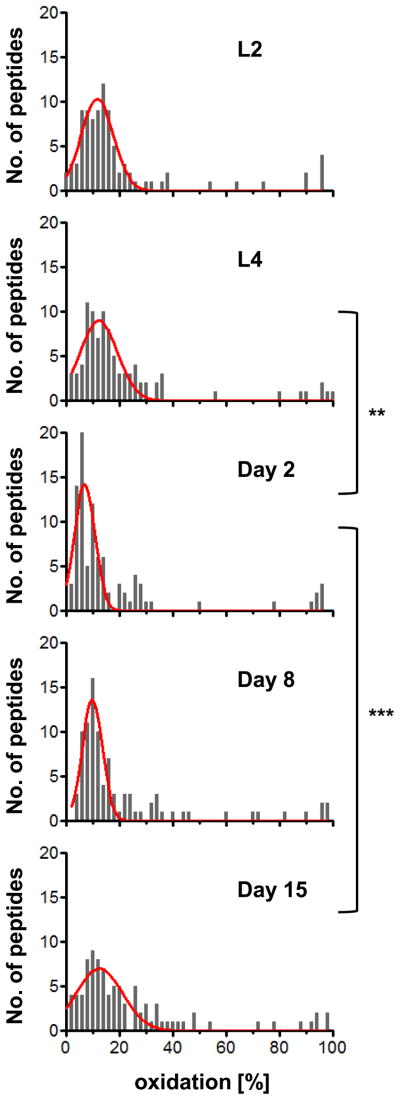
We restricted our subsequent analysis to those ~ 100 protein thiols, which were reproducibly identified at all time points tested, and plotted them against their respective average oxidation state at each time point (Fig. 2). We thus generated histograms that allowed us to directly compare the in vivo oxidation state of a common set of protein thiols during the lifespan of wild-type C. elegans (Table S2). In agreement with the free radical theory of aging, we found that the oxidation status of protein thiols generally increased with age. This was particularly noticeable in aged worms (day 15), which had significantly more oxidized protein thiols (p<0.001) than young adults (day 2). To our surprise, however, we also observed a significantly higher level of thiol oxidation in developing L4 larval worms. This elevated overall protein oxidation significantly decreased (p<0.01) upon entering the reproductive period (day 2). These results suggest that animals are exposed to elevated levels of oxidants early in life, either as part of ROS signaling, increased metabolic activity, or a combination thereof. It is of note that not all protein thiols that we found to be more oxidized during development are also more oxidized in aging worms and vice versa, suggesting more localized oxidation events or antioxidant activities during development and aging.
Fig. 2. Monitoring Thiol Oxidation during the Lifespan of C. elegans.
The oxidation status of protein thiols in a synchronized population of wild-type N2 worms cultivated at 15°C was determined at the larval stage L2 and L4 as well as at days 2, 8, and 15 of adulthood. Shown is the frequency distribution of average oxidation levels determined for all of the protein thiols that were reproducibly identified for each time point. The histograms were fitted to a single or double Gaussian model (red line) using GraphPad Prism, with R2 values of 0.8–0.9. The complete list of proteins and their respective oxidation states can be found in Table S2. Statistical analysis was done using Kruskal-Wallis one-way analysis of variance followed by Dunn’s post test. P values <0.05 are indicated with *; p values <0.01 are indicated with **.
Some of the most significantly oxidized protein thiols during development in C. elegans compared to young adult worms were found in the Hsp70 homologue HSP-1, the protease neprylisin, MEL-32 and UNC-87 (Table 1). Many of these proteins were also found highly oxidized in aging C. elegans. Proteins that were heavily oxidized in aging organisms versus young adults included the phosphoinositide 3 kinase AGE-1, NDPK, and translation initiation factor 5A. Many of our identified proteins (e.g., HSP-1, UNC-87, ribosomal proteins S28a, S12, L22, translation initiation factor 5A) have previously been observed to contain oxidation sensitive cysteines, either in peroxide-treated C. elegans (Kumsta et al., 2010) or in other oxidatively-stressed eukaryotic organisms (Fiaschi et al., 2006; Joe et al., 2008) (Table 1). Several proteins, however, have not been shown before to contain oxidation-sensitive cysteines, including AGE-1, which when deleted, confers lifespan extension (Dorman et al., 1995).
Localization analysis (www.wormbase.org) of the proteins that showed the most significant oxidation (Table 1) indicated that many of these proteins are ubiquitously distributed. We did observe that a number of our proteins that become substantially oxidized in developing and/or aging worms are exclusively localized to the intestine (neprilysin, vitellogenin-3, vitellogenin-6) or are localized to the intestine and one other cell type (MEL-32, AGE-1) (Table 1 and Table S1). Moreover, UNC-87 and DIM-1, which are both substantially oxidized during development and aging are localized exclusively to the body wall muscle cells. These results suggest that, at a minimum, increased ROS production occurs in body wall muscle cells and the intestine, cell types known for their high metabolic activity and involvement in aging (Libina et al., 2003).
ILS Pathway Mutants Are Affected in Oxidative Stress Recovery
To assess whether the onset, extent, and pattern of endogenous ROS accumulation is altered in mutant strains that show either shortened or prolonged lifespan, we decided to conduct our OxICAT experiments in worms defective in the insulin/IGF-1 signaling (ILS) pathway. The ILS pathway is evolutionarily conserved, and genetic manipulation of the signaling cascade has been shown to affect the lifespan of C. elegans as early as in young adults (Dillin et al., 2002a). DAF-2, the insulin/IGF-1- receptor, negatively regulates the forkhead transcription factor DAF-16, which controls expression of numerous antioxidant genes (Murphy et al., 2003). Reduced DAF-2 function promotes DAF-16-mediated gene expression, and increases lifespan (Kenyon et al., 1993) while deletion of DAF-16 reduces lifespan (Lin et al., 2001).
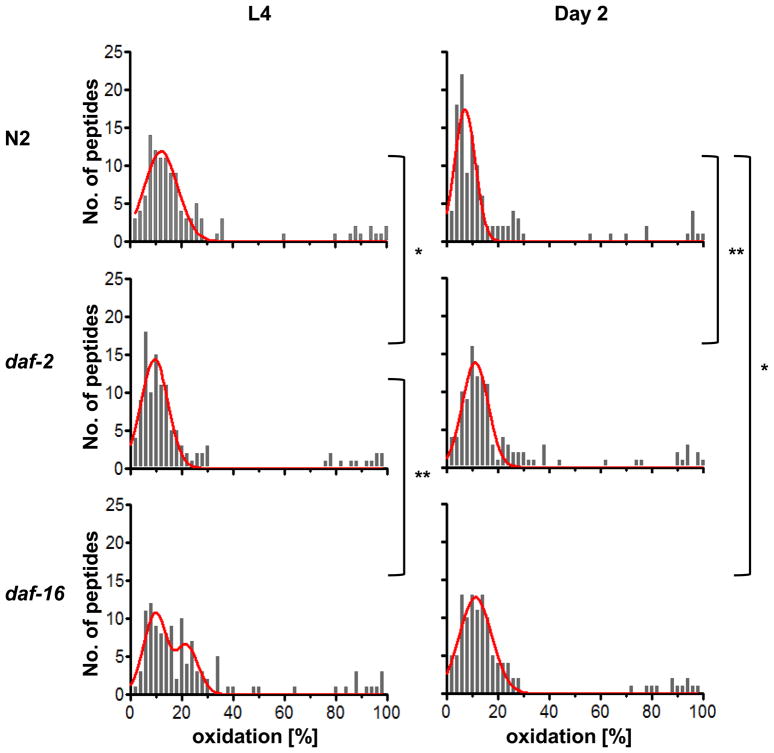
We synchronized worms and took aliquots at the same time points as in the previous experiments. Many of the peptides that we identified in wild-type N2 were also identified in daf-2 and daf-16 mutant worms, particularly at early stages in C. elegans life, making a statistical analysis of the protein thiol oxidation status possible for worms transitioning from development to early adulthood (Table S2) (Fig. 3). We did not observe any significant difference in the overall oxidation status of protein thiols isolated from daf-2 or daf-16 worms at the early larval stage L2, suggesting that disruption of the ILS pathway during early development does not globally affect cellular redox conditions (Table S2) (Fig. 3). However, differences between the strains were significant by the time C. elegans reached the last larval stage (i.e., L4). In the long-lived daf-2 mutants, protein thiol oxidation was already shifted towards lower levels whereas in the short-lived daf-16 deletion strain, protein thiols remained in a more oxidized state. Neither daf-16 nor daf-2 mutants changed their oxidation status significantly upon transition from the L4 state to the young adult state suggesting that lack of DAF-16 prevents the worm’s recovery from high oxidant levels encountered during development. It is of note that no statistically significant difference was observed between the protein thiol oxidation status of wild-type and mutant worms at either day 8 or day 15 (Table S2). This, however, could be due to the fact that at the later time points fewer of our peptides were simultaneously identified in all three strains. Analysis of the oxidation status of the same peptides is, however, crucial for our statistical evaluation.
Fig. 3. Protein Oxidation during the Lifespan of WT, daf-2, and daf-16 Worms.
Relative distribution of protein thiol oxidation in synchronized populations of wild-type N2, daf-2 and daf-16 mutant strains during development (L4) and early adulthood (day 2). Protein oxidation was determined using OxICAT (complete list of proteins and their oxidation states at time points L2, L4, Day 2, 8 and 15 can be found in Table S2). Shown is the frequency distribution of average oxidation levels determined for all of the protein thiols that were reproducibly identified for each time point. The histograms were fitted to a single or double Gaussian model (red line) using GraphPad Prism, with R2 values of 0.8–0.9. Statistical analysis was done using Kruskal-Wallis one-way analysis of variance followed by Dunn’s post test. p values <0.05 are indicated with *; p values from 0.001 to 0.01 are indicated with **.
Using HyPer to Determine Endogenous Peroxide Levels in C. elegans
Many of our proteins, whose thiol redox status was found to vary over the lifespan of C. elegans, have been previously shown to be sensitive to peroxide-mediated thiol oxidation (Table 1). To test whether accumulation of peroxide could be, at least in part, responsible for the observed protein oxidation, we tested the in vivo oxidation status of peroxiredoxin 2 (PRDX-2). This peroxide-detoxifying enzyme undergoes intermolecular disulfide bond formation with a second PRDX-2 molecule as part of its catalytic cycle (Olahova et al., 2008). We found significantly higher amounts of peroxide-induced PRDX-2 disulfides in C. elegans larvae as compared to young adults, suggesting that developing animals are indeed exposed to higher peroxide levels (Fig. S2). We thus decided to make use of HyPer, a recently developed peroxide-specific sensor protein, which has been previously used to monitor in vivo peroxide levels (Belousov et al., 2006). We reasoned that expressing HyPer in C. elegans would allow us to monitor and track peroxide levels in individual worms, and to determine whether the levels of H2O2 could possibly correlate with the lifespan of the animals. The peroxide-sensor HyPer consists of a circularly permuted yellow fluorescent protein fused to the H2O2 sensing domain of E. coli OxyR (Belousov et al., 2006). The sensor protein possesses two excitation maxima at ~420 nm and ~500 nm, and a single emission maximum at 516 nm. Upon exposure of HyPer to peroxide, one intramolecular disulfide bond forms within the OxyR domain, causing conformational changes that result in a ratiometric shift. The emission upon excitation at 500 nm increases, whereas the emission after excitation at 420 nm decreases proportionally, leading to an overall increase in the 500 nm/420 nm ratio (i.e., HyPer ratio) with rising peroxide levels.
To use HyPer as an endogenous peroxide sensor in C. elegans, we cloned the HyPer gene under the control of the UNC-54 promoter, which targets HyPer expression to the body wall muscle cells, which is one tissue where significant oxidation occurred according to our OxICAT studies (Table 1). We generated a stable transgenic line of wild-type N2 worms expressing the peroxide sensor in the body wall muscle cells (Fig. S3A). We backcrossed the strain several times, verified that the expression of HyPer did not affect the lifespan of the worms (Fig. S3B), and confirmed that the HyPer ratio was indeed independent of the amount of HyPer protein expressed (Fig. S3C). These features make HyPer well-suited to monitor endogenous peroxide levels over the lifespan of C. elegans.
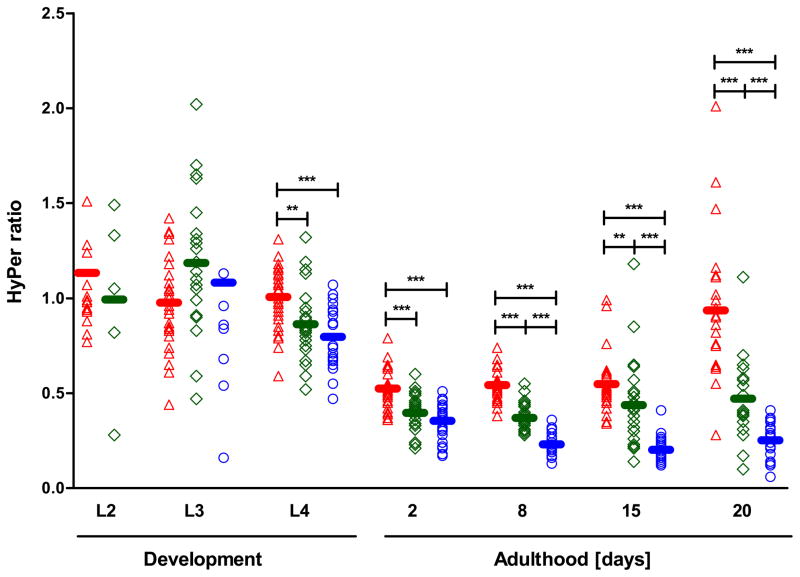
Monitoring Endogenous Peroxide Levels with temporal and spatial resolution
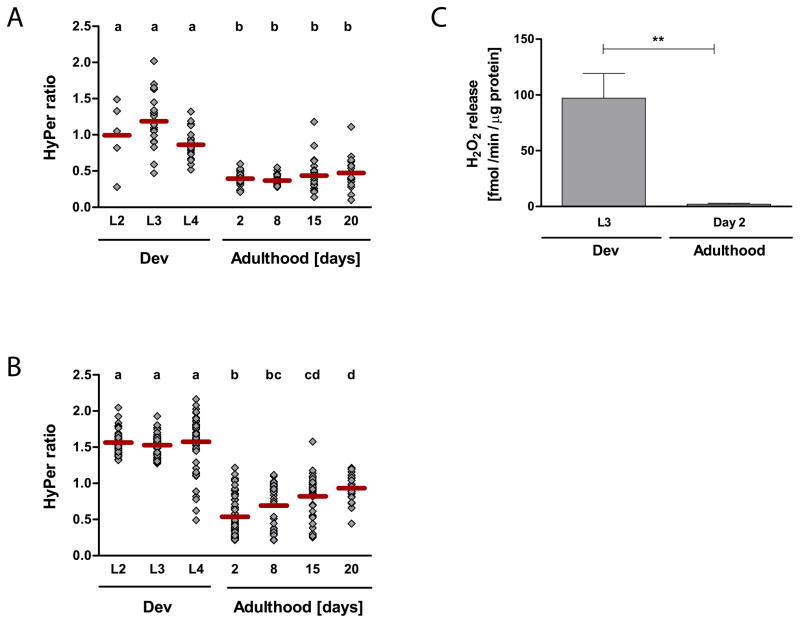
To monitor potential fluctuations in the levels of peroxide during the lifespan of C. elegans, we took about 30 worms of a synchronized population during development (larval stages L2, L3, and L4) and at defined days during adulthood (days 2, 8, 15, and 20) and determined the HyPer ratio for each individual animal. As shown in Fig. 4A, we observed high levels of endogenous peroxide during larval development. These results were in good agreement with our OxICAT studies, and suggested that accumulation of peroxide was either directly responsible for increased protein oxidation or indirectly by changing the cellular redox potential towards more oxidizing conditions. Peroxide levels rapidly decreased as the worms reached their reproductive period and remained low during the fertile phase (Fig. 4A). To exclude that the high peroxide levels observed during development were due to the synchronization procedure, which utilizes a hypochlorite solution to release the eggs from gravid adults, we also determined the HyPer ratio in animals that were manually synchronized (Fig. S3D). Again, we found a significantly higher HyPer ratio in developing animals as compared to fertile, mature adults, making it unlikely that the high HyPer ratio was caused by the synchronization procedure.
Fig. 4. Monitoring Endogenous Peroxide Levels during the Lifespan of C. elegans.
The HyPer ratios (A and B) and the H2O2 release (C) were determined as an in vivo read-out for endogenous hydrogen peroxide levels. To determine the HyPer ratio in the body wall muscle cells of wild-type N2 [unc-54::HyPer] (A) or in the head region of wild-type N2 jrIs1[Prpl-17::HyPer] (B) animals at different stages during their lifespan, worms were cultivated at 15°C and imaged at the indicated time points. Every symbol represents the HyPer ratio of an individual animal; the bar illustrates the average HyPer ratio per day. Experiments were performed a minimum of three times and representative graphs are shown here (A & B). A one-way ANOVA followed by the Tukey multiple comparison test was performed on the log-transformed HyPer ratio, and means that are not significantly different from each other (p > 0.05) share the same letter. The hydrogen peroxide release of wild-type N2 animals (C) was measured with the Amplex®UltraRed reagent using a H2O2 standard curve. The average hydrogen peroxide release of seven independent experiments and the SEM are shown. The means are significantly different (p = 0.0011, unpaired t-test).
Although our global OxICAT analysis suggested that protein oxidation is likely a system-wide process, we still wanted to test whether other C. elegans tissues experience a similar surge in peroxide production during development. We thus conducted an equivalent time course study using a separate transgenic C. elegans strain that expresses HyPer under the control of the ubiquitous RPL-21 promoter (Back et al., 2012). This strain was recently used in monoxenic solution studies to assess the peroxide levels in dietary restricted C. elegans. We focused on the endogenous peroxide levels particularly in the head region (i.e., pharynx, neurons) of the worms (Fig. S3E) as this region revealed the strongest HyPer fluorescence signal. As before, we found significantly higher peroxide levels during development as compared to early adulthood (Fig. 4B). These results strongly suggest that developmental peroxide accumulation indeed involves different tissues of C. elegans.
Upon transition to reproductive young adults, peroxide levels appeared to massively decrease and remain low during the majority of the reproductive period. Peroxide levels then slowly increased again as the population aged (Fig. 4A and 4B). This increase was not significant in the body wall muscle cells of N2 [unc-54::HyPer] worms, potentially because the HyPer fluorescence of immobile worms was very low and could not be quantified. This might have biased the average HyPer ratio of an aged population towards healthier worms. A significant age-accompanying increase in HyPer ratio was, however, observed when we determined the HyPer ratio in the head region of N2 [rpl-17::HyPer] worms. These results agreed with recent studies, which showed a global increase in peroxide levels in older worms (Back et al., 2012).
The HyPer sensor allows for a comparative assessment of endogenous peroxide levels but not for their absolute quantification. Moreover, the HyPer ratio has been shown to be also sensitive to pH-changes (Belousov et al., 2006). To obtain a more quantitative assessment of endogenous peroxide level in wild-type worms and to exclude that pH changes are responsible for the observed decrease in HyPer ratio during the L4 - day-2 transition, we decided to monitor the release of peroxide from larval and adult worms using the peroxide-specific Amplex®UltraRed reagent (Molecular Probes), which is pH-insensitive between pH 5–10. As peroxide is freely diffusible, the release of peroxide from C. elegans should, at a minimum, correlate to the peroxide levels within the worms (Zarse et al., 2012). As before, wild-type animals were synchronized and the peroxide release of developing animals (L3) and young adults (day 2) was compared over a 1 hour time period. Later time points were not assessed by this method as Amplex®UltraRed measurements had to be done in parallel to allow direct comparison of peroxide release. This, however, involved time-delayed cultivation of the strains, which becomes more error-prone the longer the time period between start of cultivation and measurement. As shown in Fig. 4C, the peroxide release of worms during development was more than 40-fold higher than of young adult worms, indicating that peroxide production and/or secretion of H2O2 is dramatically increased in developing animals. These results indicate that peroxide production is strongly enhanced in developing C. elegans followed by a rapid cease in production and/or increased clearance as animals reach their reproductive period. These studies, together with our HyPer measurements strongly suggest that C. elegans experiences ROS accumulation at least twice in life, during development and aging.
Correlation between Early Oxidative Stress Recovery and Lifespan
Our OxICAT analysis revealed a general increase in the thiol oxidation status of protein during late development and also suggested that differences exist in the extent to which animals defective in the ILS pathway might be able to restore redox homeostasis. To address this aspect in more detail, we generated HyPer-expressing daf-2 and daf-16 strains. As before, we imaged aliquots of synchronized populations during development and adulthood to compare the HyPer ratios between the three strains. In agreement with our OxICAT analysis, we did not detect any significant differences in HyPer ratios between wild-type N2 [unc-54::HyPer], daf-16 [unc-54::HyPer], and daf-2 [unc-54::HyPer] mutant animals in early development (L2 and L3 larvae)(Fig. 5). Animals appeared to be exposed to a significant bolus of H2O2 in their body wall muscle cells (Fig. 5) implying that at this stage, signaling through the ILS pathway is not involved in ROS generation or ROS detoxification. However, a clear correlation between the time of recovery from oxidative stress and the strain background was detected. By day 2, daf-2 mutant animals had reached significantly lower steady-state levels of peroxide than the short-lived daf-16 mutants, and they maintained these low peroxide levels at least until day 20 of their lifespan. Worms lacking DAF-16, however, never fully recovered from the oxidative stress levels encountered during late development, and peroxide levels remained significantly higher throughout their entire adult lifespan. Peroxide levels increased dramatically with age, eventually reaching the same levels as observed during development (Fig. 5). These results suggest that the ability to deal with and recover from oxidative stress encountered at very early stages in life might affect redox homeostasis during adulthood and might correlate with the lifespan of C. elegans.
Fig. 5. Hydrogen Peroxide Levels in Wild-Type N2, Short-Lived daf-16, and Long-Lived daf-2 Mutants during Development and Adulthood.
The H2O2 sensor HyPer was used to monitor endogenous hydrogen peroxide levels in wild-type and mutant worms. Every symbol represents the HyPer ratio of an individual animal; daf-16 [unc-54::HyPer] is shown in red, N2 [unc-54::HyPer] in green, and daf-2 [unc-54::HyPer] in blue, and the bar depicts the average HyPer ratio per strain and day. The HyPer expression levels in daf-2 [unc-54::HyPer] L2 larvae were too low to allow accurate quantification of the HyPer ratio. Two data points for daf-2 [unc-54::HyPer] L3 larvae are outside the axis limits. Experiments were performed at least three times and a representative graph is shown. A one-way ANOVA followed by the Tukey multiple comparison test was performed on the log-transformed ratio to compare the means between genotypes within a day. P values < 0.01 are indicated with **; p values <, 0.001 are indicated with ***.
DISCUSSION
In this study, we used a set of quantitative tools to determine at what point in life and to what extent multicellular organisms like C. elegans are exposed to what type(s) of ROS. We first used the redox proteomics technique OxICAT, which provides a quantitative read-out of the thiol oxidation status of proteins. The decision to use OxICAT was based on previous redox proteomic studies that showed that typically 10% to 30% of identified thiol-containing proteins get more thiol oxidized in response to increased levels of specific oxidants and/or a more oxidizing redox potential (Held and Gibson, 2012). These proteins contain either cysteines that are exquisitely sensitive to oxidants, such as the active site cysteine of peroxiredoxin, or contain disulfide bonds with redox potentials that are close to the physiological redox potential and thus rapidly adjust their oxidation status to the prevailing redox environment. We hence reasoned that quantitative analysis of the cellular thiol redox proteome will potentially serve two purposes; to function as an endogenous monitoring device of cellular redox conditions, and if changes are observed, reveal the subset of proteins and/or pathways that might be affected by oxidation. Potential shortcomings of this approach include data representation and statistics, as quantitative mass spectrometric techniques are still error-prone and the same peptides are not necessarily identified in all samples at all times. The last aspect is particularly crucial as distinct protein thiols typically respond differently to changes in their redox environment. This, however, might decrease the number of peptides below the point of statistical analysis. In response to these challenges, we took a histogram approach, simply comparing the redox status of all those protein thiols that we identified reproducibly at all time points. By using this approach, we found that the redox proteome of wild-type C. elegans is statistically significantly more oxidized during development and during aging than during the reproductive phase of young adults. Many proteins of the identified proteins that change their oxidation status during development, adulthood and aging have been previously shown to be peroxide-sensitive (Kumsta et al., 2010). Although this finding suggested that increased peroxide levels might be responsible for the increased oxidation of these proteins, it did not serve as proof, particularly as not all previously identified peroxide-sensitive C. elegans proteins were found to be oxidized. To independently test the peroxide levels in developing larvae and young adults, we thus i) monitored the oxidation levels of peroxide-specific peroxiredoxin, ii) determined the in vivo oxidation ratio of the H2O2-specific sensor protein HyPer and iii) quantified the peroxide release using the peroxide-specific Amplex®UltraRed reagent. All three methods concurred with our OxICAT results, and showed that animals accumulate high levels of peroxide during development in contrast to very low level of peroxide during early adulthood. Importantly, we found that peroxide levels differed significantly in the ILS pathway mutants daf-2 and daf-16, a difference that first became evident in late development and persisted throughout adulthood. These results support the exciting possibility that animals with improved ability to recover from early oxidative stress might have significant advantages later in life.
Our observations concur with previous studies in C. elegans and rodents, which led the authors to conclude that events early in life might dictate lifespan (Ben-Zvi et al., 2009; Dillin et al., 2002b; Sun et al., 2009). Morimoto and coworkers, for instance, reported recently that the capacity to maintain a functional proteome (i.e., proteostasis) decreases with age and that the onset of the proteostasis collapse, which is particularly obvious in muscle cells and neurons, becomes apparent as early as day 2 or 3 of adulthood (Ben-Zvi et al., 2009). Overexpression of stress transcription factors such as heat shock factor HSF-1 or DAF-16 delayed the collapse and in turn increased lifespan, whereas deletion of either of these factors accelerated the collapse and decreased lifespan. It is well known that proteins are one of the main cellular targets of ROS. Once non-specifically modified by ROS, they often aggregate and require removal by the proteasome (Bader and Grune, 2006). Protein aggregates that are resistant to proteolytic degradation can accumulate in the cell, interfere with basic cellular functions, and ultimately lead to cell death. Based on our results that oxidative stress precedes the proteostasis collapse, it is tempting to speculate that the observed lack of recovery from developmental oxidative stress in daf-16 deletion mutants might contribute to the accelerated collapse of proteostasis and shorter lifespan. Conversely, rapidly restoring the redox conditions and keeping ROS levels low, as observed in daf-2 mutants, might delay the collapse of proteostasis.
Many processes have the potential to increase the levels of peroxide during development. One process that might contribute to increased ROS levels specifically in C. elegans is larval molting, as it has been demonstrated that the dual oxidase DUOX, a H2O2 generating enzyme, is required for correct cuticle formation (Edens et al., 2001). As peroxide is freely diffusible, this enhanced peroxide production might not only contribute to the massively increased levels of secreted peroxide that we observed during C. elegans development (Fig. 4C) but might also contribute to elevated peroxide levels in tissues bordering the hypodermis. In addition, increased ROS levels are likely also caused by higher metabolic rates, which have been reported to occur specifically during C. elegans’ development and are reflected by a peak in ATP generation between the L2 and L4 larval stage (Wadsworth and Riddle, 1989). Moreover, oxidants have been shown to play crucial roles as second messengers in signal transduction, playing a role in development and differentiation (Cross and Templeton, 2006).
Our in vivo redox sensors revealed that C. elegans has evolved effective mechanisms to recover from the developmental oxidative burst by the time reproductive age is reached. At least one pathway that we identified to be involved in this recovery process is the ILS pathway (Kenyon, 2010), which functions to mediate stress resistance and lifespan in adult C. elegans (Dillin et al., 2002a). The long-lived daf-2 mutant lacks the negative regulation of the transcription factor DAF-16, which positively controls expression of stress response genes like superoxide dismutase and catalase and thus mediates increased antioxidant capacity. It is interesting to note that we did not observe any significant differences in early developmental ROS levels between short- and long-lived ILS mutants, suggesting that at this stage, DAF-16-mediated expression of antioxidant genes might not occur, thus enabling ROS signaling. It is hence tempting to speculate that growth pathways such as the ILS pathway might support growth in part by allowing increased ROS signaling to occur. We did, however, find a significant difference in the ability of daf-16 mutants to recover from oxidative stress and to achieve the low ROS levels that appear to accompany adult lifespan. While daf-2 mutants recovered from oxidative stress during early adulthood and maintained low ROS levels throughout much of their mature life, daf-16 mutants failed to fully recover and showed significantly increased ROS levels throughout their life. These results are in good agreement with ILS timing studies, which showed that the ILS pathway influences lifespan particularly when disrupted in the early days of adulthood (Dillin et al., 2002a). Other antioxidant systems that might play a role in the detoxification of peroxide upon transition from development to adulthood are peroxiredoxin-2 (PRDX-2), which has been previously shown to promote C. elegans’ recovery from exogenous peroxide treatment (Kumsta et al., 2010) and catalases (CTL-2). Deletion of either one of these two main peroxide-detoxifying enzymes causes progeric phenotypes that become apparent as early as in young adults, and significantly shortens the lifespan of C. elegans (Kumsta et al., 2010; Petriv and Rachubinski, 2004). In contrast, absence of superoxide dismutase (SOD) isoforms in C. elegans alone or in combination has none or only minor effects on the lifespan of C. elegans (Doonan et al., 2008), and mutants lacking all five SOD-isoforms show a normal lifespan (Van Raamsdonk and Hekimi, 2012). With OxICAT, HyPer, and other related redox sensors, we now have the unique opportunity to precisely track reactive oxygen species in vivo and to distinguish between ROS that might or might not be relevant for lifespan.
A recent study demonstrated that stochastic variances in the expression levels of a stress-inducible gene in early adulthood of C. elegans predicts life expectancy in an isogenic population, thus serving as biomarkers of aging (Rea et al., 2005). Our observation that in vivo peroxide levels vary dramatically within an isogenic population of the same chronological age (Fig. 4 and 5) supports now the exciting possibility that we have discovered not only another biomarker of aging but possibly one of the main factors contributing to stochastic variances. As transcriptional and epigenetic control mechanisms involve redox-regulated proteins (Cyr and Domann, 2010), early variances in ROS level might play the important role of individualizing gene expression and ultimately determining lifespan.
EXPERIMENTAL PROCEDURES
Strains, Culture Conditions, and Lifespan Analysis
The C. elegans wild-type strain N2 and GR1307 (daf-16(mgDf50)I) were provided by the Caenorhabditis Genetics Center. Strain CF1041 [(daf-2(e1370)] was provided by A. Hsu. The HyPer expressing strain N2 (jrIs1[Prpl-17::HyPer]) was provided by B. Braeckman (Back et al., 2012). Strains were cultured on Nematode Growth Media (NGM) agar plates at 15 °C using 1010 cells/ml OP50 as food source and synchronized according to (Kumsta et al., 2010). To prevent hatching of progeny, fertile adults were cultured on plates containing 20 mg/l FUdR (Sigma). The L4 molt stage was considered day 0 of adulthood. Animals were considered dead when they did not move or respond to prodding. Animals that crawled off the plate were censored at the time of the event. Generation of the transgenic animals and chromosomal integration are described in detail in the Supplemental Information.
Sample Preparation for OxICAT
A synchronized population of wild-type or mutant worms was cultivated at 15°C and aliquots of worms were taken at the indicated time points. Samples for OxICAT were prepared as previously described (Kumsta et al., 2010; Leichert et al., 2008).
Worm Image Acquisition and Image Quantification
Animals of a synchronized population were taken for image acquisition at different time points during larval development and adult life. Worms were mounted on a 2% agarose pad and immobilized using 2 mM levamisole hydrochloride. Up to 60 animals were imaged per day and group. A detailed description of the image acquisition and quantification process can be found in the Supplemental Information.
Amplex®UltraRed Assay
To monitor the release of hydrogen peroxide from C. elegans over time, the Amplex®UltraRed reagent (Molecular Probes) was used according to the manufacturer’s protocol with small modifications. A detailed description of this assay can be found in the Supplemental Material. In brief, synchronized N2 wild-type worms were arrested in L1 and allowed to resume growth in a time-delayed manner allowing the analysis of peroxide release from developing worms (L3 larva) and young adults (day 2) at the same day. The worms were removed from NGM plates, washed with M9 buffer, then with 1X Reaction Buffer (Molecular probes) supplemented with 0.05% Triton X-100, and resuspended in 1X Reaction Buffer/0.05% Triton X-100. 50 μl of the worm solution was assayed in triplicates according to the manufacturer protocol, and peroxide release per minute was calculated based on a H2O2 standard curve and normalized to the protein concentration.
Statistical Analysis
GraphPad Prism 5 (Version 5.01) was used for statistical analysis. Lifespan data were analyzed using the log-rank test (Mantel Cox) or the Gehan-Breslow-Wilcoxon test; p values < 0.05 are considered significant. The HyPer ratio values were log-transformed and analyzed with one-way ANOVA followed by the Tukey multiple comparison test. The mean values from the PRDX-2 redox state and the Amplex®UltraRed experiments were compared with an unpaired t-test with p < 0.05 indicated as* and p < 0.01 indicated as **. OxICAT data were analyzed using Kruskal-Wallis one-way ANOVA followed by Dunn’s post test.
Supplementary Material
HIGHLIGHTS.
Redox sensors monitor protein oxidation states and peroxide levels during lifespan
C. elegans accumulates oxidants twice in life; during development and during aging
Short-lived mutant retain higher oxidant levels throughout adulthood
Early oxidative stress might have severe impact on life span
Acknowledgments
We thank C. Kumsta for initiating some of the work and D. Reichmann for help with the OxICAT analysis. We are indebted to B. Braeckman and P. Back for providing the HyPer strain N2 (jrIs1[Prpl-17::HyPer]). We are grateful to J. Bardwell for many helpful discussions and thank A. Kaploe and G. Bode for database searches. We thank the Caenorhabditis Genetics Center (CGC) and A.-L. Hsu and V. Belousov for providing us with strains and HyPer constructs. We thank A. Fire for providing the Addgene plasmids, the Michigan Proteome Consortium for mass spectrometric analysis, W. Bae and Union Biometrica for conducting the worm sorting experiments, and R. Denver and R. Miller for statistical advice. This work was supported by the National Institute of Aging grant AG027349 and an OVPR grant from the University of Michigan (to U.J.) and by the NIH National Center for Research Resources, which funds CGC. N.J.N was as supported by NIH T-32-GM007315.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Back P, De Vos WH, Depuydt GG, Matthijssens F, Vanfleteren JR, Braeckman BP. Exploring real-time in vivo redox biology of developing and aging Caenorhabditis elegans. FRBM. 2012;52:850–859. doi: 10.1016/j.freeradbiomed.2011.11.037. [DOI] [PubMed] [Google Scholar]
- Bader N, Grune T. Protein oxidation and proteolysis. Biol Chem. 2006;387:1351–1355. doi: 10.1515/BC.2006.169. [DOI] [PubMed] [Google Scholar]
- Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. PNAS. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini G, Donati A, Gori Z, Bergamini E. Towards an understanding of the anti-aging mechanism of caloric restriction. Curr Aging Sci. 2008;1:4–9. doi: 10.2174/1874609810801010004. [DOI] [PubMed] [Google Scholar]
- Cross JV, Templeton DJ. Regulation of signal transduction through protein cysteine oxidation. ARS. 2006;8:1819–1827. doi: 10.1089/ars.2006.8.1819. [DOI] [PubMed] [Google Scholar]
- Cyr A, Domann F. The Redox Basis of Epigenetic Modifications: From Mechanisms to Functional Consequences. ARS. 2010;15:551–589. doi: 10.1089/ars.2010.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002a;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002b;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Gene Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens WA, Sharling L, Cheng G, Shapira R, Kinkade JM, Lee T, Edens HA, Tang X, Sullards C, Flaherty DB, et al. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. JCB l. 2001;154:879–891. doi: 10.1083/jcb.200103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaschi T, Cozzi G, Raugei G, Formigli L, Ramponi G, Chiarugi P. Redox regulation of beta-actin during integrin-mediated cell adhesion. JBC. 2006;281:22983–22991. doi: 10.1074/jbc.M603040200. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Held JM, Gibson BW. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol Cell Prot. 2012;11:R111 013037. doi: 10.1074/mcp.R111.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe PA, Banerjee A, Luduena RF. The roles of cys124 and ser239 in the functional properties of human betaIII tubulin. Cell Motil Cytoskeleton. 2008;65:476–486. doi: 10.1002/cm.20274. [DOI] [PubMed] [Google Scholar]
- Johnson TE. Caenorhabditis elegans 2007: the premier model for the study of aging. Exp Gerontol. 2008;43:1–4. doi: 10.1016/j.exger.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kumsta C, Thamsen M, Jakob U. Effects of Oxidative Stress on Behavior, Physiology, and the Redox Thiol Proteome of Caenorhabditis elegans. ARS. 2010;14:1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. PNAS. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nature Genetics. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Maeda K, Finnie C, Svensson B. Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. The Biochem J. 2004;378:497–507. doi: 10.1042/BJ20031634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Olahova M, Taylor SR, Khazaipoul S, Wang JL, Morgan BA, Matsumoto K, Blackwell TK, Veal EA. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. PNAS. 2008;105:19839–19844. doi: 10.1073/pnas.0805507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez VI, Bokov A, Van Remmen H, Mele J, Ran Q, Ikeno Y, Richardson A. Is the oxidative stress theory of aging dead? Biochim Biophys Acta. 2009;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petriv OI, Rachubinski RA. Lack of peroxisomal catalase causes a progeric phenotype in Caenorhabditis elegans. JBC. 2004;279:19996–20001. doi: 10.1074/jbc.M400207200. [DOI] [PubMed] [Google Scholar]
- Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Phy Endocrinol Metab. 2005;289:E23–29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Sun L, Sadighi Akha AA, Miller RA, Harper JM. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J Gerontol A Biol Sci Med Sci. 2009;64:711–722. doi: 10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. PNAS. 2012;109:5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth WG, Riddle DL. Developmental regulation of energy metabolism in Caenorhabditis elegans. Dev Biol. 1989;132:167–173. doi: 10.1016/0012-1606(89)90214-5. [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired Insulin/IGF1 Signaling Extends Life Span by Promoting Mitochondrial L-Proline Catabolism to Induce a Transient ROS Signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.