Abstract
Agnoprotein is required for the successful completion of the JC virus (JCV) life cycle and was previously shown to interact with JCV large T-antigen (LT-Ag). Here, we further characterized agnoprotein’s involvement in viral DNA replication. Agnoprotein enhances the DNA binding activity of LT-Ag to the viral origin (Ori) without directly interacting with DNA. The predicted amphipathic α-helix of agnoprotein plays a major role in this enhancement. All three phenylalanine (Phe) residues of agnoprotein localize to this α-helix and Phe residues in general are known to play critical roles in protein-protein interaction, protein folding and stability. The functional relevance of all Phe residues was investigated by mutagenesis. When all were mutated to alanine (Ala), the mutant virus (F31AF35AF39A) replicated significantly less efficiently than each individual Phe mutant virus alone, indicating the importance of Phe residues for agnoprotein function. Collectively, these studies indicate a close involvement of agnoprotein in viral DNA replication.
Keywords: Polyomavirus JC, SV40, BKV, large T antigen, agnoprotein, transcription, DNA replication, progressive multifocal leukoencephalopathy
Introduction
The integrity and proper functioning of living cells depend on highly organized and sophisticated interactions among their macromolecules including proteins. Upon infection, viruses often hijack these interactions for their own benefit. To understand the roles of JCV agnoprotein in an infected cellular environment, it is central to identify and characterize its interaction parameters with its partners. Agnoprotein encoded by JCV late genome was previously shown to interact with a number of viral and cellular proteins, including JCV large T-antigen (LT-Ag) (Safak et al., 2001), small t-antigen (Sm t-Ag) (Sariyer, Khalili, and Safak, 2008), p53 (Darbinyan et al., 2002), YB-1 (Safak et al., 2002), FEZ1 (Suzuki et al., 2005) and HP1-α (Okada et al., 2005). Agnoprotein is a small (71 aa), highly basic phosphoprotein (Khalili et al., 2005; Sariyer et al., 2006) and has been implicated in many different aspects of the JCV life cycle, including viral replication (Safak et al., 2001), transcription (Safak et al., 2001), encapsidation (Sariyer et al., 2006), functioning as viroporin (Suzuki et al., 2010b) and promotion of the transport of virions from nucleus to cytoplasm (Okada et al., 2005; Suzuki et al., 2005) indicating that it is a multifunctional protein. In addition, it has also been shown to deregulate cell cycle progression in that cells which stably express agnoprotein largely accumulate at the G2/M phase of the cell cycle (Darbinyan et al., 2002). Despite these previous reports and intense research, the regulatory role of this protein in viral replication cycle has yet to be fully understood.
The agnoprotein sequences from other polyomaviruses, including SV40 and BK virus (BKV) show high identity and similarity to that of JCV agnoprotein (Safak et al., 2001). For example, there is 82% identity and 93% similarity between JCV and BKV agnoprotein sequences; and these are 60% and 79% respectively between JCV and SV40 agnoproteins (Altschul et al., 1997). Like JCV agnoprotein, the agnoprotein of SV40 and BKV was also previously implicated to be involved in different aspects of viral replication cycle, including transcription, translation, virion production and maturation of the viral particles (Alwine, 1982; Haggerty, Walker, and Frisque, 1989; Hay, Skolnik-David, and Aloni, 1982; Hou-Jong, Larsen, and Roman, 1987; Johannessen et al., 2008; Margolskee and Nathans, 1983; Moens, Ludvigsen, and Van Ghelue, 2011; Myhre et al., 2010; Ng et al., 1985; Unterstab et al., 2010).
Besides agnoprotein, JCV encodes a limited number of regulatory proteins, including LT-Ag, Sm t-Ag and T′-proteins (Bollag et al., 2000; Frisque, Bream, and Cannella, 1984; Saribas et al., 2010) and investigation of their regulatory roles is critically important for understanding the progression of the fatal human brain disease, progressive multifocal encephalopathy (PML). PML results from the lytic infection of oligodendrocytes, a subset of glial cells in the central nervous system (CNS), by JCV. This virus is a ubiquitous polyomavirus and most of the human population (70–80%) is asymptomatically infected by this virus in early childhood worldwide. The virus undergoes a latency period after initial infection and reactivation mainly occurs in patients with immunosuppressive conditions (1–5%), including in those with AIDS (Berger, 2011; Major et al., 1992). However, recently scientific community was also surprised by the emergence of PML in a very small percent of the immune disorder patients including those of multiple sclerosis (MS) and severe psoriasis (Crohn’s disease) who underwent an immunosupressive immune therapy – treating patients with specific monoclonal antibodies (natalizumab and efalizumab respectively) directed against certain cell surface receptors of T and B cells (Kleinschmidt-DeMasters and Tyler, 2005; Langer-Gould et al., 2005; Van Assche et al., 2005). Occurrence of all these new PML cases in such patient population indicates that alterations in either arm of immune system may provide with a supporting environment for reactivation of JCV virus in certain MS and Crohn’s disease patients. The rationale behind using these antibodies in MS and Crohn’s disease cases was to prevent extravasation of T cells into the brain and infiltration of B cells into the layers of the skin respectively by blocking several cell surface receptors on each cell type. It was expected to down-regulate the harmful effects of these immune cells in target organs. Natalizumab, for example, targets α4 integrin, which forms heterodimer complexes when complexed with integrin b1 (α4β1, also known as VLA-4) or with integrin β7 (α4β7) on both B and T cells. Both complexes serve as attachment ligands for vascular cell adhesion molecules (VCAM) on endothelial cells, thus preventing T cells from extravasation into the brain or gut. Another monoclonal antibody, efalizumab, binds to CD11α, an integrin molecule on B and T cells and blocks the attachment of both cell types to the intercellular adhesion molecules (ICAM) on endothelial cells and prevents their infiltration into the layers of the skin (Kleinschmidt-DeMasters and Tyler, 2005; Langer-Gould et al., 2005; Saribas et al., 2010; Van Assche et al., 2005). As a result, treatment of T and B cells with specific antibodies appears to cause harmful effects on these immune cell populations and subsequently fosters conditions for reactivation of JCV in certain immunocompromized patients and leads to the onset of PML.
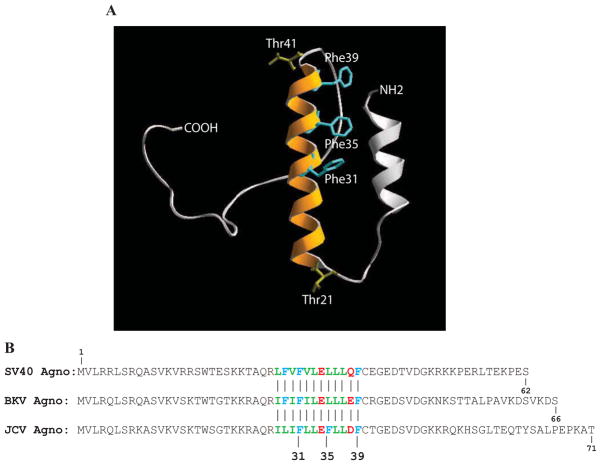
Agnoprotein is a mainly cytoplasmic protein with high concentrations accumulated around the perinuclear area of the infected cells and a small amount of it is consistently found in the nucleus of the infected cells (Safak et al., 2002). This suggests regulatory roles for this protein in the nucleus. Characterization of agnoprotein’s interactions with its targets is essential for understanding of its roles in viral replication cycle. Such detailed characterizations will provide us new opportunities to develop novel therapeutics against this protein. Our previous studies characterized the interaction of agnoprotein with LT-Ag and demonstrated that agnoprotein targets the helicase domain of JCV LT-Ag (Safak et al., 2001) and does not require DNA or RNA for interaction with LT-Ag. Recently, we also showed that the JCV and SV40 with null mutants of agnoprotein cannot sustain their replication cycle although the mutant viruses were shown to be successfully released from the infected cells (Sariyer et al., 2011). It was interesting to observe, however, that the majority of the released virions were deficient in DNA content, which may explain, at least in part, why agnoprotein null mutants were unable to continue to their viral propagation cycle (Sariyer et al., 2011). More recently, we discovered that agnoprotein forms highly stable, SDS-resistant homodimers and oligomers; and the 17–42 amino acid region of agnoprotein is responsible for this property (Saribas et al., 2011). The crystal structure of JCV agnoprotein is unknown. However, 3D computer modeling studies suggests that the 17–42 amino acid region is involved in forming an amphipathic α-helical structure (Saribas et al., 2011). This region contains a Leu/Ile/Phe-rich domain (aa 28–39), mainly consisting of Leu, Ile and Phe. Interestingly, all three Phe residues (Phe31, Phe35 and Phe39) of agnoprotein localize to this Leu/Ile/Phe-rich domain. The negatively charged residues, Glu34 and Asp38, are also interspersed within this domain.
Phenylalanine residues have been reported to play diverse but important regulatory roles in functions of many different proteins through hydrophobic interactions [pi (π)-pi(π) stacking] (Bowden et al., 2008; Brinda, Kannan, and Vishveshwara, 2002; Dhe-Paganon et al., 2004; King et al., 2011; Milardi et al., 2011). These residues were also found to be involved in “cation-π” interactions which could take place between Phe and charged residues (Arg or Lys or His) (Gallivan and Dougherty, 1999; Pless et al., 2008; Pletneva et al., 2001; Shi et al., 2002). Recent reports also indicate that Phe residues can be involved in “anion- π” interactions (Jackson et al., 2007; Philip et al., 2011). All these interactions mediated by Phe residues are known to contribute to (i) the protein-protein interactions at the protein interfaces, (ii) protein folding and stability (Gallivan and Dougherty, 1999; Pless et al., 2008; Pletneva et al., 2001; Shi et al., 2002). Based on the reported contribution of the phenylalanine residues in the literature and the positioning of all three Phe residues within the hydrophobic pocket of agnoprotein, we reasoned that they may also play important structural and functional roles in the biology of agnoprotein in the infected cells.
In this report, we show that agnoprotein induces DNA binding activity of LT-Ag to Ori without directly interacting with DNA and that the predicted main α-helix domain of the protein plays a major role in this induction. Upon mutation of each Phe residue to Ala, agnoprotein mostly lost its ability to enhance DNA binding activity of LT-Ag. Protein-protein interaction studies (GST-pull down) demonstrated that interaction of each agnoprotein mutant (F31A, F35A and F39A) with LT-Ag significantly decreased compared to that of WT, which is consistent with our findings from the DNA binding studies. More importantly, the level of the viral DNA replication significantly diminished when all three Phe residues were simultaneously mutated to Ala compared to a slight decrease, which was observed for individual mutants, indicating the importance of a combinatorial effect of Phe residues on agnoprotein function. Additionally, results from immunocytochemistry studies suggest that Phe residues also contribute to agnoprotein function by assisting to its strategic distribution in the infected cells, mostly accumulating around the perinuclear region.
Materials and Methods
Cell lines
SVG-A is a human cell line established by transformation of primary human fetal glial cells with an origin-defective SV40 mutant (Major et al., 1985). These transformed cells do not express either SV40 viral capsid proteins (VPs) or agnoprotein, but express SV40 LT-Ag. Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics [penicillin/streptomycin (100 μg/ml), ciprofloxacin (10 μg/ml]. They were maintained at 37°C in a humidified atmosphere supplemented with 7% CO2.
Plasmid constructs
The cloning of pGEX1λT-Agno (1–71) or GST fusion Agno deletion mutants [pGEX1λT-Agno (1–54), pGEX1λT-Agno (18–71), pGEX1λT-Agno (37–71), pGEX1λT-Agno (55–71)] were previously described (Safak et al., 2001). Agnoprotein-F31, -F35 and -F39 residues were individually mutated to Ala (A) in the viral background (JCV Mad-1) using the Quik Change™ site–directed mutagenesis kit (Agilent) and designated as JCV Mad-1 Agno-F31A, JCV Mad-1 Agno-F35A and JCV Mad-1 Agno-F39A mutant viruses. F31, F35 and F39 residues were also altogether mutated to Ala in the viral background and designated as triple Phe mutant [JCV Mad-1 Agno (F31AF35AF39A)]. Each single mutant of agnogene was also subcloned into pGEX1λT vector at the BamHI/EcoRI sites by PCR-based cloning and designated as pGEX1λT-Agno-F31A, pGEX1λT-Agno-F35A, pGEX1λT-Agno-F39A. The integrity of all plasmids was determined by DNA sequencing.
Expression and purification of recombinant proteins
A hundred milliliter overnight cultures of Escherichia coli DH5α cells transformed with plasmids expressing either Glutathione-S-Transferase (GST) or full length agnoprotein fused to GST [(GST-Agno (1–71)] or different deletion mutants of agnoprotein fused to GST [GST-Agno (1–54), GST-Agno (18–71), GST-Agno (37–71), GST-Agno (55–71)] or different substitution mutants of agnoprotein fused to GST [GST-Agno (F31A), GST-Agno (F35A), GST-Agno (F39A)] or pMAL-C5X-Agno (F31AF35AF39A) were first diluted 1:10 in fresh Luria-Bertani broth in 1L supplemented with ampicillin (100 μg/ml) and grown at 37°C until at an optical density of 0.5. Bacterial cultures were then induced with 0.3 mM isopropyl-β-thiogalactopyranoside (IPTG) and incubated for an additional 2 h at 28°C. Bacterial cells were harvested by centrifugation at 4°C and pellets were resuspended in 20–40 ml of PENT lysis buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 supplemented with a cocktail of protease inhibitors (Sigma). Bacterial cells were first sonicated and clear cell lysates were prepared by centrifugation at 12,000g. Lysates were then incubated with 200 μl of 50% Glutathione-Sepharose 4B resin (GE Health Care) in phosphate buffered saline (PBS) overnight at 4°C. GST fusion proteins bound to beads were purified by three cycles of washing and centrifugation with 5 ml of PENT lysis buffer, eluted with glutathione (GSH) elution buffer (50 mM Tris-HCl, 10 mM reduced GSH, pH 8.0), dialyzed overnight in band shift sample buffer [12 mm HEPES (pH 7.9), 4 mM Tris-HCl (pH 7.5), 60 mM KCl, 5 mM MgCl2 and 1.0 mM DTT]. JCV LT-Ag was expressed in Sf9 insect cells using a baculovirus expression system and affinity purified as previously described (Bollag, Mackeen, and Frisque, 1996). Finally, JCV LT-Ag, GST and GST-Agno fusion proteins were analyzed by SDS-polyacrylamide gel electrophoresis followed by coomassie blue staining to determine the protein quality.
Band shift assays
Band shift assays were carried out as previously described (Kim et al., 2003; Sadowska et al., 2003). Briefly, a double-stranded synthetic oligonucleotide encompassing a part of JCV Mad-1 origin (nt 5064–5115) was end-labeled with [γ-32P] ATP using T4 polynucleotide kinase and gel purified. Bacterially-produced and purified GST alone or GST-Agno (full length or its mutants) or baculovirus-produced and purified JCV LT-Ag were incubated with labeled probe (40,000-cpm/lane) in different combinations, as described in the respective figure legends in 50 μl reaction volume, in binding buffer containing 1.0 μg poly(dI–dC), 12 mM HEPES (pH 7.9), 4 mM Tris-HCl (pH 7.5), 60 mM KCl, 5 mM MgCl2 and 1.0 mM DTT. The reaction mixture was incubated at 4°C for 45 min to allow the formation of DNA-protein complexes. The complexes were then separated on a 6% polyacrylamide gel in 0.5×TBE [1×TBE is 89 mM Tris-HCl, (pH 8.0), 89 mM boric acid, and 2 mM EDTA, (pH 8.0)]. The gel was dried and DNA-protein complexes were detected by autoradiography. Detailed experimental conditions for all band shift assays are described in the respective figure legends.
GST affinity chromatography assay (GST pull-down)
GST pull-down assay was performed as previously described (Safak et al., 2001). Briefly, 2 μg of either GST alone, or GST-Agno WT or agnoprotein point mutants fused to GST [GST-Agno (F31A), GST-Agno (F35A) and GST-Agno (F39A)] immobilized on Sepharose beads was incubated with 0.3 mg of whole-cell extracts prepared from HJC-15b cells (Raj, 1995) constitutively expressing JCV LT-Ag in a 300 μl reaction volume for 2 h at 4°C in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.5% Nonidet P-40. The protein-complexed beads were washed extensively with lysis buffer and resolved by SDS–10% PAGE followed by Western blot analysis using anti-LT-Ag antibody (Ab-2). Ab-2 monoclonal antibody has been raised against SV40 LT-Ag but cross-reacts with JCV LT-Ag as well.
Indirect immunofluorescence microscopy
Indirect immunofluorescence microscopy studies were performed as previously described (Sadowska et al., 2003; Sariyer et al., 2011). Briefly, SVG-A cells were transfected/infected with respective viral DNA and seeded at subconfluency on polylysine-coated glass chamber slides at specific dates posttransfection indicated in the respective figure legends. The next day, cells were washed twice with PBS and fixed in cold acetone. Fixed cells were then blocked with 5% bovine serum albumin in PBS for 2 h, divided into two groups and incubated with two different antibody combinations. In the first group, fixed cells were incubated with anti-agno primary polyclonal rabbit antibody (1–200 dilution) (Del Valle et al., 2002) plus anti-JCV LT-Ag primary monoclonal mouse antibody (Ab 2000) (Bollag and Frisque, 1992) (1:200 dilution) overnight. Ab 2000 antibody specifically detects only JCV LT-Ag, not SV40 and BKV LT-Ag proteins. In the second group, cells were incubated with anti-agnoprotein primary polyclonal antibody (1–200 dilution) plus anti-VP1 primary monoclonal (pAB597) antibody (Saribas et al., 2011) (1:200 dilution) overnight. Antibody dilutions and incubations were performed in incubation buffer [PBST = (PBS-0.01% Tween 20)] buffer. Cells were then washed three times with PBST buffer for 10-min intervals and subsequently incubated with a Rhodamine-conjugated goat anti-mouse plus fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibodies (BD biosciences) for 45 min. Finally, the slides were washed three times with PBST buffer, mounted, and examined under fluorescence microscope for detection of JCV agnoprotein, JCV LT-Ag and JCV VP1. The immunocytochemistry analysis of JCV Mad-1 Agno-F31A, JCV Mad-1 Agno-F35A, JCV Mad-1 Agno-F39A and JCV Mad-1 Agno (F31AF35AF39A) mutant viruses is described in the relevant figure legends.
Replication assays
Replication assays were carried out as previously described (Sariyer et al., 2011). Briefly, SVG-A cells (2×106 cells/75cm2 flask) were separately transfected/infected either with JCV Mad-1 WT, JCV Mad-1 Agno-F31A, JCV Mad-1 Agno-F35A, JCV Mad-1 Agno-F39A, or JCV Mad-1 Agno (F31AF35AF39A) viral genomes (8μg each) using lipofectamine 2000 according to Manufacturer’s recommendations (Invitrogen). After 5h, cells were washed with PBS and fed with complete DMEM supplemented with 10% FBS and antibiotics [penicillin/streptomycin (100 μg/ml), ciprofloxacin (10 μg/ml]. Next day, transfected cells were trypsinized and transferred into a 175cm2 flasks; and fed with 40 ml complete media supplemented with antibiotics. Every three days, half of the media (20 ml) from each flask was discarded and replenished with new 20 ml complete media. Note that transfected/infected cells were maintained in the same flask without splitting until harvest, which is important for obtaining productive and consistent results. At the indicated time points, low-molecular-weight DNA containing both input and replicated viral DNA was isolated using Qiagen spin columns (Ziegler et al., 2004), digested with BamHI and DpnI enzymes, resolved on a 1% agarose gel and analyzed by Southern blotting.
Preparation of whole-cell extracts for Western blotting
SVG-A cells (2×106 cells/75cm2 flask) were transfected/infected separately with JCV Mad-1 WT and JCV Mad-1 Agno (F31AF35AF39A) genomic DNA (8μg each) using lipofectamine 2000 according to Manufacturer’s recommendations (Invitrogen) as previously described (Sariyer et al., 2011). Whole cell extracts were prepared at the indicated time points (see the respective figure legends) as previously described (Sariyer et al., 2011), separated on a 15% SDS-PAGE and blotted on a nitrocellulose membrane (Bio-Rad) for 10 min at 250 mA. The membrane was then incubated with a primary polyclonal anti-Agno antibody (1:1000 dilution) for 2 h (Del Valle et al., 2002) and washed with PBST buffer three times to remove the nonspecific binding. The membrane was finally incubated with a HRP-conjugated secondary antibody and developed using an ECL-kit (GE HealthCare) according to the manufacture’s recommendations (Sadowska et al., 2003).
Results
Localization of agnoprotein in infected cells
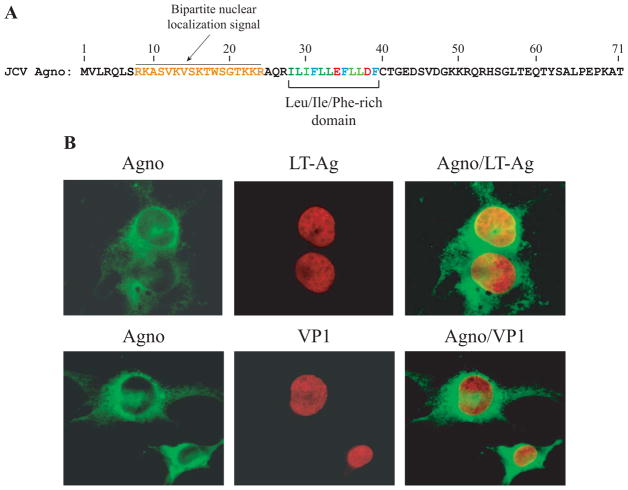
Agnoprotein is a mainly cytoplasmic protein with high concentrations accumulating in the perinuclear region of the infected cells. However, it has been consistently observed that a small amount of this protein can be detected in the nucleus with conventional fluorescence microscopy studies (Safak et al., 2002). Prediction studies show that agnoprotein has a weak bipartite nuclear localization signal (Dingwall and Laskey, 1986; Dingwall and Laskey, 1991; Sigrist et al., 2010)localized to the N-terminus region of the protein which supports our previous observations (Safak et al., 2002) (Fig. 1A). To confirm its nuclear localization, we examined the co-localization of agnoprotein with VP1 and LT-Ag by employing deconvolution microscopy. This technique allows us to examine different cross sections of the infected cells similar to those obtained using confocal fluorescence microscopy studies. For this purpose, SVG-A cells were infected with JCV Mad-1 strain, fixed with cold acetone at 15d posttransfection, probed with appropriate primary and secondary antibodies as indicated in the figure legend and examined under a deconvolution microscope. As shown in Fig. 1B, agnoprotein localizes mostly to the perinuclear and cytoplasmic region of the cells but a small portion of the protein (approximately 15–20%, determined by the statistical analysis software which is incorporated into the operating software of deconvolution microscope) was also found to localize to the nucleus with LT-Ag (Fig 1B), which is consistent with our previous studies (Safak et al., 2002). In support of these observations, recent confocal microscopy and cell-membrane fractionation studies by Suzuki et al., also demonstrated a heavy perinuclear localization of agnoprotein in JCV-infected cells, co-localizing with endoplasmic reticulum (ER) markers, including calreticulin and BiP (Suzuki et al., 2010a). These investigators have also examined the impact of several agnoprotein N-terminal mutants on agnoprotein distribution. Some of these mutants include those from the nuclear localization signal (NLS) region of the protein. Mutation of the Arg8Lys9 residues to AlaAla did not affect the targeting of the protein to ER. That is, this mutant maintained some of its localization to ER. However, mutation of the Lys22Lys23Arg24 residues to AlaAlaGly resulted in loss of such an activity, suggesting that some of the basic residues located within the NLS region of agnoprotein are important for ER targeting (Suzuki et al., 2010a). We also examined the co-localization of agnoprotein with VP1 in a similar manner using appropriate primary and secondary antibodies. Taken together, our results confirmed previous reports in that a small portion of agnoprotein expressed in the infected cells also localizes to the nucleus, suggesting a possible role for agnoprotein in the nucleus.
Figure 1.
(A) Primary structure of JCV agnoprotein. A predicted weak bipartite nuclear localization signal and the Leu/Ile/Phe-rich domain involved in dimer and oligomer formation are indicated. (B) Localization of agnoprotein in infected cells. SVG-A cells were transfected/infected with JCV Mad-1 strain, plated on glass chamber slides and fixed with cold acetone at day 15 posttransfection. Cells were incubated with 5% bovine serum albumin (BSA) prepared in incubation buffer (PBS, 0.1% Tween 20, PBST) for 2h to reduce the primary antibody binding. Then, cells were washed with the same incubation buffer for 3 times with 10 min intervals and subsequently incubated with a combination of anti-Agno (rabbit polyclonal, 1:200 dilution) (Del Valle et al., 2002) plus anti-JCV LT-Ag (Ab2000, mouse monoclonal, 1:50 dilution) (Bollag and Frisque, 1992) or anti-Agno (rabbit polyclonal, 1:200 dilution) plus VP1 (PAB597, mouse monoclonal, 1:200 dilution) (Saribas et al., 2011) primary antibodies for overnight as described in Materials and Methods. Cells were subsequently washed with incubation buffer three times in 10 min intervals. Cells were then incubated with a combination of FITC-conjugated anti-rabbit goat IgG plus Rhodamine-conjugated anti-mouse goat IgG for 45 min, washed three times with incubation buffer, mounted using Vectashield mounting media (Vector Laboratories Inc., Burlingame, CA) and analyzed under a deconvolution microscope (Nikon Eclipse TE300).
Agnoprotein enhances LT-Ag binding to Ori
LT-Ag is the major regulatory protein of JCV and involved in both viral DNA replication (Lynch and Frisque, 1990; Lynch and Frisque, 1991; Lynch, Haggerty, and Frisque, 1994; Tavis and Frisque, 1991) and viral gene transcription (Khalili, Feigenbaum, and Khoury, 1987; Lashgari et al., 1989; Safak et al., 2001). It shows significant sequence homology to the LT-Ag of SV40 and BKV, with the greatest divergence occurring within the carboxy-terminal region (Fanning and Knippers, 1992; Frisque, Bream, and Cannella, 1984).
Biochemical and genetic analyses have shown that SV40 LT-Ag interacts with specific nucleotide sequences within the viral origin of DNA replication (Ori) by forming a double hexamer in an ATP-dependent manner, unwinds the Ori DNA sequences (Borowiec et al., 1990; Fanning and Knippers, 1992) and thereby creates a replication bubble. Then cellular DNA polymerase α-primase is recruited to the DNA replication initiation sites along with other replication factors. The elongation process is regulated by the helicase-ATPase activity of LT-Ag (DePamphilis, 1986; Stillman, 1994). JCV LT-Ag appears to behave similarly during the viral DNA replication as described for SV40 LT-Ag (Bollag, Mackeen, and Frisque, 1996; Lynch and Frisque, 1990; Lynch and Frisque, 1991; Lynch, Haggerty, and Frisque, 1994).
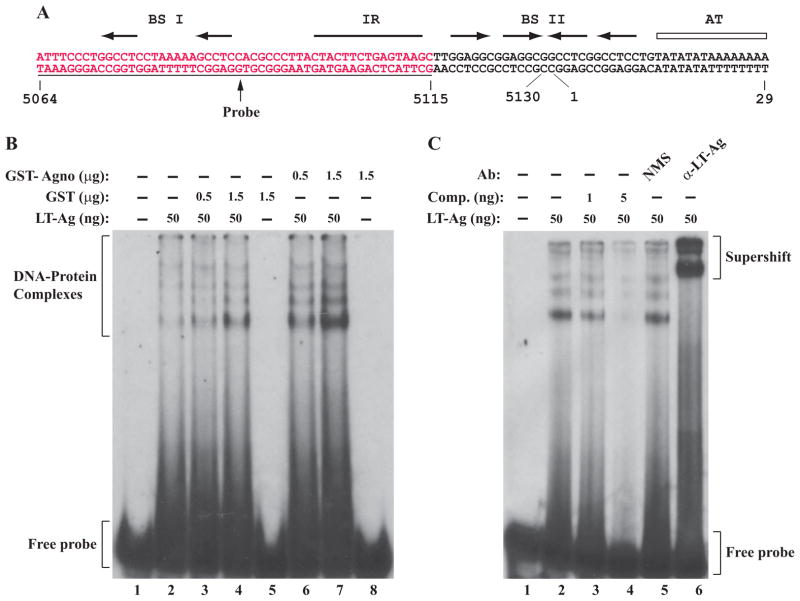
Presence of a small amount of agnoprotein in the nucleus of the infected cells (Fig. 1B) and previously reported interaction of agnoprotein with the helicase domain of JCV LT-Ag (Safak et al., 2001) suggested that agnoprotein may be involved in regulation of LT-Ag function, perhaps through influencing LT-Ag binding to Ori. We explored this possibility in vitro by employing a band shift assay. In this assay, we used recombinant proteins (agnoprotein and LT-Ag) and a double-stranded nucleotide probe, which encompasses 5064–5115 nucleotide region of JCV which completely overlaps with BS I (binding site I) and IR (inverted repeat) regions of JCV Ori respectively as indicated in Fig. 2A (Lynch and Frisque, 1990). As expected, LT-Ag formed complexes with Ori sequences (Fig. 2B, lane 2). The multiple banding patterns that we observed are most likely due to the interaction of monomeric and different multimeric forms of LT-Ag with DNA (Borowiec et al., 1990; Fanning and Knippers, 1992).
Figure 2.
JCV agnoprotein enhances LT-Ag binding to Ori sequences. (A) Structure of the putative JCV core origin of replication. Arrows indicates the positions of JCV LT-Ag recognition sites. The positions of binding sites I (BS I) and II (BS II) are also indicated. The inverted repeat region (IR) is marked by a solid bar and the A+T-rich tract (AT) is identified by an open bar. Position of the probe used in the band shift assays is marked by a thin line which encompasses the nucleotides of 5064–5115 (JCV Mad-1, NC_001699). (B) Band shift assay. The double-stranded nucleotide encompassing nt 5064–5115 Mad-1 was end-labeled with γ-[32P]-dATP as described in Materials and Methods and gel purified. The labeled probe (40,000 cpm/lane) was incubated with different combinations and amounts of JCV LT-Ag, GST and GST-Agno proteins as indicated. DNA-protein complexes were then separated on a 6% native polyacrylamide gel and visualized by autoradiography. In lane 1, probe alone was loaded on the gel. (C) Competitive band shift and antibody super-shift assay. Labeled probe was incubated with fixed amount of LT-Ag (50 ng/lane) as indicated plus with different amounts of unlabeled oligonucleotide (Comp.) (1 ng and 5 ng, lanes 3 and 4 respectively). In addition, the reaction mixture was also incubated with either normal mouse serum (NMS) (2 μg, lane 5) or α-LT-Ag (2 μg, Ab-2) antibody as indicated. In lane 1, probe alone was loaded on the gel. DNA-protein-antibody complexes were separated on a 6% polyacrylamide gel under non-denaturing conditions and analyzed by autoradiography.
Next, we analyzed whether agnoprotein influences the DNA binding activity of LT-Ag to its target sequences on the Ori probe. Surprisingly, simultaneous addition of the increasing concentration of recombinant GST-Agno to the reaction mixture significantly enhanced the binding activity of LT-Ag to Ori (Fig. 2B, compare lane 2 with 6 and 7) in a dose dependent manner, although agnoprotein alone did not show any indication of binding to Ori (lane 8), emphasizing the influence of agnoprotein on LT-Ag binding to DNA. However, addition of agnoprotein to the reaction mixture did not alter the banding pattern or mobility of the LT-Ag/Ori complexes other than enhancing the binding activity of LT-Ag. This is consistent with our previous findings, where we demonstrated that Agno/LT-Ag interaction occurs off the DNA by co-immunoprecipitation assays (Safak et al., 2001). We believe that a similar interaction is most likely taking place in our current band shift assays as well. That is, these two proteins interact with each other off the DNA and this interaction may result in a conformational change on LT-Ag which subsequently leads to more efficient binding of LT-Ag to its target sequences on DNA (Fig. 8). Under identical reaction conditions, we tested the specificity of the agnoprotein-mediated enhancement of LT-Ag binding to DNA by using the GST protein in the binding reactions. In this regard, addition of GST alone to the binding reaction did not result in a considerable level of induction (Fig. 2, lanes 3 and 4) compared to that observed for GST-Agno (Fig. 2, lanes 6 and 7). This suggests that the binding activity of LT-Ag to its target sequences is specifically stimulated by agnoprotein but not GST (compare lane 4, with lane 7). GST alone did not interact with the probe (Fig. 2, lane 5). Collectively, these results demonstrate that the binding activity of JCV LT-Ag to its target sequences within Ori is specifically enhanced by JCV agnoprotein.
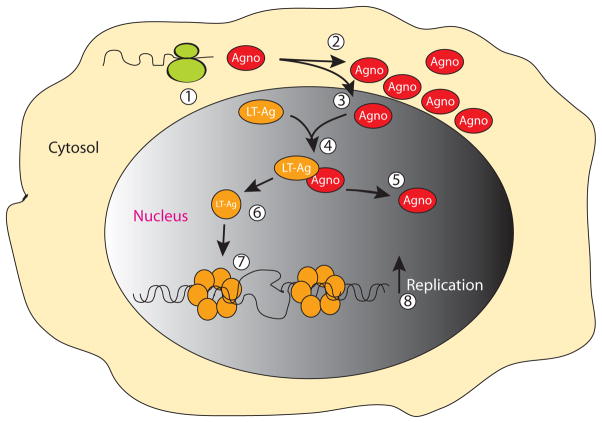
Figure 8.
Model for agnoprotein/LT-Ag interaction in the nucleus. Upon translation of the JCV late transcripts (1), agnoprotein mostly localizes to cytoplasm, largely accumulating around the perinuclear area (2) (Fig. 1B) and a small amount of the protein, however, translocates into the nucleus (3), where it interacts with LT-Ag off the DNA (Safak et al., 2001) and perhaps induces conformational changes on LT-Ag too (6). Subsequently, agnoprotein separates from the LT/Agno complex (4) and liberated LT-Ag then binds to Ori (Fig. 2A) more efficiently in a double hexamer manner (7), unwinds DNA and initiates viral DNA replication which may lead to an increased viral DNA replication (8).
We further examined the specificity of interaction of LT-Ag with the probe by competitive band shift and antibody super-shift assays. The Ori probe was incubated with LT-Ag protein in the absence or presence of 50-and 250-fold molar excesses of unlabeled competitor DNA (Fig 2C, lanes 3 and 4). It was observed that, wild-type Ori DNA efficiently competes with the labeled Ori probe for LT-Ag binding (Fig, 2C, lanes 3 and 4). Reaction mixture was also incubated with either a specific antibody directed against LT-Ag or with normal mouse serum (NMS) as indicated. While α-LT-Ag antibody super-shifted the protein-DNA complexes (compare lane 5 with 6), the normal mouse serum (NMS) did not have a noticeable effect on the migration pattern of LT-Ag-Ori complexes. As a result, both the competitive band shift and the antibody super-shift assays confirm the specificity of LT-Ag binding to Ori.
The region between amino acids 18–36 is important for stimulation of LT-Ag binding to Ori
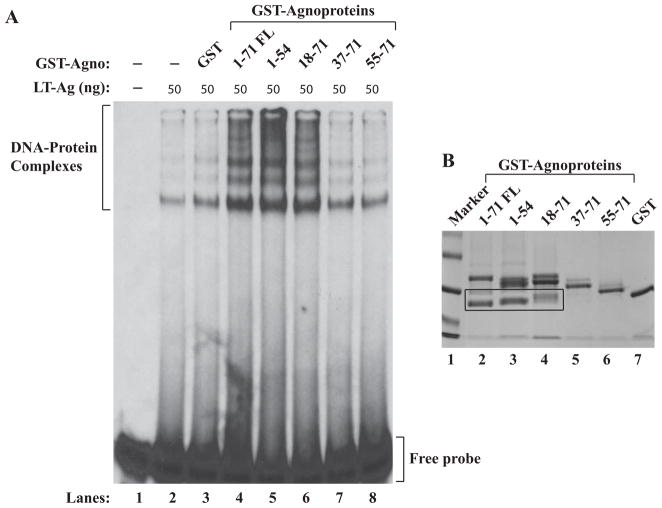
Next, we attempted to identify the region(s) of agnoprotein responsible for the enhancement of LT-Ag binding to Ori. For this purpose, a previously created series of deletion mutants of agnoprotein were expressed as GST fusion proteins in bacteria (Safak et al., 2001), affinity purified, dialyzed against DNA binding buffer as described in Materials and Methods and used in a band shift assay. A recombinant JCV LT-Ag alone was incubated with a double-stranded probe (Fig. 3A, lane 2). A labeled DNA probe plus recombinant LT-Ag was also incubated in combination with purified GST protein alone (lane 3) or with GST-Agno (WT) (lane 4, 1–71 FL) or Agno deletion mutants fused to GST (lanes 5–8) as indicated. Formed complexes were then separated on a 6% native non-denaturing gel and analyzed by autoradiography. As shown in Fig. 3A, consistent with the observations from Fig. 2B, LT-Ag binds to Ori sequences but the addition of GST alone to the binding mixture does not significantly affect the LT-Ag binding to DNA (lane 3). Similarly, two N-terminal deletion mutants of agnoprotein (aa 37–71 and aa 55–71) did not show any significant effect on the enhancement of LT-Ag binding either (lanes 7 and 8). However, the other two deletion mutants that retained N-terminal 54 amino acids (aa 1–54, lane 5) and C-terminal 53 amino acids (aa 18–71, lane 6) enhanced the binding activity of LT-Ag to the Ori, at least, as strongly as WT did. These findings suggest that the agnoprotein region between aa 18 and 36 is important for the stimulation of LT-Ag binding to Ori. Interestingly, this region of agnoprotein, as previously reported, is also involved in interaction with JCV LT-Ag (Safak et al., 2001), JCV Sm t-Ag (Sariyer, Khalili, and Safak, 2008), p53 (Darbinyan et al., 2002) and Yb-1 (Safak et al., 2002); and in formation of SDS-resistant dimeric and oligomeric structures (Saribas et al., 2011). SDS-PAGE analysis of the JCV agnoprotein mutants followed by coomassie staining is shown in Fig. 3B.
Figure 3.
The region between amino acids 18–36 of agnoprotein is important for induced binding of LT-Ag to Ori. Labeled probe was incubated either with LT-Ag alone (lane 2,) or LT-Ag in combination with GST (0.5 μg, lane 3) or LT-Ag in combination with GST-Agno FL (0.5 μg lane 4) or LT-Ag plus agnoprotein deletion mutants fused to GST (0.5 μg each, lanes 5–8) as indicated. DNA-protein complexes were separated on a 6 % polyacrylamide gel and analyzed by autoradiography as described under the legend for figure 2B. In lane 1, probe alone was loaded on the gel. (B) Analysis of GST-Agno FL and agnoprotein deletion mutants fused to GST by SDS-polyacrylamide gel (12%) electrophoresis followed by coomassie blue staining. Degradation products of GST-Agno proteins were encased by a rectangular box.
Analysis of the individual Phe mutants (F31A, F35A or F39A) by immunocytochemistry and band shift assays
Our recent work demonstrated that 17–42 amino acid region of agnoprotein participates in SDS-resistant dimer/oligomer formation and adopts a predicted amphipathic α-helix structure (Saribas et al., 2011) and Fig. 4A illustrates the predicted 3D structure of agnoprotein by computer modeling (Roy, Kucukural, and Zhang, 2010). Interestingly, this α-helix contains a Leu/Ile/Phe-rich domain (aa 28–39), consisting of multiple Leu, Ile and Phe residues and two charged residues, Glu34 and Asp38, interspersed within amino acids of the Leu/Ile/Phe-rich domain (Fig. 1A and 4B). The predicted amphipathic nature of the α-helix can accommodate the spatial conformation of the negatively charged residues (Glu34 and Asp38) in the Leu/Ile/Phe-rich domain. An amphipathic nature of the α-helix was also reported for BKV agnoprotein (Unterstab et al., 2010). All Phe residues (Phe31, Phe35 and Phe39) of agnoprotein also localize to this α-helical domain (Fig. 4B). While F35 and F39 residues are conserved among agnoproteins from JCV, BKV and SV40, the position of JCV F31 is shifted to the position 29 in BKV and SV40 (Fig 4B). Importantly, Phe residues, in general, are known to play diverse but important roles in the function of many different proteins through hydrophobic interactions and possibly through “cation-π” and “anion-π” interactions (Gallivan and Dougherty, 1999; Jackson et al., 2007; Philip et al., 2011; Pless et al., 2008; Pletneva et al., 2001; Shi et al., 2002). Such interactions mediate protein-protein interactions at protein interfaces and also aid to the folding and stability of many different proteins (Bowden et al., 2008; Gallivan and Dougherty, 1999; Jackson et al., 2007; King et al., 2011; Milardi et al., 2011; Philip et al., 2011; Pless et al., 2008; Pletneva et al., 2001; Shi et al., 2002). As such, it is conceivable that the Phe residues of agnoprotein may participate in such interactions to form homodimers between its monomers or heterodimers between agnoprotein and other cellular or viral proteins and thereby contribute to the function of agnoprotein during the viral replication cycle.
Figure 4.
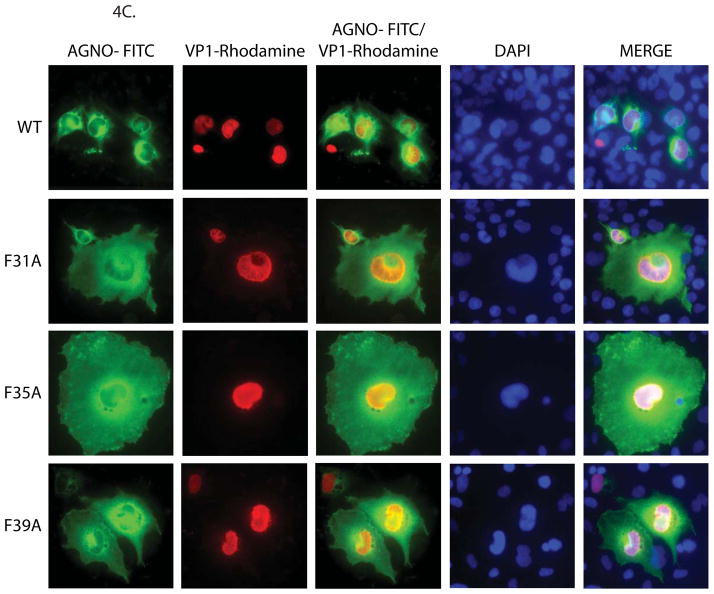
Structural model of agnoprotein and immunocytochemical analysis of F31A, F35A and F39A mutants. (A) Three-D model of JCV agnoprotein was predicted using I-Tasser program (Roy, Kucukural, and Zhang, 2010). Positions of the three Phe residues are indicated on the α-helical structure. (B) Alignment of SV40, BKV and JCV agnoprotein sequences. (C) Immunocytochemical analysis of F31A, F35A and F39A mutants of agnoprotein in the infected cells. SVG-A cells were transfected/infected either with JCV Mad-1 WT or JCV Mad-1 Agno-F31A or JCV Mad-1 Agno-F35A or JCV Mad-1 Agno-F39A mutant genome and at day 5, cells were fixed with cold acetone and blocked with 5% BSA prepared in PBST for 2h. Cells were then incubated with a combination of α-Agno (rabbit polyclonal, 1:200 dilution) (Del Valle et al., 2002) and α-VP1 (PAB597, mouse monoclonal, 1:200 dilution) (Saribas et al., 2011) overnight. Cells were first washed with PBST three times with (10 min intervals) and incubated with the combination of FITC-conjugated anti-rabbit goat and Rhodamine-conjugated anti-mouse goat secondary antibodies for 45 min. Cells were finally washed with PBST three times with 10 min intervals, mounted with mounting media and examined under a fluorescence microscope.
We then assessed the contribution of each Phe residue to agnoprotein function by site-directed mutagenesis followed by immunocytochemistry assays. As shown in Fig. 4C, contrary to our findings for WT agnoprotein (mostly shows a perinuclear distribution), mutant agnoproteins were found to be more evenly distributed within the cytoplasm and lost their high level of accumulation around the perinuclear area (Fig. 4C). This contrasting observation suggests that these Phe residues may play a role in a preferential and strategic distribution of agnoprotein around the perinuclear region. These altered distribution patterns also suggest that agnoprotein mutants maybe lost or gained some new abilities to interact with their partners, which subsequently modified their distribution patterns in the infected cells. Another peculiarity regarding these individual Phe mutants was that the majority of cells infected with the mutant viruses were found to have a larger cell volume than those infected with WT. We currently do not have an explanation for this interesting observation.
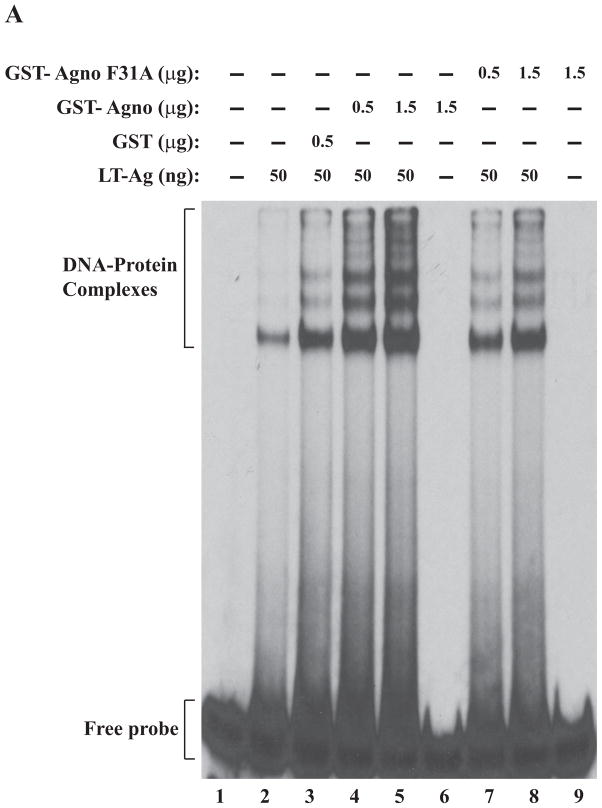
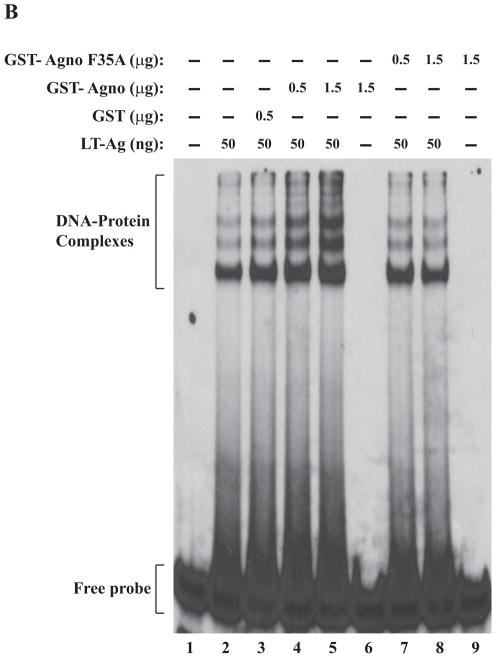
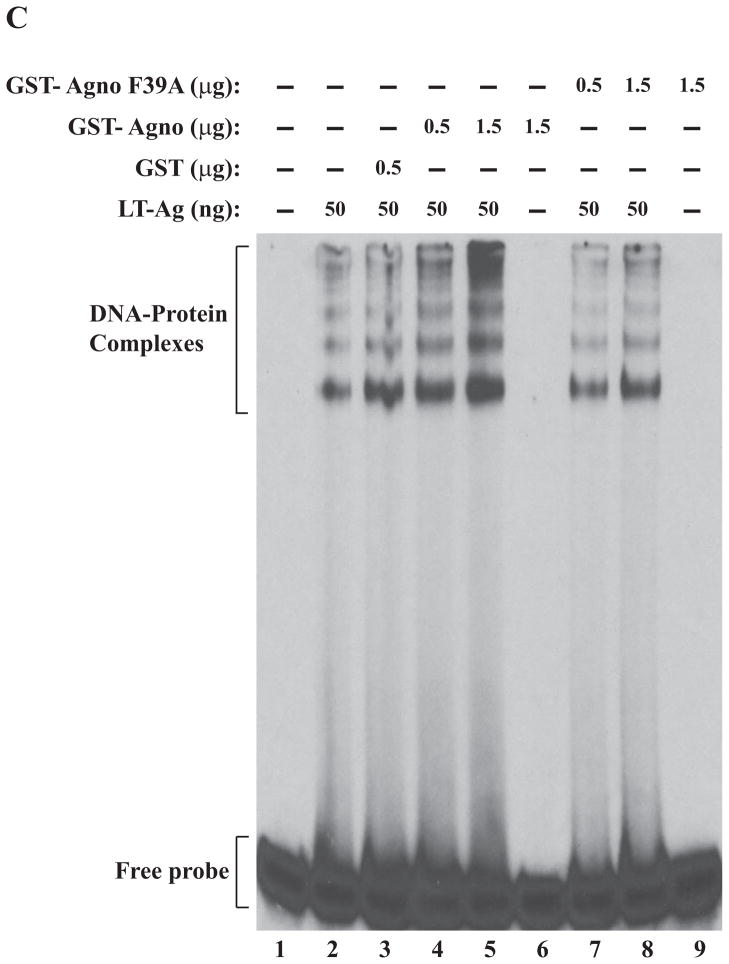
We also examined the effect of Ala substitutions for all three individual Phe residues on agnoprotein-enhanced DNA binding activity of LT-Ag by gel shift assays. All three individual Phe mutants (F31A, F35A, F39A) and WT agnoprotein were produced as GST-fusions in bacteria and affinity purified as described in Materials and Methods. These mutant proteins were then used in band shift assays together with baculovirus-produced recombinant LT-Ag as described in figure legend 5. All individual substitution mutants of agnoprotein failed to stimulate LT-Ag binding activity to Ori at varying degrees (Fig. 5A, 5B and 5C). Comparison of the band intensities of the DNA-protein complexes of WT agnoprotein with those of mutants clearly demonstrated this difference (compare lane 5 with 8 in each panel).
Figure 5.
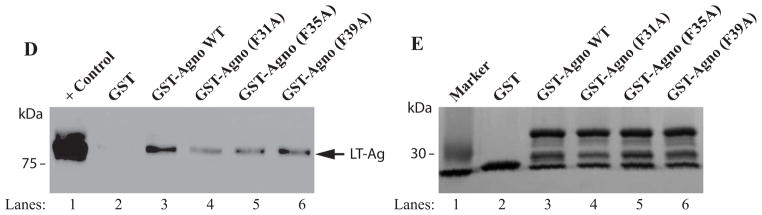
F31A, F35A and F39A mutants of agnoprotein failed to enhance LT-Ag binding to Ori. (A, B and C) F31, F35 and F39 were substituted with Ala by Quik-Change™ mutagenesis kit and mutant DNA was subcloned into pGEX1λT vector at BamHI/EcoRI sites. Subsequently, the mutant proteins were produced in E. coli and purified as described in Materials and Methods and used in band shift assays as indicated. The band shift assay conditions were identical to those described under the legend for figure 2A. In lane 1, probe alone was loaded on the gel. (D) Analysis of the interaction of agnoprotein point mutants [GST-Agno (F31A), GST-Agno (F35A) and GST-Agno (F39A)] with JCV LT-Ag by a GST pull-down assay. Whole-cell extracts prepared from HJC-15b cells, which express JCV LT-Ag constitutively (Raj, 1995), were incubated with either GST alone or GST-Agnoprotein WT or agnoprotein point mutants fused to GST [GST-Agno (F31A), GST-Agno (F35A) and GST-Agno (F39A)] as described in Materials and Methods. Beads were washed extensively and proteins interacting with GST or GST-Agno Phe mutants [GST-Agno (F31A), GST-Agno (F35A) and GST-Agno (F39A)] were resolved by SDS-PAGE and analyzed by Western blotting using anti-LT-Ag antibody (Ab-2). In lane 1, 15 μg of whole-cell extract were loaded as a positive (+) control. (E) Analysis of the GST and GST-Agno WT and agnoprotein point mutants [GST-Agno (F31A), GST-Agno (F35A) and GST-Agno (F39A)] by a 10%-SDS-PAGE followed by coomassie staining.
In parallel to DNA binding assays, we also examined the ability of interaction of each agnoprotein point mutant (F31A, F35A and F39A) with LT-Ag by employing a GST-pull down assay as described in Materials and Methods. This assay is a powerful technique to test the interaction between two or more proteins. As previously reported (Safak et al., 2001), WT agnoprotein strongly interacts with LT-Ag off the DNA (Fig. 5D, lane 3). The strength of this interaction significantly decreased when each agnoprotein point mutant (F31A, F35A and F39A) was incubated with LT-Ag (> 3-fold decrease, compare lane 2 with lanes 4–6), which is consistent with our findings from our DNA binding assays (Fig. 5A–C). In DNA binding assays, we observed that WT agnoprotein strongly induces LT-Ag binding to Ori (Fig. 2A) but each agnoprotein point mutant (F31A, F35A and F39A) lost much of this ability (Fig. 5A–C). The lack of interaction between LT-Ag and GST alone (lane 2) demonstrates the specificity of the interaction between agnoprotein and LT-Ag and serves as a negative control in this protein-protein interaction (GST-pull down) assay.
Functional analysis of agnoprotein Phe mutants
We next examined the impact of each Phe mutant (F31A, F35A and F39A) on the viral DNA replication cycle in the viral background measured by DpnI assay. Mutant genomes were individually transfected/infected into SVG-A cells and low molecular weight DNA was isolated at 5 and 15 day posttransfection. DNA samples were then digested with BamHI/DpnI restriction enzymes. BamHI linearizes the input and replicated DNA; and DpnI is known to specifically digest out the transfected DNA (bacterially produced and methylated input DNA) while keeping the newly-replicated viral DNA intact (Hirt, 1967). DNA samples were then analyzed by Southern blotting as described in the Materials and Methods. As shown in Fig. 6A, the levels of the viral DNA replication for JCV Agno mutants, F35A and F39A, modestly decreased (~30% and ~26% decrease respectively) (compare lane 6 to lanes 8 and 9), while that for F31A remained similar to WT (~11% decrease) (compare lane 6 to 7). These findings are consistent with those found with DNA binding assays (Fig. 5A, 5B and 5C), where it was observed that both F35A and F39A mutants almost completely lost their ability to enhance LT-Ag binding to Ori (Fig. 5B and 5C), whereas F31A mutant slightly retained such an ability (Fig. 5A).
Figure 6.
Effect of Phe mutations on the JCV DNA replication. (A) DpnI assay. SVG-A cells (2×106 cells/75cm2 flask) were separately transfected/infected with JCV Mad-1 WT, JCV Mad-1 Agno-F31A, JCV Mad-1 Agno-F35A or JCV Mad-1 Agno-F39A viral DNA genomes (8 μg each). The low-molecular-weight DNA containing both input and replicated viral DNA was isolated using Qiagen spin columns (Ziegler et al., 2004), and digested with BamHI and DpnI restriction enzymes. Digested DNA was separated on a 1% agarose gel, transferred onto a nitrocellulose membrane (Bio-Rad) and probed for detection of the newly replicated DNA using a probe prepared from the JCV Mad-1 WT as a template. In lane 1, 2 ng of JCV Mad-1 WT linearized by BamHI digestion was loaded as positive control (+ Cont.). Replication assays were repeated several times and a representative result is shown here. (B) Quantitation analysis of Southern blots by a semi-quantitative densitometry method (using NIH Image J program) and presentation of the results in arbitrary units. The replication efficiency of each data point was presented relative to that of WT.
Combinatorial effect of all three Phe mutants on agnoprotein function
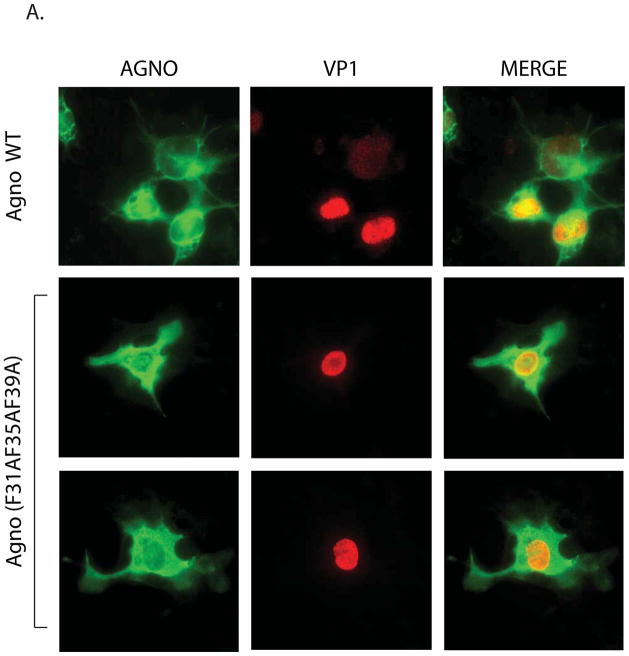
In order to get more insight into the roles of Phe residues in agnoprotein function, a cumulative effect of all Phe residues, where all three Phe residues were converted into Ala (triple Phe mutant), was analyzed by immunocytochemistry and replication assays. It was interesting to observe that each individual Phe mutant influenced the distribution pattern of agnoprotein in the infected cells. That is, in contrast to the distribution pattern of WT agnoprotein - mostly accumulating around the perinuclear area and localizing to nucleus in small amounts -, all the individual mutants, however, showed a more uniform distribution pattern throughout the infected cells. We also wanted to analyze whether a triple Phe mutant has a similar differential effect on agnoprotein distribution. To test this possibility, JCV Mad-1 WT and JCV Mad-1 Agno triple Phe mutant was separately transfected/infected into SVG-A cells and cells were fixed at 15 day posttransfection and analyzed by immunocytochemistry as described for Fig. 4C. As expected, WT agnoprotein showed high level of accumulation around the perinuclear area of the infected cells. However, the triple Phe mutant lost such ability and showed a more uniform distribution pattern throughout the infected cells (Fig. 7A), similar to those observed for each individual Phe mutant (Fig. 4C).
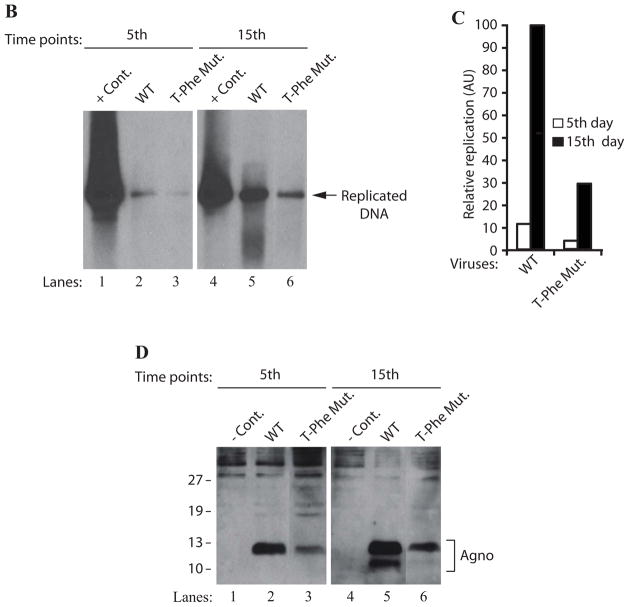
Figure 7.
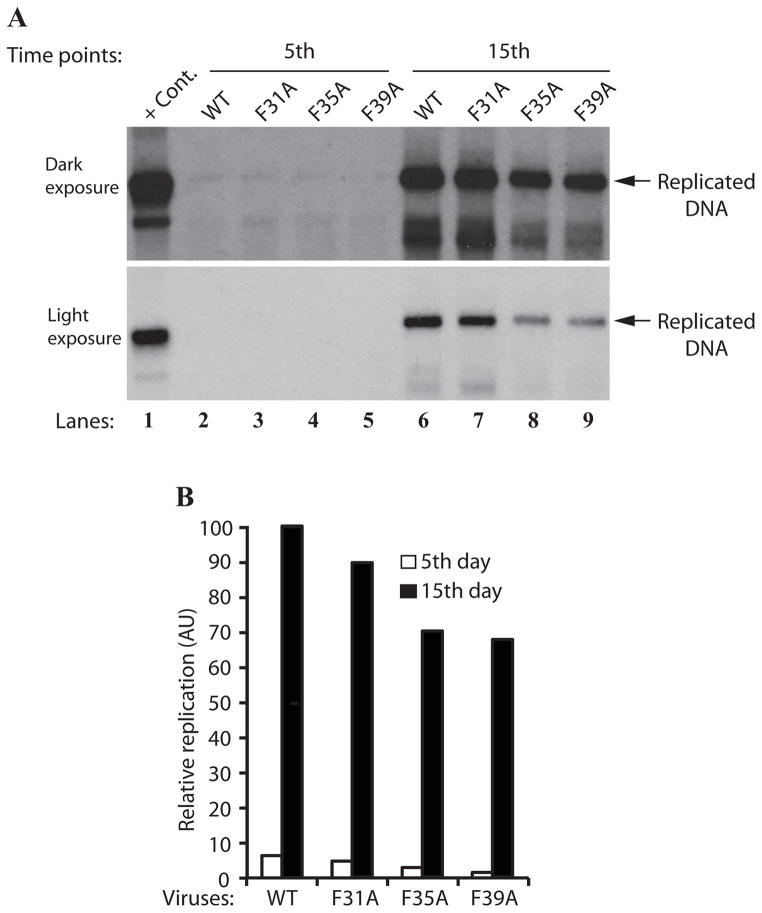
Effect of a triple Phe mutant (F31AF35AF39A) on agnoprotein expression and the viral DNA replication in the viral background. (A) Immunocytochemical analysis of a triple Phe mutant (F31AF35AF39A). SVG-A cells were transfected/infected either with JCV Mad-1 WT or the triple Phe mutant and at day 5, cells were fixed with cold acetone and blocked with 5% BSA prepared in PBST for 2h. Then cells were processed for immunocytochemical detection of agnoprotein as described under the legend for figure 4C. (B) A cumulative effect of all three Phe mutations of agnoprotein on viral DNA replication. DpnI assay for a triple Phe mutant [F31A F35AF39A)=(T-Phe Mut.). JCV Mad-1 Agno WT and JCV Mad-1 Agno-(F31A F35A F39A) mutant genomes were transfected/infected into SVG-A cells and low molecular DNA was isolated at the indicated data points and analyzed by Southern blotting as described for Figure 6A. Replication assays were repeated several times and a representative result is shown here. (C) Quantitation analysis of Southern blots by a semi-quantitative densitometry method (using NIH Image J program) and presentation of the results in arbitrary units. The replication efficiency of the mutant virus was presented relative to that of WT. (D) Western blot analysis. In parallel to Southern blot analysis in 7B, whole-cell extracts were prepared at day 5 and 15 posttransfection from SVG-A cells transfected/infected with either JCV Mad-1 WT or JCV Mad-1 Agno-(T-Phe Mut.) as indicated, and analyzed by Western blotting. In this regard, 40 μgs of whole-cell extracts were separated on a 15% SDS-PAGE, transferred onto a nitrocellulose membrane and incubated with a primary (anti-Agno, polyclonal) (Del Valle et al., 2002) and secondary antibodies. The protein of interest was then detected by using ECL reagent as described in Materials and Methods. In lane 1 and 4, whole-cell extracts prepared from untransfected SVGA cells were loaded as negative controls (- Cont.).
In parallel, we also analyzed the effect of this triple Phe mutant on the viral DNA replication cycle in the viral background, employing DpnI assay. The low molecular weight DNA was isolated at 5 and 15 day posttransfection. DNA samples were then analyzed by southern blotting to measure the level of newly replicated mutant viral DNA relative to that of WT. As shown in Fig. 7B, the level of the viral DNA replication for the triple Phe mutant decreased more drastically (3.5 fold, Fig. 7B and 7C) than those observed for the individual Phe mutants (Fig. 6A) by 15 day posttransfection. This observation clearly demonstrates a cumulative negative effect of all the three Phe residues on the viral DNA replication. In addition, the expression level of agnoprotein triple mutant was also significantly decreased compared to WT (Fig. 7D), which is consistent with the replication assay data (Fig. 7B). Taken together, these findings suggest that Phe residues may play significant roles in agnoprotein function and perhaps in its structure.
Discussion
Agnoprotein is a major regulatory protein of JC virus in addition to LT-Ag, Sm t-Ag and the T′-proteins (Bollag et al., 2010; Bollag et al., 2000; Frisque, Bream, and Cannella, 1984; Safak et al., 2001; Sariyer, Khalili, and Safak, 2008). In the absence of agnoprotein expression, the replication cycle of JCV, BKV and SV40 does not proceed efficiently (Myhre et al., 2010; Saribas et al., 2011; Sariyer et al., 2011). Many studies indicate that agnoprotein is a multifunctional protein and is involved in different aspects of viral replication cycle, including transcription and replication (Safak et al., 2001; Safak et al., 2002). While much has been learned, the precise function of this protein in JCV infection is not fully understood.
Our DNA binding studies demonstrated that agnoprotein enhances the DNA binding activity of LT-Ag to Ori without directly interacting with DNA (Fig. 2A). This is a novel finding suggesting that agnoprotein may be involved in viral DNA replication through interaction with LT-Ag. However, agnoprotein does not seem to form an apparent ternary complex with LT-Ag/DNA complex under our band shift assay conditions, because the migration pattern of DNA-protein complexes was not altered upon addition of agnoprotein into the binding mixture (Fig. 2B). This is consistent with our previous reports, where it was shown that agnoprotein interacts with the helicase domain of LT-Ag without requiring the presence of DNA molecule for the interaction (Safak et al., 2001). This observation suggests that (i) interaction of agnoprotein with LT-Ag off the DNA is sufficient to induce a conformational change on LT-Ag protein so that LT-Ag binds to its target sequences more efficiently. Agnoprotein is then most likely liberated from LT-Ag/Agno complex before LT-Ag binds to DNA (Fig. 8). This is, at least, one of the reasons that agnoprotein is not the part of LT-Ag/DNA complex. (ii) Alternatively, the interaction between agnoprotein and LT-Ag/DNA complex is weak and therefore is unstable under our gel shift running conditions. Nonetheless, the ability of one protein to influence the DNA binding capacity of another one has been well-established. There are many similar interactions reported in the literature, including the induction of Tst-1 binding to DNA by HMG-1/Y (Leger et al., 1995), stimulation of the serum response factor binding to DNA by Phox1 (Grueneberg et al., 1992) and induction of YB-1 binding to the 23-bp element of JCV archetype by LT-Ag (Safak et al., 1999).
The observation that agnoprotein does not directly interact with DNA contrasts with a previously reported case of SV40 agnoprotein (Gilbert et al., 1981). Gilbert et al., reported that SV40 agnoprotein is a DNA binding protein based on the premise that this protein has a highly basic character containing many positively charged residues, including Arg and Lys, which may influence its nucleic acid interaction (Gilbert et al., 1981). The experimental settings employed by Gilbert et al., were somewhat different than those used in our DNA binding assays. This may account for the difference in our and their findings and conclusions. We have used a purified recombinant protein (GST-Agno) and a short-labeled synthetic oligonucleotide corresponding to the BS I and IR region of Ori (Fig. 2A) in our binding assays and showed that purified agnoprotein does not directly interact with DNA (Fig. 2B). In the report by Gilbert et al, however, metabolically labeled whole-cell extracts prepared from African Green Monkey Kidney (AGMK) cells infected with SV40 were loaded onto either a single-stranded or a double-stranded calf thymus DNA-cellulose columns (Gilbert et al., 1981). The columns were then consecutively washed with a low salt and high salt buffers; and various fractions were analyzed by SDS-PAGE followed by autoradiography. Labeled agnoprotein was only detected in fractions eluted with high salt but not with low salt buffer. Interestingly, many other labeled-proteins, in addition to agnoprotein, were also detected in the elution fractions, thus making the conclusions somewhat complicated. Because these results also suggest a strong possibility that agnoprotein does not necessarily directly bind to DNA but rather interacts with the cellular proteins that were retained in the DNA columns due to their intrinsic DNA binding activities. Therefore, it is possible that agnoprotein was eluted out along with those proteins and detected in those fractions.
Agnoprotein has been previously shown to interact with cellular, (YB-1, FEZ1 and HP1–α, p53) and viral (LT-Ag, Sm t-Ag) proteins (Darbinyan et al., 2002; Okada et al., 2005; Safak et al., 2001; Safak et al., 2002; Sariyer, Khalili, and Safak, 2008) and the 18–36 amino acid region of the protein has been consistently observed to be critical for such interactions. We have recently reported that the same region of agnoprotein plays an important role in the viral replication cycle perhaps through forming highly stable SDS-resistant dimeric/oligomeric structures (Saribas et al., 2011). In addition, 3D computer modeling suggests that this region is also involved in forming an α-helical structure (Saribas et al., 2011). This helix region contains a Leu/Ile/Phe-rich domain (aa 28–39) and two negatively charged residues, Glu34 and Asp38, are also found to be present in the same region (Fig. 1A, Fig. 4B). Interestingly, all three Phe residues (Phe31, Phe35 and Phe39) of JCV agnoprotein are located in this Leu/Ile/Phe-rich domain. All three Phe residues of BKV and SV40 agnoproteins also localize to the respective Leu/Ile/Phe-rich domain of each protein but there are differences among their positioning compare to those present in JCV agnoprotein (Fig. 4B). The position of Phe31 and Phe39 is conserved among the agnoprotein of all three polyomaviruses, JCV, BKV and SV40, but the position of Phe35 of JCV agnoprotein shifted to position Phe29 in BKV and SV40 agnoprotein (Fig 4B), indicating possible differential structural and functional roles for them in their respective viral replication cycles.
Phenylalanine residues are known to play important roles in functions of many different proteins, including in mediation of protein-protein interactions, protein folding and stability (Bowden et al., 2008; Gallivan and Dougherty, 1999; King et al., 2011; Milardi et al., 2011; Pless et al., 2008; Pletneva et al., 2001; Shi et al., 2002). Phe residues accomplish such functions through hydrophobic interactions mediated by the π–π stacking of the aromatic ring of Phe or “cation-π” interactions (Bowden et al., 2008; Gallivan and Dougherty, 1999; King et al., 2011; Milardi et al., 2011; Pless et al., 2008; Pletneva et al., 2001; Shi, Olson, and Kallenbach, 2002) or “anion-π interactions (Jackson et al., 2007; Philip et al., 2011) or through the combination of all these three types of interactions. Such interactions may also take place between homodimers (agno-agno interaction) or heterodimers (interaction of agnoprotein with its partners in the infected cell) of agnoprotein and significantly contribute to its functions. Based on these reports, we have then investigated the contribution of Phe residues to agnoprotein function by site-directed mutagenesis. Individual substitution of Phe residues to Ala resulted in partial or full loss of agnoprotein-mediated enhancement of LT-Ag binding to Ori (Fig. 5A, B and C). This loss was more prominent in the case of individual conversion of Phe35 and Phe39 to Ala than that of Phe31 to Ala (Fig. 5). Corroborative results were also obtained when these mutants were tested in viral DNA replication assays in vivo (Fig. 6A), where it was observed that the level of the viral DNA replication for F35A and F39A mutants decreased more prominently than that observed for F31A compared to WT (Fig. 6A). Importantly, the level of viral replication reduced more drastically compared to WT when all three Phe residues all together mutated to Ala, which further illustrates the importance of Phe residues in agnoprotein function (Fig. 7A and 7B).
Relatively high level accumulation of agnoprotein around the prenuclear area and smaller amounts being detected in the nucleus are the characteristics of agnoprotein distribution pattern in the infected cells (Fig. 1B, and 4C). However, this distribution pattern was altered when Phe mutants of agnoprotein was analyzed by immunocytochemistry. It was found that all individual as well as the triple Phe mutants of agnoprotein more evenly distribute throughout the infected cells rather than primarily accumulating around the perinucleus area as regularly observed for WT agnoprotein. This suggests that Phe residues may direct agnoprotein strategically to the perinuclear area of infected cells or differentially interacts with new targets. Additionally, these residues may also help agnoprotein to homodimerize by itself and/or heterodimerize with other cellular and viral partners to fulfill its functions.
Polyomaviruses including JCV, BKV and SV40 encode only a limited number of regulatory proteins, one of which is agnoprotein, and yet these viruses successfully go through their replication cycles. This suggests that these regulatory proteins perform more than one function to optimize the viral life cycle. Published reports indicate that JCV agnoprotein is involved in many aspects of viral replication cycle, including viral DNA replication (Safak et al., 2001; Saribas et al., 2011) and transcription (Safak et al., 2001; Safak et al., 2002; Sariyer et al., 2011). If this is the case, then, the question arises “how is such a small protein like agnoprotein able to achieve so many different functions despite its small size?” It appears that agnoprotein sequence contains many functional domains strategically packed into a short structure. Any disturbances in its sequences may result in severe consequences. For instance, it was previously reported that conversion of Thr21 [protein kinase C (PKC)-phosphorylation site] alone into Ala on agnoprotein results in a phenotype that is unable to sustain the viral replication cycle (Sariyer et al., 2006). It was also recently found that deletion of either internal amino acids 17 to 42 (Saribas et al., 2011) or C-terminal sequences from 51 to 71 (Akan et al., 2006) also resulted in phenotypes that are replication incompetent. All these observations suggest that there is a close relationship between the structure and the function of agnoprotein, which has to be further investigated to determine the details of such a relationship.
There are many other examples of the small viral proteins from the literature that have multifunctional characteristics, including HIV-1 Rev and Vpr. Rev is a 91 aa phosphorylated nuclear protein and like JCV agnoprotein, it also forms dimers and oligomers as evidenced by X-ray crystallography studies (Daugherty et al., 2010; Daugherty, Liu, and Frankel, 2010). It is involved in the transport of incomplete spliced RNA molecules from nucleus to cytoplasm. Rev binds to Rev response elements present in the intron region of the viral transcripts and functions as both stable dimers and oligomers as evidenced by 3D structural studies (Daugherty et al., 2010; Daugherty, Liu, and Frankel, 2010; DiMattia et al., 2010). Vpr, another small protein (96 aa) of HIV, also forms stable dimers as shown by NMR structural studies (Bourbigot et al., 2005; Morellet et al., 2003), arrests cells at G2/M phase transition and induces apoptosis (Bolton and Lenardo, 2007; Cui et al., 2006; Fritz et al., 2008; Fritz et al., 2010; Godet et al., 2010; Iordanskiy et al., 2004; Poon, Chang, and Chen, 2007), indicating its multifunctional nature.
Agnoproteins of JCV, BKV and SV40 all form highly stable dimeric and oligomeric structures (Saribas et al., 2011), functions of which are currently unknown. However, it is conceivable that such structures may provide considerable flexibility to agnoprotein to diversify its biological functions in the infected cells. Rough mapping studies of JCV agnoprotein showed that aa 17–42 are important for dimer and oligomer formation property of agnoprotein (Saribas et al., 2011). In the current study, we have advanced our understanding of agnoprotein function one step further by examining its influence on viral DNA replication and by particularly focusing on the functional analysis of its Phe residues. Our future studies will be directed to further dissect out 17–42 aa region of JCV agnoprotein and analyze the contribution of this region to agnoprotein function. Such detailed studies may provide us with important clues to design effective inhibitors against agnoprotein to curb the progression of PML in affected individuals.
Acknowledgments
We would like to thank past and present members of the Department of Neuroscience and Center for Neurovirology for their insightful discussion and sharing of ideas and reagents. This work was made possible by grants awarded by NIH to MS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akan I, Sariyer IK, Biffi R, Palermo V, Woolridge S, White MK, Amini S, Khalili K, Safak M. Human polyomavirus JCV late leader peptide region contains important regulatory elements. Virology. 2006;349(1):66–78. doi: 10.1016/j.virol.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine JC. Evidence for simian virus 40 late transcriptional control: mixed infections of wild-type simian virus 40 and a late leader deletion mutant exhibit trans effects on late viral RNA synthesis. J Virol. 1982;42(3):798–803. doi: 10.1128/jvi.42.3.798-803.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JR. The basis for modeling progressive multifocal leukoencephalopathy pathogenesis. Curr Opin Neurol. 2011;24(3):262–7. doi: 10.1097/WCO.0b013e328346d2a3. [DOI] [PubMed] [Google Scholar]
- Bollag B, Frisque RJ. PAb 2000 specifically recognizes the large T and small t proteins of JC virus. Virus Res. 1992;25(3):223–39. doi: 10.1016/0168-1702(92)90136-w. [DOI] [PubMed] [Google Scholar]
- Bollag B, Hofstetter CA, Reviriego-Mendoza MM, Frisque RJ. JC virus small T antigen binds phosphatase PP2A and Rb family proteins and is required for efficient viral DNA replication activity. PLoS One. 2010;5(5):e10606. doi: 10.1371/journal.pone.0010606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag B, Mackeen PC, Frisque RJ. Purified JC virus T antigen derived from insect cells preferentially interacts with binding site II of the viral core origin under replication conditions. Virology. 1996;218(1):81–93. doi: 10.1006/viro.1996.0168. [DOI] [PubMed] [Google Scholar]
- Bollag B, Prins C, Snyder EL, Frisque RJ. Purified JC virus T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology. 2000;274(1):165–78. doi: 10.1006/viro.2000.0451. [DOI] [PubMed] [Google Scholar]
- Bolton DL, Lenardo MJ. Vpr cytopathicity independent of G2/M cell cycle arrest in human immunodeficiency virus type 1-infected CD4+ T cells. J Virol. 2007;81(17):8878–90. doi: 10.1128/JVI.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec JA, Dean FB, Bullock PA, Hurwitz J. Binding and unwinding--how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60(2):181–4. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- Bourbigot S, Beltz H, Denis J, Morellet N, Roques BP, Mely Y, Bouaziz S. The C-terminal domain of the HIV-1 regulatory protein Vpr adopts an antiparallel dimeric structure in solution via its leucine-zipper-like domain. Biochem J. 2005;387(Pt 2):333–41. doi: 10.1042/BJ20041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, Stuart DI. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15(6):567–72. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- Brinda KV, Kannan N, Vishveshwara S. Analysis of homodimeric protein interfaces by graph-spectral methods. Protein Eng. 2002;15(4):265–77. doi: 10.1093/protein/15.4.265. [DOI] [PubMed] [Google Scholar]
- Cui J, Tungaturthi PK, Ayyavoo V, Ghafouri M, Ariga H, Khalili K, Srinivasan A, Amini S, Sawaya BE. The role of Vpr in the regulation of HIV-1 gene expression. Cell Cycle. 2006;5(22):2626–38. doi: 10.4161/cc.5.22.3442. [DOI] [PubMed] [Google Scholar]
- Darbinyan A, Darbinian N, Safak M, Radhakrishnan S, Giordano A, Khalili K. Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene. 2002;21(36):5574–81. doi: 10.1038/sj.onc.1205744. [DOI] [PubMed] [Google Scholar]
- Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc Natl Acad Sci U S A. 2010;107(28):12481–6. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol. 2010;17(11):1337–42. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle L, Gordon J, Enam S, Delbue S, Croul S, Abraham S, Radhakrishnan S, Assimakopoulou M, Katsetos CD, Khalili K. Expression of human neurotropic polyomavirus JCV late gene product agnoprotein in human medulloblastoma. J Natl Cancer Inst. 2002;94(4):267–73. doi: 10.1093/jnci/94.4.267. [DOI] [PubMed] [Google Scholar]
- DePamphilis MLaMKB. Replication of SV40 and polyomavirus chromosomas. In: Salzman E, editor. The Polyomaviridae. Vol. 1. Plenum Press; New York: 1986. [Google Scholar]
- Dhe-Paganon S, Werner ED, Nishi M, Hansen L, Chi YI, Shoelson SE. A phenylalanine zipper mediates APS dimerization. Nat Struct Mol Biol. 2004;11(10):968–74. doi: 10.1038/nsmb829. [DOI] [PubMed] [Google Scholar]
- DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A. 2010;107(13):5810–4. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Protein import into the cell nucleus. Annu Rev Cell Biol. 1986;2:367–90. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991;16(12):478–81. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. J Virol. 1984;51(2):458–69. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JV, Didier P, Clamme JP, Schaub E, Muriaux D, Cabanne C, Morellet N, Bouaziz S, Darlix JL, Mely Y, de Rocquigny H. Direct Vpr-Vpr interaction in cells monitored by two photon fluorescence correlation spectroscopy and fluorescence lifetime imaging. Retrovirology. 2008;5:87. doi: 10.1186/1742-4690-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JV, Dujardin D, Godet J, Didier P, De Mey J, Darlix JL, Mely Y, de Rocquigny H. HIV-1 Vpr oligomerization but not that of Gag directs the interaction between Vpr and Gag. J Virol. 2010;84(3):1585–96. doi: 10.1128/JVI.01691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 1999;96(17):9459–64. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J, Nomura S, Anderson CW, George K. Identification of the SV40 agnoproduct: a DNA binding protein. Nature. 1981;291:346–349. doi: 10.1038/291346a0. [DOI] [PubMed] [Google Scholar]
- Godet AN, Guergnon J, Croset A, Cayla X, Falanga PB, Colle JH, Garcia A. PP2A1 binding, cell transducing and apoptotic properties of Vpr(77–92): a new functional domain of HIV-1 Vpr proteins. PLoS One. 2010;5(11):e13760. doi: 10.1371/journal.pone.0013760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueneberg DA, Natesan S, Alexandre C, Gilman MZ. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992;257(5073):1089–95. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- Haggerty S, Walker DL, Frisque RJ. JC virus-simian virus 40 genomes containing heterologous regulatory signals and chimeric early regions: identification of regions restricting transformation by JC virus. J Virol. 1989;63(5):2180–90. doi: 10.1128/jvi.63.5.2180-2190.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Skolnik-David H, Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982;29(1):183–93. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26(2):365–9. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hou-Jong MH, Larsen SH, Roman A. Role of the agnoprotein in regulation of simian virus 40 replication and maturation pathways. J Virol. 1987;61(3):937–9. doi: 10.1128/jvi.61.3.937-939.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanskiy S, Zhao Y, DiMarzio P, Agostini I, Dubrovsky L, Bukrinsky M. Heat-shock protein 70 exerts opposing effects on Vpr-dependent and Vpr-independent HIV-1 replication in macrophages. Blood. 2004;104(6):1867–72. doi: 10.1182/blood-2004-01-0081. [DOI] [PubMed] [Google Scholar]
- Jackson MR, Beahm R, Duvvuru S, Narasimhan C, Wu J, Wang HN, Philip VM, Hinde RJ, Howell EE. A preference for edgewise interactions between aromatic rings and carboxylate anions: the biological relevance of anion-quadrupole interactions. J Phys Chem B. 2007;111(28):8242–9. doi: 10.1021/jp0661995. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Myhre MR, Dragset M, Tummler C, Moens U. Phosphorylation of human polyomavirus BK agnoprotein at Ser-11 is mediated by PKC and has an important regulative function. Virology. 2008;379(1):97–109. doi: 10.1016/j.virol.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Khalili K, Feigenbaum L, Khoury G. Evidence for a shift in 5′-termini of early viral RNA during the lytic cycle of JC virus. Virology. 1987;158(2):469–72. doi: 10.1016/0042-6822(87)90224-8. [DOI] [PubMed] [Google Scholar]
- Khalili K, White MK, Sawa H, Nagashima K, Safak M. The agnoprotein of polyomaviruses: a multifunctional auxiliary protein. J Cell Physiol. 2005;204(1):1–7. doi: 10.1002/jcp.20266. [DOI] [PubMed] [Google Scholar]
- Kim J, Woolridge S, Biffi R, Borghi E, Lassak A, Ferrante P, Amini S, Khalili K, Safak M. Members of the AP-1 family, c-Jun and c-Fos, functionally interact with JC virus early regulatory protein large T antigen. J Virol. 2003;77(9):5241–52. doi: 10.1128/JVI.77.9.5241-5252.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G, Oates J, Patel D, van den Berg HA, Dixon AM. Towards a structural understanding of the smallest known oncoprotein: investigation of the bovine papillomavirus E5 protein using solution-state NMR. Biochim Biophys Acta. 2011;1808(6):1493–501. doi: 10.1016/j.bbamem.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt-DeMasters BK, Tyler KL. Progressive multifocal leukoencephalopathy complicating treatment with natalizumab and interferon beta-1a for multiple sclerosis. N Engl J Med. 2005;353(4):369–74. doi: 10.1056/NEJMoa051782. [DOI] [PubMed] [Google Scholar]
- Langer-Gould A, Atlas SW, Green AJ, Bollen AW, Pelletier D. Progressive multifocal leukoencephalopathy in a patient treated with natalizumab. N Engl J Med. 2005;353(4):375–81. doi: 10.1056/NEJMoa051847. [DOI] [PubMed] [Google Scholar]
- Lashgari MS, Tada H, Amini S, Khalili K. Regulation of JCVL promoter function: transactivation of JCVL promoter by JCV and SV40 early proteins. Virology. 1989;170(1):292–5. doi: 10.1016/0042-6822(89)90381-4. [DOI] [PubMed] [Google Scholar]
- Leger H, Sock E, Renner K, Grummt F, Wegner M. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol Cell Biol. 1995;15(7):3738–47. doi: 10.1128/mcb.15.7.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KJ, Frisque RJ. Identification of critical elements within the JC virus DNA replication origin. J Virol. 1990;64(12):5812–22. doi: 10.1128/jvi.64.12.5812-5822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KJ, Frisque RJ. Factors contributing to the restricted DNA replicating activity of JC virus. Virology. 1991;180(1):306–17. doi: 10.1016/0042-6822(91)90035-a. [DOI] [PubMed] [Google Scholar]
- Lynch KJ, Haggerty S, Frisque RJ. DNA replication of chimeric JC virus-simian virus 40 genomes. Virology. 1994;204(2):819–22. doi: 10.1006/viro.1994.1600. [DOI] [PubMed] [Google Scholar]
- Major EO, Amemiya K, Tornatore CS, Houff SA, Berger JR. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5(1):49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci U S A. 1985;82(4):1257–61. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolskee RF, Nathans D. Suppression of a VP1 mutant of simian virus 40 by missense mutations in serine codons of the viral agnogene. J Virol. 1983;48(2):405–9. doi: 10.1128/jvi.48.2.405-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milardi D, Sciacca MF, Pappalardo M, Grasso DM, La Rosa C. The role of aromatic side-chains in amyloid growth and membrane interaction of the islet amyloid polypeptide fragment LANFLVH. Eur Biophys J. 2011;40(1):1–12. doi: 10.1007/s00249-010-0623-x. [DOI] [PubMed] [Google Scholar]
- Moens U, Ludvigsen M, Van Ghelue M. Human polyomaviruses in skin diseases. Patholog Res Int. 2011;2011:123491. doi: 10.4061/2011/123491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N, Bouaziz S, Petitjean P, Roques BP. NMR structure of the HIV-1 regulatory protein VPR. J Mol Biol. 2003;327(1):215–27. doi: 10.1016/s0022-2836(03)00060-3. [DOI] [PubMed] [Google Scholar]
- Myhre MR, Olsen GH, Gosert R, Hirsch HH, Rinaldo CH. Clinical polyomavirus BK variants with agnogene deletion are non-functional but rescued by trans-complementation. Virology. 2010;398(1):12–20. doi: 10.1016/j.virol.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Ng SC, Mertz JE, Sanden-Will S, Bina M. Simian virus 40 maturation in cells harboring mutants deleted in the agnogene. J Biol Chem. 1985;260(2):1127–32. [PubMed] [Google Scholar]
- Okada Y, Suzuki T, Sunden Y, Orba Y, Kose S, Imamoto N, Takahashi H, Tanaka S, Hall WW, Nagashima K, Sawa H. Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: role in nuclear egress of viral particles. EMBO Rep. 2005;6(5):452–7. doi: 10.1038/sj.embor.7400406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip V, Harris J, Adams R, Nguyen D, Spiers J, Baudry J, Howell EE, Hinde RJ. A survey of aspartate-phenylalanine and glutamate-phenylalanine interactions in the protein data bank: searching for anion-pi pairs. Biochemistry. 2011;50(14):2939–50. doi: 10.1021/bi200066k. [DOI] [PubMed] [Google Scholar]
- Pless SA, Millen KS, Hanek AP, Lynch JW, Lester HA, Lummis SC, Dougherty DA. A cation-pi interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J Neurosci. 2008;28(43):10937–42. doi: 10.1523/JNEUROSCI.2540-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletneva EV, Laederach AT, Fulton DB, Kostic NM. The role of cation-pi interactions in biomolecular association. Design of peptides favoring interactions between cationic and aromatic amino acid side chains. J Am Chem Soc. 2001;123(26):6232–45. doi: 10.1021/ja010401u. [DOI] [PubMed] [Google Scholar]
- Poon B, Chang MA, Chen IS. Vpr is required for efficient Nef expression from unintegrated human immunodeficiency virus type 1 DNA. J Virol. 2007;81(19):10515–23. doi: 10.1128/JVI.00947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj GV, Gordon J, Logan TJ, Hall DJ, Deluca A, Giordano A, Khalili K. Characterization of glioma cells derived from polyomavirus-induced brain tumors in hamsters. Int J Oncol. 1995;7:801–808. doi: 10.3892/ijo.7.4.801. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowska B, Barrucco R, Khalili K, Safak M. Regulation of human polyomavirus JC virus gene transcription by AP-1 in glial cells. J Virol. 2003;77(1):665–72. doi: 10.1128/JVI.77.1.665-672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J Virol. 2001;75(3):1476–86. doi: 10.1128/JVI.75.3.1476-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Gallia GL, Ansari SA, Khalili K. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J Virol. 1999;73(12):10146–57. doi: 10.1128/jvi.73.12.10146-10157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Sadowska B, Barrucco R, Khalili K. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J Virol. 2002;76(8):3828–38. doi: 10.1128/JVI.76.8.3828-3838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Arachea BT, White MK, Viola RE, Safak M. Human polyomavirus JC small regulatory agnoprotein forms highly stable dimers and oligomers: implications for their roles in agnoprotein function. Virology. 2011;420(1):51–65. doi: 10.1016/j.virol.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Ozdemir A, Lam C, Safak M. JC virus-induced Progressive Multifocal Leukoencephalopathy. Future Virol. 2010;5(3):313–323. doi: 10.2217/fvl.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, Safak M. Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. J Virol. 2006;80(8):3893–903. doi: 10.1128/JVI.80.8.3893-3903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Khalili K, Safak M. Dephosphorylation of JC virus agnoprotein by protein phosphatase 2A: inhibition by small t antigen. Virology. 2008;375(2):464–79. doi: 10.1016/j.virol.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Saribas AS, White MK, Safak M. Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virol J. 2011;8:255. doi: 10.1186/1743-422X-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Olson CA, Bell AJ, Jr, Kallenbach NR. Non-classical helix-stabilizing interactions: C-H…O H-bonding between Phe and Glu side chains in alpha-helical peptides. Biophys Chem. 2002;101–102:267–79. doi: 10.1016/s0301-4622(02)00171-0. [DOI] [PubMed] [Google Scholar]
- Shi Z, Olson CA, Kallenbach NR. Cation-pi interaction in model alpha-helical peptides. J Am Chem Soc. 2002;124(13):3284–91. doi: 10.1021/ja0174938. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38(Database issue):D161–6. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Smart machines at the DNA replication fork. Cell. 1994;78(5):725–8. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010a;107(16):7461–6. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Okada Y, Semba S, Orba Y, Yamanouchi S, Endo S, Tanaka S, Fujita T, Kuroda S, Nagashima K, Sawa H. Identification of FEZ1 as a protein that interacts with JC virus agnoprotein and microtubules: role of agnoprotein-induced dissociation of FEZ1 from microtubules in viral propagation. J Biol Chem. 2005;280(26):24948–56. doi: 10.1074/jbc.M411499200. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, Nagashima K, Hall WW, Sawa H. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 2010b;6(3):e1000801. doi: 10.1371/journal.ppat.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis JE, Frisque RJ. Altered DNA binding and replication activities of JC virus T-antigen mutants. Virology. 1991;183(1):239–50. doi: 10.1016/0042-6822(91)90136-y. [DOI] [PubMed] [Google Scholar]
- Unterstab G, Gosert R, Leuenberger D, Lorentz P, Rinaldo CH, Hirsch HH. The polyomavirus BK agnoprotein co-localizes with lipid droplets. Virology. 2010;399(2):322–31. doi: 10.1016/j.virol.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Van Assche G, Van Ranst M, Sciot R, Dubois B, Vermeire S, Noman M, Verbeeck J, Geboes K, Robberecht W, Rutgeerts P. Progressive multifocal leukoencephalopathy after natalizumab therapy for Crohn’s disease. N Engl J Med. 2005;353(4):362–8. doi: 10.1056/NEJMoa051586. [DOI] [PubMed] [Google Scholar]
- Ziegler K, Bui T, Frisque RJ, Grandinetti A, Nerurkar VR. A rapid in vitro polyomavirus DNA replication assay. J Virol Methods. 2004;122(1):123–7. doi: 10.1016/j.jviromet.2004.08.012. [DOI] [PubMed] [Google Scholar]