Abstract
Proprotein convertases (PCs), furin and proprotein convertase 1/3 (PC1), cleave substrates at dibasic residues along the eukaryotic secretory/endocytic pathway. PCs are evolutionarily related to bacterial subtilisin and are synthesized as zymogens. They contain N-terminal propeptides (PRO) that function as dedicated catalysts which facilitate folding and regulate activation of cognate proteases through multiple-ordered cleavages. Previous studies identified a histidine residue (His69) that functions as a pH sensor in the propeptide of furin (PROFUR), which regulates furin activation at pH~6.5 within the trans Golgi network. Although this residue is conserved in the PC1 propeptide (PROPC1), PC1 nonetheless activates at pH~5.5 within the dense core secretory granules. Here we analyze the mechanism by which PROFUR regulates furin activation and examine why PROFUR and PROPC1 differ in their pH-dependent activation. Sequence analyses establish that while both PROFUR and PROPC1 are enriched in histidines when compared with cognate catalytic-domains and prokaryotic orthologs, histidine content in PROFUR is ~two-fold greater than PROPC1, which may augment its pH sensitivity. Spectroscopy and molecular dynamics establish that histidine-protonation significantly unfolds PROFUR when compared to PROPC1 to enhance autoproteolysis. We further demonstrate that PROFUR and PROPC1 are sufficient to confer organelle-sensing on folding and activation of their cognate proteases. Swapping propeptides between furin and PC1 transfers pH-dependent protease activation in a propeptide-dictated manner in vitro and in cells. Since prokaryotes lack organelles and eukaryotic PCs evolved from propeptide-dependent, not propeptide-independent prokaryotic subtilases, our results suggest that histidine enrichment may have enabled propeptides to evolve to exploit pH-gradients to activate within specific organelles.
Keywords: Protease activation, subtilisin, folding, molecular dynamics, pH-sensor
INTRODUCTION
Proprotein convertases (PCs) are endoproteases that mediate diverse regulatory and protective processes by controlled proteolysis of their substrates1; 2; 3. The PC-family includes seven mammalian Ca2+ dependent endoproteases: furin, PC1/PC3, PC2, PC4, PACE4, PC5/PC6, and PC7/LPC/PC83; 4; 5; 6; 7. More recently, SKI/S1P8; 9 and NARC-1/PCSK910 have also been identified as enzymes that share sequence similarity with PCs11. Structures of the catalytic domains of furin12 and yeast kexin13 have been solved using X-ray crystallography, and homology models for PCs that were derived from these structures provide the basis for substrate specificity12; 13; 14; 15. Although PCs potentially share overlapping cleavage specificity and function, each PC catalyzes limited proteolysis of proprotein and prohormone substrates at a pair of basic residues to excise bioactive proteins and peptides within specific compartments of the TGN/endosomal system that are characteristic of eukaryotic cells1; 11; 16. The necessity for such precise spatiotemporal cleavage of substrates mandates the activity of PCs to be likewise stringently controlled17. Dysregulation of PC activity has been observed in various diseases such as cancer18; 19, obesity20; 21, diabetes22 and heart disease23. Consistent with these observations, small-molecule inhibitors of the constitutively expressed furin can inhibit cancer cell motility and invasiveness24.
The activity of PCs is regulated by N-terminal propeptide-domains25 which function initially as folding catalysts that facilitate folding of cognate protease domains, and subsequently serve as temporary inhibitors that mask the protease active site17. Given their ability to chaperone single-turnover folding, these propeptides are often referred to as intramolecular chaperones (IMCs)26. IMCs constitute diverse, substrate-specific, single-turnover, energy-independent chaperones27; 28; 29 whose primary sequences have diverged faster than their target client substrates30 and are distinct from substrate-promiscuous, multi-turnover, energy-dependent inter-molecular chaperones17; 30. PCs are homologs of prokaryotic subtilases31; 32; 33, proteins in which the roles of propeptides have been thoroughly investigated. Analysis of prokaryotic subtilases and other proteases suggests that propeptides evolved to regulate protease folding within harsh extracellular environments such as soil or vegetation 27; 34; 35; 36; 37; 38, and are absent from paralogs functioning in milder intracellular environments34.
Subsequent to guiding protease-domain folding, propeptide-dependent subtilases undergo ordered proteolytic cleavages within their propeptide-domains. The first cleavage forms catalytically inactive propeptide:protease inhibition complexes wherein propeptides non-covalently bind to protease active-sites, while subsequent cleavages activate proteases by facilitating propeptide dissociation, enabling the now unmasked catalytic domain to cleave substrates in trans11; 28; 29; 39. While these obligatory cleavages are extracellular events in prokaryotes that delay onset of protease activity until after protein export, they control secretory pathway compartment-specific activation of substrate-specific eukaryotic proprotein convertases (PCs)11. Since eukaryotic PCs evolved from propeptide-dependent, and not propeptide-independent prokaryotic subtilases34, it is tempting to speculate that propeptides confer functional advantages through speciation, namely to regulate organelle-specific activation of secretory-pathway proteases, a complexity absent in unicellular prokaryotes that is essential to maintain physiological homeostasis within eukaryotic cells40; 41; 42. For example, the activation of furin is regulated in a pH-dependent manner as it transits the secretory pathway28. In the neutral pH in the ER, the propeptide is cleaved to form a stoichiometric propeptide:furin inhibition complex. Upon reaching the early Trans-Golgi network (TGN; pH 6.5) the furin propeptide (PROFUR) undergoes a second cleavage which removes the inhibitory propeptide and thus activates furin28. While PC1 transits the secretory pathway in much the same way, the PC1 propeptide (PROPC1) remains in a stoichiometric complex with the PC1 protease domain until undergoing its activating second cleavage upon reaching the dense core secretory granules (DCSGs; pH 5.5). A study by Feliciangeli et al.29 demonstrated that in furin, mutating residue His69 in the propeptide to leucine blocks activation of the complex in the TGN while allowing for correct folding, while a His69Lys substitution results in accumulation of unprocessed furin precursor in the ER29. On this basis, they suggested that the His69 in the propeptide is not only important for folding of furin, but is also a vital pH-sensor that regulates furin activation in the pH of the TGN. However, mechanisms by which the His69 functions as a pH sensor in furin are unknown. Moreover, while the residue corresponding to His69 (in furin) is strictly conserved within the PC-family, PC1 and furin undergo their activating second cleavages at different pH within the TGN and DCSGs, respectively. This suggests that additional factors may play a role in regulating activation of the protease domains.
In this manuscript we demonstrate through various biophysical, biochemical, cell-based and computational approaches that the propeptide domains of furin and PC1 (PROFUR and PROPC1, respectively) contain sufficient information to confer organelle-sensing on the folding and activation of cognate proteases. Circular dichroism spectroscopy as a function of pH establishes that the pH-dependent stability of propeptide domains coincides with the optimum pH for compartment specific activation. Monitored by ellipticity at 222 nm the PROFUR undergoes a transition in structure, the midpoint of which occurs at pH 6.5, while the midpoint in structural transition for PROPC1 occurs at a lower pH (pH~5.5). Furthermore, swapping propeptides between eukaryotic paralogs —furin and PC1— transfers pH-dependent protease activation in a propeptide-dictated manner in vitro and in cells. Our results suggest that PROFUR and PROPC1 encode information essential for regulating compartment specific activation of cognate proteases and that other residues in addition to the conserved pH sensor His69 are necessary to enable subtle differentiation in pH-dependent activation between furin and PC1. Using molecular dynamics simulations, we also demonstrate that histidine protonation leads to conformational changes in PROFUR but not in PROPC1. Together, our results provide insights into the structural mechanisms by which propeptides can regulate the pH-dependent activation of their cognate PCs.
RESULTS
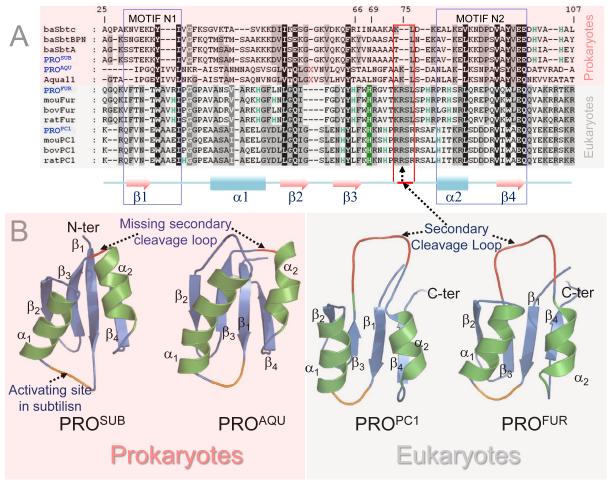
Eukaryotic propeptides harbor an internal cleavage site loop that is missing within their prokaryotic paralogs
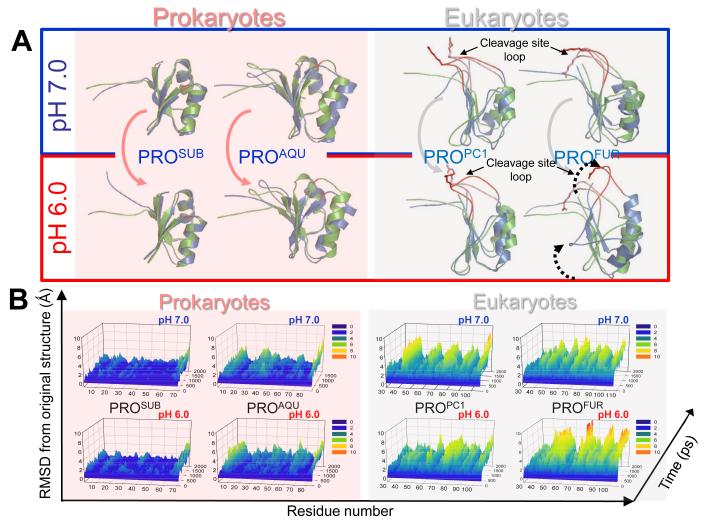
To understand how eukaryotic propeptides can mediate compartment specific activation of their cognate protease domains, we compared sequences and structures of prokaryotic propeptides— subtilisin (PROSUB) and aqualysin I (PROAQU) —with eukaryotic propeptides— pro-protein convertase 1 (PROPC1) and furin (PROFUR). While several laboratories have analyzed the sequences and structures of propeptides, to date no detailed comparison between the sequences and structures of the propeptides of prokaryotic and eukaryotic proteins has been conducted. PROAQU was selected because unlike its intrinsically unfolded prokaryotic homologue PROSUB, PROAQU adopts a well-defined structure and chaperones folding of its cognate protease domain43. From the PC-family members, we selected PROPC1 and PROFUR because despite significant sequence and structural similarity with prokaryotic orthologs (Fig.1A and 1B), they activate in different organelles along the proton-gradient of the secretory pathway, a complexity missing in prokaryotes. Furin is optimally active at pH 6.5, consistent with its role in cleaving proprotein substrates in the mildly acidic environment of the TGN/endosomal system. PC1 is optimally active at pH 5.5, consistent with its role in cleaving prohormone molecules in secretory granules.
Fig.1. Comparison of sequences, structures, evolution and composition biases of propeptides in prokaryotic and eukaryotic subtilases.
The pink and grey background in panel A through D indicates prokaryotes and eukaryotes, respectively. (A) Multiple Sequence Alignment (MSA) displaying conservation between eukaryotic subtilases and prokaryotic orthologs. Numbering is based on furin. Residues shaded black are 100% conserved, dark grey >80%, and light grey >50%. The conserved pH-sensor in furin is shaded green and the secondary cleavage loop is indicated by the red box. Red X’s represents an insertion of 5 residues in aqualysin. Pink shading represents prokaryotes while the light gray represents eukaryotes. Secondary structures displayed below MSA are based on PROPC1 (1KN6). Motifs N1 and N2 depict folding nucleation sites for MATSUB. (B) Structures of propeptides displayed as ribbon diagrams. PROSUB structure was extracted from the propeptide:subtilisin (1SCJ), while PROAQU structure is a homology model based on 1SCJ and 2W2M. The structure of PROPC1 is derived from the NMR (1KN6) while PROFUR represents a homology model of PROPC129. (C) Heat map displaying amino acid content within the propeptides and catalytic domains of prokaryotic subtilisin and aqualysin and eukaryotic PCs, furin and PC1. Protein sequences for furin (n=26), PC1 (n=14), subtilisin (n=69), and aqualysin (n=7) families were obtained from the 50% sequence identity clusters UniRef50_P09958, UniRef50_P29120, UniRef50_P00782, and UniRef50_P08594 in the UniRef database, respectively. Amino acid content for each family of propeptides and protease domains were calculated and averaged. Contents of amino acids belonging to individual groups were added and divided by the sum of their content in the whole UniProt database (release 2011_12) to obtain the fold change. Within an individual group, fold values greater than one indicates residue enrichment, values less than one indicate residue depletion while a value of one indicates no change.
Amino acids absent between residues 75-81 in PROSUB (red box; Fig. 1A) coincide with organelle-specific cleavage-sites 29 within eukaryotes (red loop; Fig. 1B). In prokaryotic subtilases, the secondary cleavage site is fairly promiscuous and presumably occurs in the flexible region between β1 and α1 (Fig 1B). Additionally, there are significant differences in residues 100-107 within the propeptide-domains between prokaryotic subtilisins and eukaryotic PCs. This C-terminal region harbors the primary cleavage site within propeptides and interacts with the substrate binding regions within cognate proteases to initiate activation. It is noteworthy that cellular substrates of PCs contain the consensus sequence [R/K]-Xn-[R/K]↓, identical to the primary cleavage site within propeptides44. Given the promiscuous specificity of bacterial subtilases when compared to the stringent substrate specificity of eukaryotic PCs, the differences between residues 100-107 reflects the requirement of PCs to cleave at highly conserved dibasic residues. This region reflects the divergence of propeptides from prokaryotes and eukaryotes to function with more cleavage specificity, likely due to the difference in cellular environment, namely, the inclusion of membrane bound organelles in eukaryotes17.
PROFUR and PROPC1 are rich in histidine residues when compared with PROSUB and PROAQU
The pH within an organelle can dramatically affect the ionization states of charged residues in a protein sequence, by altering its structure, stability and function. Hence, we next analyzed the fold increase in amino acid residues within the propeptides and cognate proteases within prokaryotic and eukaryotic subtilases using the averaged amino acid distribution within the Uniprot database as our baseline (Fig. 1C). The individual amino acid content for each family of propeptides and proteases were calculated and averaged. The contents of amino acids belonging to individual groups were added and divided by the sum of their content in the whole UniProt database (release 2011_12) to obtain the fold change as described in the Methods Section. Fold values greater than one (varying shades of red) indicate residue enrichment in propeptide domains within an individual group, values less than one (varying shades of green) indicate depletion of specific residues within propeptides, while a value of one (white) indicates no change. This graphical representation of the fold increase in specific groups of amino acid residues (Fig. 1C) demonstrates that the His content in PROFUR and PROPC1 from eukaryotes is significantly greater that their cognate catalytic domains and prokaryotic paralogs. Furthermore, protease domains of prokaryotes are biased towards acidic and basic residues as demonstrated by Inouye and co-workers30; 45; 46 which was hypothesized to enhance kinetic stability within their catalytic domains34. The average composition of proteins in the Uniprot database establishes histidine (2.27%) as the third least abundant residue, and is ~four-fold less than leucine (9.67%), the most abundant residue. While propeptide domains generally display a bias for charged and polar residues when compared to proteases30, it is noteworthy that within subtilases, only eukaryotic propeptides are rich in histidine-content (Fig. 1C) when compared with prokaryotic propeptides and cognate catalytic-domains. Similar results are observed when propeptides within the PC-family are compared with their cognate catalytic domains and prokaryotic orthologs (data not shown). Histidine is a unique residue because the pKa of its imidazole side-chain (pH~6.0) is close to physiological pH, and relatively small shifts along the proton gradient can change the net charge, and subsequently alter pH-dependent conformational stability of propeptides.
PROFUR regulates furin activation by acting as a pH-sensor that delays the internal propeptide cleavage until after the PROFUR:furin complex is trafficked to the mildly acidic TGN/endosomal system. Protonation of His69 forms a cleavable furin site at Arg75 which releases the bound propeptide from the catalytic domain29. Based on the demonstration of a histidine driven pH-sensor in PROFUR 29 and the histidine bias within PROFUR and PROPC1 from eukaryotes when compared with PROSUB and PROAQU from prokaryotes, we propose that histidine protonation may regulate organelle-specific propeptide-release/degradation to regulate furin and PC1 activation in endosomal/lysosomal compartments elaborated in eukaryotic cells.
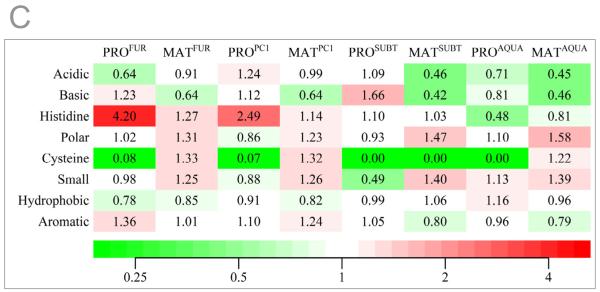
Circular dichroism spectroscopy demonstrates pH dependent structural changes in eukaryotic propeptides
Since the pKa (~6.0) of the imidazole side-chain of histidine is close to physiological pH, we next investigated whether small changes in proton concentration alter pH-dependent structural stability of propeptides in prokaryotes and eukaryotes. The secondary structures measured using circular dichroism spectroscopy measured at pH 7.0 demonstrates that PROFUR and PROPC1 adopt structures similar to PROAQU and PROSUB-C complexed to subtilisin (Fig. 2A). Since isolated PROSUB is intrinsically unstructured47, PROSUB-C structure was obtained by a difference spectra between the cleaved PROSUB:S221C-subtilisin complex and mature subtilisin as described earlier48.
Fig. 2. pH dependent structure and function propeptides.
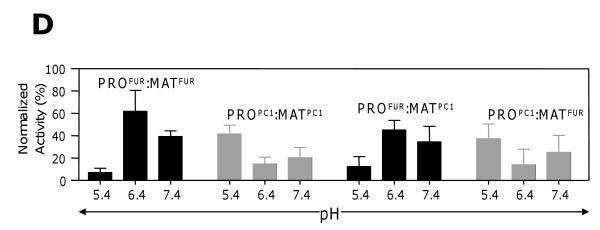
(A) Secondary structures determined using CD spectroscopy performed at a pH 7.0 and plotted as molar ellipticity [θ] deg.cm2.dmol−1. (B) Structural stability of propeptides monitored by changes in ellipticity at 222 nm as a function of pH. (C) The secondary structure of PROFUR at pH 7.4 and 5.0, compared with completely denatured furin. The arrow marks 222 nm on the scale. (D) Type of eukaryotic propeptide dictates pH-optimum for activation of propeptide:protease complex. The activation optimum for MATFUR shifts from pH~6.5 in the presence of PROFUR to pH~5.5 when PROPC1 forms the complex. Conversely, MATPC1 activation shifts from pH~5.5 in presence of PROPC1 to pH~6.5 when PROFUR forms its complex. Values are measurements of three independent experiments.
The pH dependent structural stability of various propeptides was monitored by observing changes in negative ellipticity at 222 nm as a function pH (Fig 2B); as a representative example, we show the complete CD spectrum of PROFUR at the two ends of the pH range (pH 7.4 and pH 5.0) compared with a completely denatured PROFUR (Fig 2C). It is noteworthy that when the pH of the buffer is lowered from pH 7.4 to pH 5.0, PROFUR loses approximately 25% of its ellipticity at 222 nm when compared with the propeptide completely denatured in 8 M urea. Furthermore, changes in negative ellipticity at 222 nm as a function of pH (Fig 2B) suggest that the conformation of PROFUR tends to stabilize at approximately - 2800 deg.cm2/dmol−1 under acidic conditions, but does not reach the ellipticity of completely unfolded PROFUR (approximately -20 deg.cm2/dmol−1). This suggests that the changes in pH do not result in complete unfolding and that PROFUR may adopt a partially folded molten-globule like state similar to that observed using NMR spectroscopy under acidic conditions49. The NMR data also suggest that PROPC1 and PROFUR do not aggregate in their isolated forms.
When conformational changes of the propeptides as a function of pH are compared, it is evident that PROPC1 and PROFUR unfold at different pHs, ~5.5 and ~6.5, respectively (Fig. 2B). Although the unfolding of PROPC1 is not complete at pH 5.0, the structure of PROPC1 at a pH below 5.0 was not analyzed because it is beyond the range of the buffering capacity of our system. While this prevents the accurate determination of the mid-point of unfolding transition in the case of PROPC1, changing buffer systems to accommodate lower pH is problematic because diverse ions that can differentially influence structure, stability and/or activity of the propeptide and protease system. Nonetheless, comparing the folding transitions profiles of PROFUR and PROPC1 suggests that PROPC1 is more stable with regards to pH dependent unfolding when compared with PROFUR. Under similar conditions, PROSUB and PROAQU are stable with minor changes in conformation. Due to its intrinsically unstructured state, PROSUB would not be expected to undergo conformational changes as a function of pH. However, studies have suggested that an increase in proton concentrations can induce molten-globule like states into unfolded proteins50; 51; 52; 53. Our studies suggest that acid induced folding is not observed in case of PROSUB. It is noteworthy that the pH-associated structural transitions PROPC1 and PROFUR correlates with organelle-specific pHs necessary for activating the mature catalytic domains, MATPC1 and MATFUR 3. We next investigated whether propeptides alone are sufficient for pH-dependent activation of cognate proteases, in vitro and in tissue culture cells.
Swapping propeptides between PC1 and furin reassigns pH-dependent activation
To monitor in vitro activation of propeptide:protease inhibition complexes, we measured enzyme activity as a function of pH (see Methods). Fig. 2D demonstrates that PROFUR:MATFUR and PROPC1:MATPC1 show maximum activation at pH~6.5 and pH~5.5, respectively, consistent with the optimal activation pH of their zymogens3. However, the PROPC1:MATFUR complex (wherein PROPC1 substitutes PROFUR) forces the catalytic-domain of furin (MATFUR) to now display PC1-like activation. Similarly, replacing PROPC1 with PROFUR causes the catalytic-domain, MATPC1, to alter its activation to mimic furin (pH~6.5; Fig. 2D). Together, the CD spectroscopy, sequence/structural congruence with PROSUB, and the reassignment of activation pH by swapping PROFUR and PROPC1 support the hypothesis that eukaryotic propeptides recognize and regulate pH-dependent activation of their cognate proteases in vitro.
PROPC1 and PROFUR control folding and activation of MATFUR and MATPC1 in cells
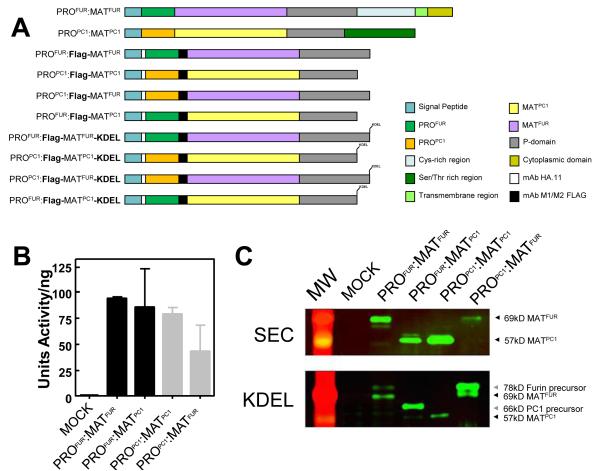
Since only correctly folded secretory proteins are efficiently transported from the ER54, we measured catalytic activities of chimeras (PROPC1:MATFUR; ~78kDa and PROFUR:MATPC1; ~66kDa; Fig. 3A) as readouts for folding/activation, using wild-type constructs (PROFUR:MATFUR and PROPC1:MATPC1) as controls. All constructs display protease activity when compared with mock transfections (Fig. 3B). To monitor primary cleavage of propeptides, we inserted FLAG epitopes between the C-termini of propeptides and N-termini of proteases. These epitopes, which do not affect trafficking, activation, or activity of furin25; 55, confirm presence of the processed MATFUR (~69kDa) and MATPC1 (~57kDa) in the conditioned media when probed using Western Blot analysis (Fig. 3C, top panel as indicated by arrowheads). Together, the catalytic activities and western blots establish that propeptides can assist folding and activation of their paralogs in cells. To isolate cleaved inhibition complexes, we assayed each variant using ER-localized constructs wherein the transmembrane- and cytosolic-domains were replaced by the ER localization motif -Lys-Asp-Glu-Leu (KDEL) (Fig. 3A). The KDEL motif restricts zymogen reporters to the neutral pH environment of the ER56, where constructs undergo primary propeptide-cleavages that form PRO:MAT complexes but are blocked from secondary cleavages28. Western blot analyses (see Methods) confirm the presence of both unprocessed and processed precursors. Fig. 3C (lower panel as indicated by arrowheads) demonstrates the presence of both the protease domain that has undergone the primary processing step to generate the 69kD and 57kD forms of furin and PC1, respectively, as well as the immature protease that has yet to undergo this processing step (78kD and 66kD for furin and PC-1, respectively). The constructs expressing the furin protease domain reliably express at higher levels in the cell culture system we have chosen, regardless of which propeptide it is in complex with, thus the ease in visualizing the two different species. In contrast, the PC1 protease does not express as strongly. Nonetheless there is evidence of both the 66kD unprocessed and 57kD processed forms of PROFUR-MATPC1 as indicated by the arrowheads in Fig. 3C, although the efficiency of this processing is less than the PROPC1-MATPC1 which appears to undergo efficient autoprocessing under similar conditions. The differences in the processing efficiency of furin and PC1 may reflect differences in the catalytic domains.
Fig. 3. Transferring propeptides between eukaryotic subtilases reassigns their optimum pH for activation.
(A) Schematic of constructs for Furin and PC1. (B) Normalized protease activity assayed in conditioned media (CM) from Cos-7 cells transfected with 2 μg of DNA (C) Western blot analysis of CM from cells expressing secreted reporter constructs (top panel; SEC), and ER fractions from cells expressing KDEL-tagged reporters probed using mAB-M2. Molecular weight of each species is indicated by the arrowheads; Unprocessed furin, 78kD; Processed furin, 69kD; Unprocessed PC1, 66kD; Processed PC1 57kD. (D) pH-dependent activation of KDEL-tagged reporters measured after incubating ER membrane fractions at designated pH29. Maximal activity was estimated by trypsinizing membrane fractions for 1 hr and inhibiting trypsin by soybean trypsin inhibitor prior to the protease assay.
We next examined pH-dependent activation of these KDEL-tagged chimeras using wild-type KDEL-tagged reporters and mock transfections as positive and negative controls, respectively. Fig. 3D confirms that while maximal activation of PROFUR:MATFUR-KDEL occurs at pH ~6.5, the activation of PROPC1:MATFUR-KDEL shifts to pH ~5.5. Conversely, activation of PROPC1:MATPC1-KDEL shifts from pH~5.5 to pH~6.5 when PROFUR is used to fold MATPC1-KDEL in COS-7 cells. Although experiments conducted using crude membrane fractions have higher background activity, they nonetheless confirm that propeptide-dictated reassignment of pH-dependent activation is consistent with the in vitro activation of inhibition complexes (Fig. 2D) and demonstrates that propeptides regulate compartment-specific pH-dependent activation of furin and PC1.
Histidine-protonation alters conformational dynamics of eukaryotic propeptides
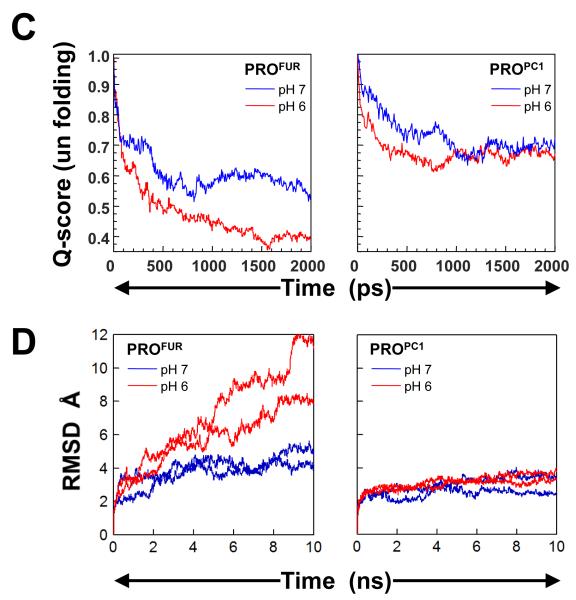
Based on experimental studies, we had hypothesized that the protonation of His69 and potentially other histidines may induce conformational changes within PROFUR to mediate pH dependent activation29. Moreover, although His69 is conserved, PROPC1 undergoes its pH dependent activation at a much lower pH (5.0). To better understand how histidine protonation may influence propeptide conformations, we conducted MD simulations on PROFUR and PROPC1 with unprotonated (pH 7) or protonated (pH 6) histidine residues, using PROSUB and PROAQU from prokaryotes as controls. MD simulations57 can provide information that complements biophysical and biochemical studies on mechanisms of propeptide-mediated protease activation in eukaryotes (see Methods). Early MD simulations of the unfolding of reduced bovine pancreatic trypsin inhibitor (BPTI) on a 500ps time scale suggest the formation of a molten-globule like state that was compact but expanded relative to the native BPTI (11-25%) that is consistent with experimental data58; 59. MD simulations have also analyzed the structure and fluctuations of “native” apomyoglobin in aqueous solution for a period of greater than 0.5 nanoseconds and has yielded a detailed model for structure and fluctuations in apomyoglobin which complements the experimental studies60. Unfolding simulations using MD methods have yielded insights into the mechanism of extreme unfolding cooperativity in the kinetically stable alpha-lytic protease, a protein that exploits the mechanism of propeptide-dependent folding61. In these studies the simulated alpha-lytic protease unfolding pathway produces a robust transition state ensemble that is observed within the 10ns simulation and is consistent with prior biochemical experiments demonstrating that unfolding proceeds through a preferential disruption of the domain interface. Furthermore, the authors demonstrate that αLP unfolds extremely cooperatively while, trypsin, a protein that folds independent of its propeptide, undergoes gradual unfolding under identical conditions of simulations. MD simulations studies have also been used to investigate the role of hydrogen bonding involving the backbone in hen egg white lysozyme, using native as well as partly and fully thionated lysozyme62. The results of the simulations show that the structural properties of fully thionated lysozyme clearly differ from those of the native protein, while for partly thionated lysozyme changes only slightly when compared to native lysozyme. In these studies, the extent of observed unfolding remains constant after 10ns. Hence in our studies are performed MD simulations on a 10ns time-scale. We compared the similarity of structures to the starting conformation by measuring the root-mean-square deviation (RMSD) values at alpha carbons in every residue of the propeptide-domain, along equally spaced snapshots of the simulation trajectory. Simulations suggest that while PROSUB and PROAQU are stable, PROPC1 and PROFUR display enhanced conformational dynamics (Fig. 4A and B). Our time-evolved, pH-dependent, residue-specific conformational dynamics suggest that although eukaryotic propeptides display local fluctuations at neutral pH, histidine protonation enhances overall movement and potentially exposes the compartment-specific second cleavage-site loop for proteolysis in PROFUR (residues 70 to 80) when compared with PROPC1, which is more stable at pH~6.0-7.0 (Fig. 4A). Under identical conditions, PROSUB and PROAQU from prokaryotes display remarkable stability towards histidine protonation (Fig 4B). To further dissect the structural changes, we plotted the global unfolding of the PROFUR and PROPC1 as a function of time and at the two different pHs (Fig 4C). Global unfolding (Qscore), which was computed using the fraction of native contacts that are retained as a function of time during the simulation at different pHs, demonstrates that PROFUR appears to undergo significant changes in the native-like contacts upon protonation of the histidine residues. Under similar conditions, PROPC1 appears to be more stable at both pHs.
Fig. 4. pH dependent structural dynamics of prokaryotic and eukaryotic propeptides.
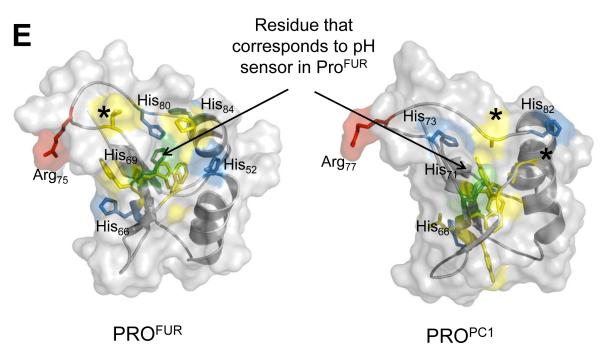
(A) Green and blue cartoons represent initial and final structures of the simulations, respectively. The second cleavage site loop (red/salmon) in PROFUR (structure on right-side) is stable when histidines are deprotonated (pH 7.0; bordered by black box) but changes conformation upon histidine-protonation (pH 6.0; bordered by red box). The dynamics of the loop are unaffected by the histidine-protonation status of PROPC1 (cartoons on left-side). Under identical conditions PROSUB and PROAQU show insignificant changes in dynamics as a function of pH (B) Protonation status dependent, time-resolved, residue-specific dynamics of PROSUB, PROAQU, PROPC1 and PROFUR. Arrow-head indicates secondary cleavage-site and color scale represents RMSD from initial structures. (C) Global unfolding (Qscore) of PROFUR and PROPC1 at different pHs. Unfolding was computed using the fraction of native contacts that are retained as a function of time during the simulation at different pHs and suggest that PROFUR undergoes global unfolding at a pH of 6.0 when compared with pH 7.0 and with PROPC1 at both, pH 7.0 and 6.0, respectively. (D) Evaluating the robustness of independent MD simulations using different models and longer time scales. We compared the similarity of structures to the starting conformation by measuring the root-mean-square deviation (RMSD) within the propeptide-domain, along equally spaced snapshots of the simulation trajectory. Our results suggest that while PROPC1 appears stable at different pHs, PROFUR displays significantly larger conformational changes, which may contribute to its increased proteolytic susceptibility at pH 6.0, and is consistent with our spectroscopic studies. (E) A comparison of the structural locations of various histidine residues in PROFUR and PROPC1. The pH sensor His69 in PROFUR (green) along with other histidine residues (blue) and their corresponding residues with PROPC1 are depicted. Hydrophobic residues surrounding His69 in PROFUR are depicted in yellow, while the asterixes denote residue substitutions at cognate histidine residues.
Since our model for PROFUR is based on a homology model derived from the NMR structure of PROPC1, it can be argued that the model may not correspond to an energetically favorable conformation and the simulations may be biased by the homology model. To address this issue we have performed two additional independent simulations on PROFUR and PROPC1 and for a longer time scale (Fig. 4D). To analyze the structural changes we plotted the RMSD of the core and the secondary cleavage site loop between the initial structure and equally spaced snapshots of the trajectory of simulation, both as a function of time and at two different pHs (Fig 4D). While PROPC1 remained stable at both pHs, PROFUR showed increasing RMSD values throughout the simulation at pH 6, while remaining stable at pH7. The results confirm our earlier simulations on a shorter time scale and suggest that protonation/deprotonation of histidines play a role in the conformational destabilization of PROFUR compared to PROPC1. While our simulations do not provide information on why PROPC1 is more stable that PROFUR towards pH dependent unfolding, they corroborate our experimental observations on the pH dependent stabilities of the propeptides. His69 in furin and the corresponding His residue in PC1 reside closely to other histidines and charged residues in the cleavage loop (Fig. 4E). The interaction of this protonated His with these other residues may provide key insights into why the activation pHs of furin and PC1 differ dramatically.
Together with our biophysical, biochemical and cell-based studies, the MD simulations suggest that upon protonation of His residues, PROFUR undergoes conformational changes that may potentially destabilize the propeptide domain to expose the internal cleavage site for proteolysis. Given that PROPC1 undergoes activation at pH ~5.5 in the DCSGs and remains stable upon His protonation, we can conclude that either additional residues must play a role in the activation of PROPC, or the timescale of the simulations is too short to capture the unfolding event.
DISCUSSION
Compartmentalizing metabolic pathways within organelles enables eukaryotic cells to process numerous spatiotemporal reactions with efficiency and precision. Optimal organelle function requires maintenance of lumenal-pH and propeptide-dependent eukaryotic proteases must have evolved from prokaryotic orthologs to exploit this proton gradient as energy currency to function only at appropriate sub-cellular compartments 40; 41. Although structures and functions of individual protein families may impose unique evolutionary constraints, an analysis of divergence patterns suggests that individual responses of most proteins are variations on a common set of selective constraints42. In protein families with low divergence, mutations within the interior are limited by strong evolutionary pressures to maintain a conserved core that removes all but a few conservative changes 41. With increasing divergence, mutations in the interior become more widespread and closer in number to what is found in the intermediate and exposed regions63. Since catalytic domains exhibit a higher degree of conservation within subtilases when compared to their propeptides17, this suggests that the catalytic and propeptide domains may have encountered different mutational frequencies and different selective constraints.
Our work provides insight as to why nature may have imposed differential selective constraints that alter both sequence and the asymmetrical distribution of histidines in two functional domains, namely the propeptides and their cognate catalytic domains within furin and PC1. In this manuscript we demonstrate that PROFUR and PROPC1 are enriched in histidine-content when compared with cognate proteases and prokaryotic orthologs (Fig 1C). Spectroscopic studies demonstrate that changes in pH can induce conformational changes only within PROFUR and PROPC1, while their prokaryotic orthologs, PROSUB and PROAQU, are largely unaffected (Fig 2A and B). Since swapping propeptides between eukaryotic paralogs transfers pH-dependent protease activation in an propeptide-dictated manner (Fig. 2D and 3D), while allowing folding and cellular localization (Fig. 3B and C), our results argue that PROFUR and PROPC1 may have evolved from prokaryotic orthologs to encode histidine-driven pH-sensors that enable furin and PC1 to recognize and adapt to cellular organelles. Our MD simulations suggest that histidine protonation may be sufficient to induce conformational changes that enable the second activating cleavage of the propeptide and are consistent with our spectroscopic analysis (Fig. 2B and C). While it would be interesting to compare the structures of the chimeras with those of the wild-type complexes and examine how their structures are affected by changes in pH, such experimentation is currently unfeasible due to the high concentrations of protein required for circular dichroism spectroscopic analysis.
It is important to note that despite histidine enrichment, the specific location of these residues within the amino acid sequences of propeptides can vary significantly (Fig 1A and 4E). Moreover, the His69 that was identified as a primary pH sensor in PROFUR 29 is also conserved in PROPC1, although the pH dependent activation of furin and PC1 differs significantly3. This suggests that additional undetermined residues and/or cellular factors must play a significant role in pH dependent activation of their cognate protease domains. Propeptides also contain several charged residues30 which may interact with protonated and non-protonated histidine residues, thereby enabling subtleties in their sensitivity to compartment specific pH. Hence, our studies emphasize the necessity of more detailed analyses of the differences between pH-sensors of PROFUR and PROPC1 using detailed site-directed mutagenesis studies, to tease out the interplay with residues in the proximity of their cognate pH sensors.
Since propeptides facilitate the folding of several eukaryotic proteases, this raises the possibility that other propeptide dependent eukaryotic proteases may also display similar bias towards His-residues. Cathepsins represent another example where preliminary results suggest similar histidine enrichment within propeptides (Elferich, unpublished data). Cathepsins also undergo compartment specific activation of their cognate catalytic domains within the acidic pH of the lysosomes (pH 4.0). However, unlike furin and PC1, which can be compared with prokaryotic orthologs from the ubiquitously expressed subtilase super-family, cathepsins do not have well characterized prokaryotic orthologs to precisely compare histidine enrichment as a function of prokaryotic versus eukaryotic evolution. Interestingly, the histidine residues localized within the propeptides are likely to modulate a wide range of pH dependent activation. Hence, at low pH typically found within the lysozome, all of the histidine residues are likely to be protonated if their pKa is not altered by their structural context. Therefore it is possible that other residues such as aspartic and glutamic acid residues may collaborate with histidines to mediate subtle changes in pH dependent activation. Other residues could either become protonated themselves to mediate activation or influence the pKa of histidine protonation. It is also possible that the pH dependence in activity for furin and PC1 could also partially reflect the pKa values of catalytic residues and would required detailed characterization of active site residues. The challenge is to understand which specific histidines interact with additional residues to provide a broad range of pH-dependent activation of secretory proteases.
MATERIALS AND METHODS
Expression and Purification of PROFUR, PROPC1, MATFUR and MATPC1
Codon optimized genes encoding human PROFUR and mouse PROPC1 were synthesized from CELTEK genes, cloned into pET11a and expressed in BL21(DE3) as described 64. Inclusion bodies containing MATFUR and MATPC1 were isolated and proteins were purified using reverse phase chromatography. Enzymatically active MATFUR and MATPC1 were obtained from recombinants expressing human VV:fur/f/ha/ΔTCK29 and mouse VV:mPC1 65 in BSC40 cells as described 29. Cos7 cells were maintained in DMEM-high glucose medium (HyClone) containing 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were incubated at 37°C in a 5% CO2 environment as described 29.
Circular Dichroism Studies
Circular dichroism (CD) measurements were performed on an AVIV model 215 CD spectrometer using a 1 mm path-length cell at 4 °C as described earlier34; 35. Briefly, propeptide samples (4 mg/ml) stored in 6M GdnHCl (to avoid side-chain modifications commonly seen when samples are stored in urea) were diluted to a final concentration of 0.4 mg/ml), and were refolded using stepwise dialysis against 50 mM cacodylate buffer, pH 7.4 containing 150 mM KCl (Buffer A) and decreasing amounts of urea. The proteins were dialyzed twice in Buffer A without urea, against Buffer A in different pH (5.0-7.0), and then subjected to ultracentrifugation in TLA-100 for 30 min to remove particulates. The CD spectra between 200-260 nm were averaged over three independent experiments and plotted as a change in ellipticity at 222 nm as a function of pH and plotted as [θ] molar ellipticity66 deg.cm2.dmol−1. The PROSUB-C structure was obtained by a difference spectra between the cleaved PROSUB:S221C-subtilisin complex and mature subtilisin as described earlier48.
Molecular Dynamic Simulations
1SCJ47, 1KN667 and homology models of furin derived from 1KN6, and aqualysin derived from 1SCJ were used for as PDB models PROSUB, PROPC1, PROFUR, and PROAQU respectively. Homology models were built using either SWISS-MODEL or MODELLER. All hydrogen and non-protein atoms were removed and hydrogen were added back using the autoPSF function in NAMD 68. Structures were solvated in cubes with TIP3P explicit water using VMD, with a minimum of 12 Å distance to the edge. All simulations were carried out with periodic boundary conditions, PME for long-range electrostatics, and a 12 Å cutoff for non-bonded interactions with the CHARMM22 force field69 using NAMD (version 2.5). Snapshots were saved every 10 ps using a time-step of 1 fs. The system was equilibrated by first constraining the protein and minimizing solvent for 1000 steps using a conjugate gradient algorithm. The solvent was initially equilibrated for 100 ps, then fully constrained and the protein minimized for 500 steps. The entire system was subsequently minimized and used in the simulations. MD simulations require defining of a potential function or a force field that describes the ways through which particles in a simulation will interact70. Force fields can be defined at many levels of physical accuracy and those used in MD-simulations often embody a classical treatment of particle-particle interactions, which can reproduce structural and conformational changes, but usually cannot reproduce precise chemical reactions. Therefore, to simulate the pH-dependent protonation reactions, we have approximated the pH environment by predetermining the protonation state in the starting structure, an approach that has been extensively employed in the field of molecular dynamics. For pH 7, we used the HSD parameters for histidine residues which represent an uncharged side chain, with a proton bound to the nitrogen atom in the delta position. To simulate an environment of pH 6 we used the HSP parameter which represents a positively charged histidine with protons bound to both nitrogen atoms. For testing the robustness of our simulations we took two different models of PROFUR and PROPC1 and repeated the simulations as described above. An adjustment of the pH to exact values would require a prediction of the pKa values of individual residues, which was not practical in the given study.
Amino acid content analysis
Protein sequences for human furin, mouse PC1, subtilisin from Bacillus subtilis, and aqualysin from Thermus aquaticus families were obtained from the 50% sequence identity clusters UniRef50_P09958, UniRef50_P29120, UniRef50_P00782, and UniRef50_P08594 in the UniRef database, respectively. Subsequences representing the propeptides and the protease domain were extracted using annotation from the Interpro database entries IPR009020 and IPR000209, respectively. Sequences that were not annotated by both entries were omitted. Amino acid content of both domains in all sequences were calculated and averaged for each domain and protein family. Contents of amino acids belonging to individual groups were added and divided by the sum of their content in the whole UniProt database (release 2011_12). The multiple sequence alignment of selected prokaryotic and eukaryotic subtilases was obtained using ClustalW and colored using Genedoc.
Secreted enzyme activity assays
For all assays, 113 μM furin substrate (Abz-RVKRGLA-Tyr[3-NO2]) in dimethyl sulfoxide was incubated with 40 μl secreted enzyme in 155 μl of 50 mM cacodylate buffer, pH 7.0 containing 1 mM CaCl2 and 50 mM KCl. Cacodylate buffer was used in all experiments to maintain consistency throughout the analyses. The assays were conducted on a SpectraMax-M2 spectrofluorometer equipped with a 96-well plate reader. Excitation wavelength was set at 320 nm while emission wavelength was set at 425 nm. Given values are averages of triplicate assays. The activity was normalized by quantifying the relative amounts of proteins secreted in the media using ImageJ software.
Isolation of in trans propeptide:protease complexes
Since propeptides are potent competitive inhibitors of protease paralogs64, PRO:MAT complexes in trans were generated by adding 10-fold excess of PROFUR and PROPC1 (~2 nM) to MATFUR or MATPC1 (~0.2 nM) in 50 mM cacodylate buffer, at different pH (5.0 to 7.4) containing 150 mM KCl in a 96-well quartz plate. Complexes were incubated for 30 min at RT and the activities assayed as described earlier29. The percent activity at each pH was calculated using the activity of uninhibited protease as a control.
Construction of Secreted and ER Localized PCDNA3.1 Expression Vectors
The plasmid p2Vneo containing human furin25 was cut with EcoRI and HindIII to release the Furin-Flag gene. The plasmid pBSSK containing the mouse PC1-Flag gene65 was cut with Ncol and BamHI. Both genes were treated with Klenow and ligated with PCDNA3.1 cut using EcoRV. Gene orientations were confirmed by digestion with BamHI and Xhol and through DNA sequencing. To obtain soluble and secreted PROFUR-Flag-MATFUR, a stop codon was introduced after Leu713 in the plasmid containing full-length furin-flag, which removes the cysteine-rich, cytoplasmic, and transmembrane domains. Similarly, the PC1-Flag plasmid was truncated at Arg618 to produce secreted PROPC1-Flag-MATPC1. The chimeras (PROFUR-Flag-MATFUR, PROFUR-Flag-MATPC1, PROPC1-Flag-MATPC1, PROPC1-Flag-MATFUR) were constructed using PCR. To localize proteases in ER, KDEL sequences were inserted into the genes expressing soluble PROFUR-Flag-MATFUR and PROPC1-Flag-MATPC1 using PCR. All constructs were confirmed through sequencing (OHSU DNA Services Core, Portland, OR).
Expression of Constructs and ER Fractionation
For secretion experiments, cells were transfected with expression vectors containing PROFUR-Flag-MATFUR, PROFUR-Flag-MATPC1, PROPC1-Flag-MATPC1, PROPC1-Flag-MATFUR or the empty PCDNA3.1 vector using LT1 transfection reagent (Invitrogen) as recommended by the manufacturer. After 5 hrs, the cells were washed with PBS, replaced with serum free DMEM media and CM was harvested 24 hrs post media change. For the ER retention experiments the KDEL-tagged constructs were transfected as described above. The media was not changed after transfection and the cells were maintained in 10 cm plates. The microsomal fraction was prepared following manufacturer’s instructions from Endoplasmic Reticulum Isolation Kit (Sigma). Constructs were probed by Western blotting as follows: primary antibody, mAB M2 flag (Sigma), was used in a 1:1000 dilution and the secondary antibody, IgG3000 anti-mouse (Fisher), was used in a 1:10,000 dilution.
KDEL Enzyme Activity Assays
Cells were transfected with constructs containing PROFUR-Flag-MATFUR-KDEL, PROFUR-Flag-MATPC1-KDEL, PROPC1-Flag-MATPC1-KDEL, PROPC1-Flag-MATFUR-KDEL29. After 24 hrs, harvested cells were lysed and incubated in a 25 °C water bath for 1 hr in 50 mM cacodylate buffer, pH 7.4 containing 1 mM CaCl2, 150 mM KCl, and a fresh protease inhibitor cocktail (Sigma). To six micro centrifuge tubes containing 95 μL of 100 mM cacodylate buffer of varying pH, 100 μL of cell lysate was added to bring the mixtures to a final pH of 5.4, 6.4, and 7.4, in duplicates. The cell lysates containing processed enzymes were incubated at 25°C for 2 hrs with one tube from each pH incubated with 0.83 nM trypsin (as a control for complete activation). Soybean trypsin inhibitor was added to the tubes (to block trypsin, which can interfere with the activity assay) and they were incubated for an additional 15 minutes. For each assay, 113 μM furin substrate was added for a final volume of 200 μL. The assays were conducted as described earlier. Each experiment was performed at least 3-times and the values given are the average of assays done in triplicate.
HIGHLIGHTS.
◻ PCs evolved from propeptide-dependent not -independent bacterial subtilases
◻ PC-propeptides are rich in histidine when compared with bacterial propeptides
◻ PC-propeptides encode pH-sensors that regulate organelle-specific activation
◻ Swapping propeptides in eukaryotic PCs transfers pH-dependent protease activation
ACKNOWLEDGEMENTS
We thank Mahta Nili and Parvathy Ramakrishnan for their help in cloning and the cell-based techniques and Nathan Brandt and Laura Figoski for protein expression. Special thanks to Jimmy Dikeakos for reading the manuscript. This work was supported by the National Science Foundation CAREER award (MCB0746589) and Grant in Aid from American Heart Foundation to U.S., an NIH training grant to D.M.W., an AHA pre-doctoral training grant to J.E, and NIH grants (DK37274 and CA151564) to G.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
FOOTNOTES: IMC, Intramolecular Chaperone; PCs, ProproteinConvertases; PC1, Proprotein Convertase 1/3 or neuroendocrine convertase 1/3; PROSUB, propeptide of subtilisin; PROAQU, propeptide of Aqualysin; PROFUR, propeptide for furin; PROPC1, propeptide for PC1; MATFUR, Protease domain for furin; MATPC1, Protease domain for PC1; CM, Conditioned Media; mAb, Monoclonal antibody; MD, Molecular Dynamics; MSA, Multiple sequence alignment; ER, Endoplasmic Reticulum
REFERENCES
- 1.Seidah NG. The proprotein convertases, 20 years later. Methods Mol Biol. 2011;768:23–57. doi: 10.1007/978-1-61779-204-5_3. [DOI] [PubMed] [Google Scholar]
- 2.Seidah NG. What lies ahead for the proprotein convertases? Ann N Y Acad Sci. 2011;1220:149–61. doi: 10.1111/j.1749-6632.2010.05883.x. [DOI] [PubMed] [Google Scholar]
- 3.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–66. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller RS, Brake A, Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci U S A. 1989;86:1434–8. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuller RS, Brake AJ, Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989;246:482–6. doi: 10.1126/science.2683070. [DOI] [PubMed] [Google Scholar]
- 6.Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–62. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- 7.Thomas G, Thorne BA, Thomas L, Allen RG, Hruby DE, Fuller R, Thorner J. Yeast KEX2 endopeptidase correctly cleaves a neuroendocrine prohormone in mammalian cells. Science. 1988;241:226–30. doi: 10.1126/science.3291117. [DOI] [PubMed] [Google Scholar]
- 8.Seidah NG, Mowla SJ, Hamelin J, Mamarbachi AM, Benjannet S, Toure BB, Basak A, Munzer JS, Marcinkiewicz J, Zhong M, Barale JC, Lazure C, Murphy RA, Chretien M, Marcinkiewicz M. Mammalian subtilisin/kexin isozyme SKI-1: A widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc Natl Acad Sci U S A. 1999;96:1321–6. doi: 10.1073/pnas.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toure BB, Munzer JS, Basak A, Benjannet S, Rochemont J, Lazure C, Chretien M, Seidah NG. Biosynthesis and enzymatic characterization of human SKI-1/S1P and the processing of its inhibitory prosegment. J Biol Chem. 2000;275:2349–58. doi: 10.1074/jbc.275.4.2349. [DOI] [PubMed] [Google Scholar]
- 10.Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci U S A. 2003;100:928–33. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, Essalmani R, Prat A. The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol. 2008;40:1111–25. doi: 10.1016/j.biocel.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Henrich S, Cameron A, Bourenkov GP, Kiefersauer R, Huber R, Lindberg I, Bode W, Than ME. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10:520–6. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 13.Holyoak T, Wilson MA, Fenn TD, Kettner CA, Petsko GA, Fuller RS, Ringe D. 2.4 A resolution crystal structure of the prototypical hormone-processing protease Kex2 in complex with an Ala-Lys-Arg boronic acid inhibitor. Biochemistry. 2003;42:6709–18. doi: 10.1021/bi034434t. [DOI] [PubMed] [Google Scholar]
- 14.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J Mol Biol. 2005;345:211–27. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 15.Holyoak T, Kettner CA, Petsko GA, Fuller RS, Ringe D. Structural basis for differences in substrate selectivity in Kex2 and furin protein convertases. Biochemistry. 2004;43:2412–21. doi: 10.1021/bi035849h. [DOI] [PubMed] [Google Scholar]
- 16.Bard F, Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol. 2006;22:439–55. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]
- 17.Shinde U, Thomas G. Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol Biol. 2011;768:59–106. doi: 10.1007/978-1-61779-204-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassi DE, Mahloogi H, Lopez De Cicco R, Klein-Szanto A. Increased furin activity enhances the malignant phenotype of human head and neck cancer cells. Am J Pathol. 2003;162:439–47. doi: 10.1016/s0002-9440(10)63838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassi DE, Mahloogi H, Klein-Szanto AJ. The proprotein convertases furin and PACE4 play a significant role in tumor progression. Mol Carcinog. 2000;28:63–9. [PubMed] [Google Scholar]
- 20.Choquet H, Stijnen P, Creemers JW. Genetic and functional characterization of PCSK1. Methods Mol Biol. 2011;768:247–53. doi: 10.1007/978-1-61779-204-5_13. [DOI] [PubMed] [Google Scholar]
- 21.Benzinou M, Creemers JW, Choquet H, Lobbens S, Dina C, Durand E, Guerardel A, Boutin P, Jouret B, Heude B, Balkau B, Tichet J, Marre M, Potoczna N, Horber F, Le Stunff C, Czernichow S, Sandbaek A, Lauritzen T, Borch-Johnsen K, Andersen G, Kiess W, Korner A, Kovacs P, Jacobson P, Carlsson LM, Walley AJ, Jorgensen T, Hansen T, Pedersen O, Meyre D, Froguel P. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat Genet. 2008;40:943–5. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 22.Steiner DF, Rouille Y, Gong Q, Martin S, Carroll R, Chan SJ. The role of prohormone convertases in insulin biosynthesis: evidence for inherited defects in their action in man and experimental animals. Diabetes Metab. 1996;22:94–104. [PubMed] [Google Scholar]
- 23.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassi DE, Lopez De Cicco R, Mahloogi H, Zucker S, Thomas G, Klein-Szanto AJ. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proc Natl Acad Sci U S A. 2001;98:10326–31. doi: 10.1073/pnas.191199198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson ED, VanSlyke JK, Thulin CD, Jean F, Thomas G. Activation of the furin endoprotease is a multiple-step process: requirements for acidification and internal propeptide cleavage. Embo J. 1997;16:1508–18. doi: 10.1093/emboj/16.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inouye M. Intramolecular chaperone: the role of the pro-peptide in protein folding. Enzyme. 1991;45:314–21. doi: 10.1159/000468904. [DOI] [PubMed] [Google Scholar]
- 27.Jaswal SS, Sohl JL, Davis JH, Agard DA. Energetic landscape of alpha-lytic protease optimizes longevity through kinetic stability. Nature. 2002;415:343–6. doi: 10.1038/415343a. [DOI] [PubMed] [Google Scholar]
- 28.Anderson ED, Molloy SS, Jean F, Fei H, Shimamura S, Thomas G. The ordered and compartment-specfific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J Biol Chem. 2002;277:12879–90. doi: 10.1074/jbc.M108740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feliciangeli SF, Thomas L, Scott GK, Subbian E, Hung CH, Molloy SS, Jean F, Shinde U, Thomas G. Identification of a pH sensor in the furin propeptide that regulates enzyme activation. J Biol Chem. 2006;281:16108–16. doi: 10.1074/jbc.M600760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinde U, Inouye M. Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin Cell Dev Biol. 2000;11:35–44. doi: 10.1006/scdb.1999.0349. [DOI] [PubMed] [Google Scholar]
- 31.Siezen RJ. Subtilases: subtilisin-like serine proteases. Adv Exp Med Biol. 1996;379:75–93. doi: 10.1007/978-1-4613-0319-0_9. [DOI] [PubMed] [Google Scholar]
- 32.Siezen RJ, Creemers JW, Van de Ven WJ. Homology modelling of the catalytic domain of human furin. A model for the eukaryotic subtilisin-like proprotein convertases. Eur J Biochem. 1994;222:255–66. doi: 10.1111/j.1432-1033.1994.tb18864.x. [DOI] [PubMed] [Google Scholar]
- 33.Siezen RJ, Leunissen JA. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997;6:501–23. doi: 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subbian E, Yabuta Y, Shinde U. Positive selection dictates the choice between kinetic and thermodynamic protein folding and stability in subtilases. Biochemistry. 2004;43:14348–60. doi: 10.1021/bi048397x. [DOI] [PubMed] [Google Scholar]
- 35.Subbian E, Yabuta Y, Shinde UP. Folding pathway mediated by an intramolecular chaperone: intrinsically unstructured propeptide modulates stochastic activation of subtilisin. J Mol Biol. 2005;347:367–83. doi: 10.1016/j.jmb.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Cunningham EL, Jaswal SS, Sohl JL, Agard DA. Kinetic stability as a mechanism for protease longevity. Proc Natl Acad Sci U S A. 1999;96:11008–14. doi: 10.1073/pnas.96.20.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sohl JL, Jaswal SS, Agard DA. Unfolded conformations of alpha-lytic protease are more stable than its native state. Nature. 1998;395:817–9. doi: 10.1038/27470. [DOI] [PubMed] [Google Scholar]
- 38.Truhlar SM, Cunningham EL, Agard DA. The folding landscape of Streptomyces griseus protease B reveals the energetic costs and benefits associated with evolving kinetic stability. Protein Sci. 2004;13:381–90. doi: 10.1110/ps.03336804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327:625–35. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 41.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–30. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 42.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–82. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 43.Marie-Claire C, Yabuta Y, Suefuji K, Matsuzawa H, Shinde U. Folding pathway mediated by an intramolecular chaperone: the structural and functional characterization of the aqualysin I propeptide. J Mol Biol. 2001;305:151–65. doi: 10.1006/jmbi.2000.4233. [DOI] [PubMed] [Google Scholar]
- 44.Shinde U, Thomas G. Insights from bacterial subtilases into the mechanisms of intramolecular chaperone mediated activation of furin. Methods in Molecular Biology. 2011 doi: 10.1007/978-1-61779-204-5_4. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen YJ, Inouye M. The intramolecular chaperone-mediated protein folding. Curr Opin Struct Biol. 2008;18:765–70. doi: 10.1016/j.sbi.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Shinde U, Inouye M. Intramolecular chaperones and protein folding. Trends Biochem Sci. 1993;18:442–6. doi: 10.1016/0968-0004(93)90146-e. [DOI] [PubMed] [Google Scholar]
- 47.Jain SC, Shinde U, Li Y, Inouye M, Berman HM. The crystal structure of an autoprocessed Ser221Cys-subtilisin E-propeptide complex at 2.0 A resolution. J Mol Biol. 1998;284:137–44. doi: 10.1006/jmbi.1998.2161. [DOI] [PubMed] [Google Scholar]
- 48.Shinde U, Li Y, Chatterjee S, Inouye M. Folding pathway mediated by an intramolecular chaperone. Proc Natl Acad Sci U S A. 1993;90:6924–8. doi: 10.1073/pnas.90.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharjya S, Xu P, Xiang H, Chretien M, Seidah NG, Ni F. pH-induced conformational transitions of a molten-globule-like state of the inhibitory prodomain of furin: implications for zymogen activation. Protein Sci. 2001;10:934–42. doi: 10.1110/ps.41301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fink AL, Calciano LJ, Goto Y, Kurotsu T, Palleros DR. Classification of acid denaturation of proteins: intermediates and unfolded states. Biochemistry. 1994;33:12504–11. doi: 10.1021/bi00207a018. [DOI] [PubMed] [Google Scholar]
- 51.Goto Y, Calciano LJ, Fink AL. Acid-induced folding of proteins. Proc Natl Acad Sci U S A. 1990;87:573–7. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uversky VN, Goto Y. Acid denaturation and anion-induced folding of globular proteins: multitude of equilibium partially folded intermediates. Curr Protein Pept Sci. 2009;10:447–55. doi: 10.2174/138920309789352029. [DOI] [PubMed] [Google Scholar]
- 53.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gething MJ, McCammon K, Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell. 1986;46:939–50. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- 55.Molloy SS, Thomas L, VanSlyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. Embo J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 57.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci U S A. 2005;102:6679–85. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daggett V, Levitt M. A model of the molten globule state from molecular dynamics simulations. Proc Natl Acad Sci U S A. 1992;89:5142–6. doi: 10.1073/pnas.89.11.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daggett V, Levitt M. Protein unfolding pathways explored through molecular dynamics simulations. J Mol Biol. 1993;232:600–19. doi: 10.1006/jmbi.1993.1414. [DOI] [PubMed] [Google Scholar]
- 60.Brooks CL., 3rd Characterization of “native” apomyoglobin by molecular dynamics simulation. J Mol Biol. 1992;227:375–80. doi: 10.1016/0022-2836(92)90893-o. [DOI] [PubMed] [Google Scholar]
- 61.Salimi NL, Ho B, Agard DA. Unfolding simulations reveal the mechanism of extreme unfolding cooperativity in the kinetically stable alpha-lytic protease. PLoS Comput Biol. 2010;6:e1000689. doi: 10.1371/journal.pcbi.1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W, Eichenberger AP, van Gunsteren WF. Molecular dynamics simulation of thionated hen egg white lysozyme. Protein Sci. 2012 doi: 10.1002/pro.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Povolotskaya IS, Kondrashov FA. Sequence space and the ongoing expansion of the protein universe. Nature. 2010;465:922–6. doi: 10.1038/nature09105. [DOI] [PubMed] [Google Scholar]
- 64.Fu X, Inouye M, Shinde U. Folding pathway mediated by an intramolecular chaperone. The inhibitory and chaperone functions of the subtilisin propeptide are not obligatorily linked. J Biol Chem. 2000;275:16871–8. doi: 10.1074/jbc.275.22.16871. [DOI] [PubMed] [Google Scholar]
- 65.Benjannet S, Rondeau N, Day R, Chretien M, Seidah NG. PC1 and PC2 are proprotein convertases capable of cleaving proopiomelanocortin at distinct pairs of basic residues. Proc Natl Acad Sci U S A. 1991;88:3564–8. doi: 10.1073/pnas.88.9.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1:2876–90. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tangrea MA, Bryan PN, Sari N, Orban J. Solution structure of the pro-hormone convertase 1 pro-domain from Mus musculus. J Mol Biol. 2002;320:801–12. doi: 10.1016/s0022-2836(02)00543-0. [DOI] [PubMed] [Google Scholar]
- 68.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roux B, Prod’hom B, Karplus M. Ion transport in the gramicidin channel: molecular dynamics study of single and double occupancy. Biophys J. 1995;68:876–92. doi: 10.1016/S0006-3495(95)80264-X. [DOI] [PMC free article] [PubMed] [Google Scholar]