Abstract
Potent and selective inhibitors of the enzyme dimethylarginine dimethylaminohydrolase (DDAH) are useful as molecular probes to better understand cellular regulation of nitric oxide. Inhibitors are also potential therapeutic agents for treatment of pathological states associated with the inappropriate overproduction of nitric oxide, such as septic shock, selected types of cancer, and other conditions. Inhibitors with structures dissimilar to substrate may overcome limitations inherent to substrate analogs. Therefore, to identify structurally-diverse inhibitor scaffolds, high-throughput screening (HTS) of a 4000-member library of fragment-sized molecules was completed using the Pseudomonas aeruginosa DDAH and human DDAH-1 isoforms. Use of a substrate concentration equal to its KM value during the primary screen allowed for the detection of inhibitors with different modes of inhibition. A series of validation tests were designed and implemented in the identification of four inhibitors of human DDAH-1 that were unknown prior to the screen. Two inhibitors share a 4-halopyridine scaffold and act as quiescent affinity labels that selectively and covalently modify the active-site Cys residue. Two inhibitors are benzimidazole-like compounds that reversibly and competitively inhibit human DDAH-1 with Ligand Efficiency values ≥ 0.3 kcal / mol / heavy (non-hydrogen) atom, indicating their suitability for further development. Both inhibitor scaffolds have available sites to derivatize for further optimization. Therefore, use of this fragment-based HTS approach is demonstrated to successfully identify two novel scaffolds for development of DDAH-1 inhibitors.
Keywords: Fragment library, High-throughput screen, Inhibitor discovery, Dimethylarginine dimethylaminohydrolase, Nitric oxide
1. Introduction
The design of inhibitors to block pathological production of nitric oxide is of significant interest. For example, chronic overproduction of nitric oxide contributes to the growth of some cancerous cells by inhibiting apoptosis and promoting angiogenesis.1 Also, acute overproduction of nitric oxide complicates septic shock by causing a drastic drop in blood pressure that can lead to multiple organ failure.2 For treating sepsis, the strategy of directly inhibiting inducible nitric oxide synthase has been attempted.3, 4 Inhibitors of nitric oxide synthase block the detrimental overproduction of nitric oxide in the endothelium, but are also expected to block the beneficial production of nitric oxide by macrophages. An alternative target has been suggested for blocking nitric oxide in a more tissue selective manner.2 The enzyme dimethylarginine dimethylaminohydrolase-1 (DDAH-1; E.C. 3.5.3.18) regulates nitric oxide production indirectly by catabolizing Nω, Nω-dimethyl-l-arginine (1) (Figure 1), which is an endogenous inhibitor of all three isoforms of nitric oxide synthase.5, 6 DDAH-1 is found in the endothelium, but is only present at low levels in immune cells.7, 8 Therefore, the development of potent and selective DDAH-1 inhibitors may enable the tissue selective inhibition of nitric oxide production by an indirect mechanism.
Figure 1.
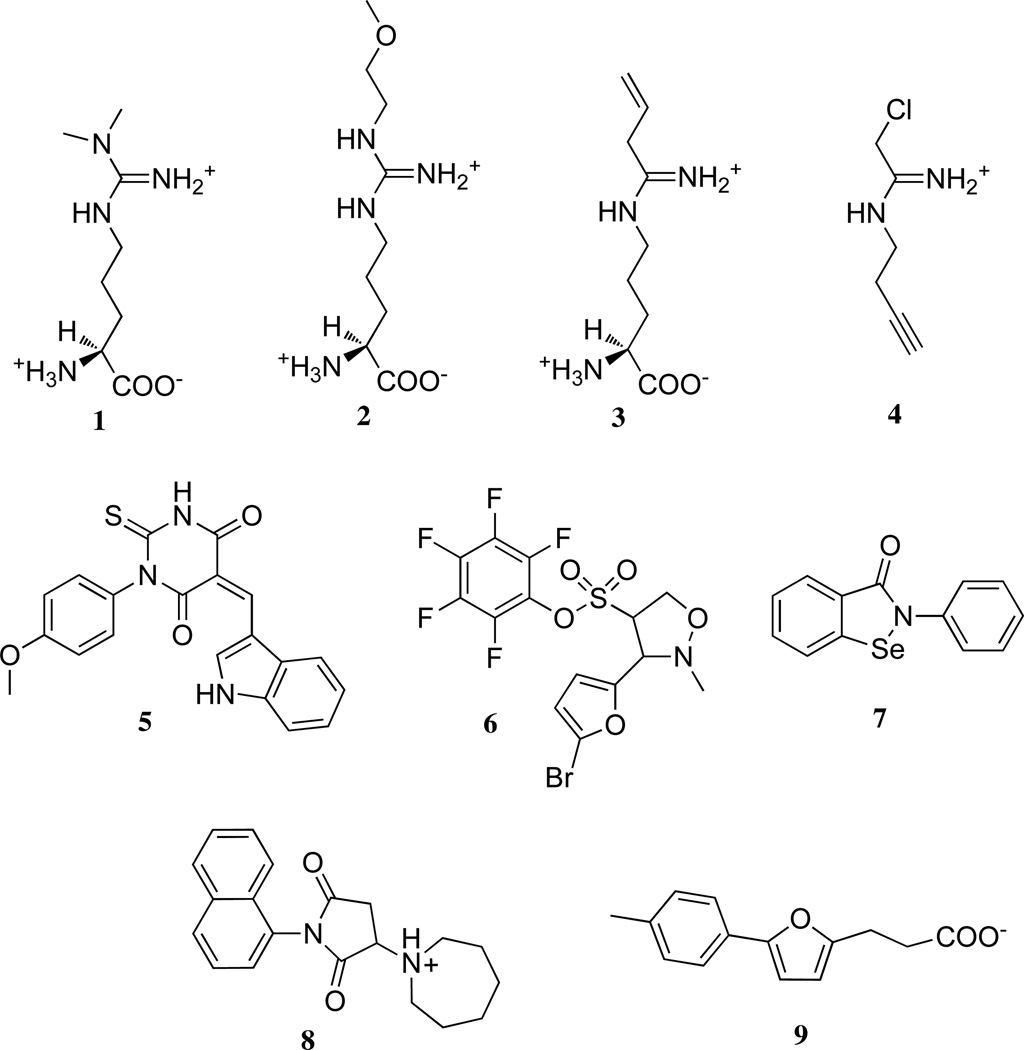
Selected examples of different structural classes of DDAH ligands. The substrate is Nω, Nω -dimethyl-l-arginine (1), substrate-like inhibitors include those with a guanidine (Nω-(2-methyoxyethyl)-l-arginine (2) or amidine (N5-(1-imino-but-3-enyl)-l-ornithine (3) and N-but-3-ynyl-2-chloroacetamidine (4)). Inhibitors with structures dissimilar to substrate include indolyl barbiturates (5), pentafuorophenyl sulfonates (6), ebselen (7), the succinimide 8, and the furan 9. See Introduction for references for each representative compound.
Most reported DDAH-1 inhibitors are structurally similar to substrate and can be grouped into categories that contain either guanidine (2)9, 10 or amidine (3, 4)11–14 substitutents that mimic the guanidinium group of Nω, Nω-dimethyl-l-arginine (Figure 1). Although these highly charged molecules might seem to be unlikely drug candidates, they have efficacy in cultured cells and in vivo, likely gaining access to their cytoplasmic DDAH-1 target through the y+ cationic amino acid transporter.15 However, this transporter imposes a second set of constraints on inhibitor design. In order to retain physiological activity, this class of inhibitors must maintain substituents that are compatible with the transporter, and must effectively compete with the substrate of nitric oxide synthase (l-arginine), the substrate of DDAH-1 (Nω, Nω-dimethyl-l-arginine, 1), and other molecules recognized by this transporter just to gain access to its target enzyme. Additionally, the unintended inhibition of human arginase is also a concern about many arginine-like inhibitors because any resulting increase in l-arginine concentration could lead to a counterproductive increase in nitric oxide production.16
Some attempts have been made to develop DDAH-1 inhibitors with structures dissimilar to substrate (Figure 1). A virtual screen for inhibitors of the DDAH from Pseudomonas aeruginosa led to the discovery of an indolyl barbiturate inhibitor (5), but this compound did not inhibit the human DDAH-1 isoform, and other hits from this screen suffered from poor solubility.11, 17 Pentafluorophenyl sulfonates (6) were reported as inhibitors of P. aeruginosa DDAH and may represent a promising scaffold, but tests with human DDAH-1 have not been reported, and it is unclear which aspects of their structures are important for affinity to the enzyme.18 Through a high-throughput screening (HTS) approach, we identified ebselen (7) as an inhibitor of human DDAH-1, but the polypharmacology of this compound complicates its use.19, 20 Recently, HTS of a 130,000 member diverse library using saturating concentrations of substrate ([S] > KM), revealed a number of human DDAH-1 inhibitors, but their structures suggest that the hits are mostly reactive electrophiles.21 Two promising inhibitors were identified (8, 9)21, but the mode of inhibition by 8 (Hill coefficient = 1.8) was not reported, and compound 9 did not inhibit human DDAH-1 in our hands (Sigma-Aldrich, (m/z) M + H+ calc’d for 231.10, found 231.10; (m/z) M + Na+ calc’d for 253.08, found 253.08; Gayle Burstein;T.W.L; W.F; University of Texas, Austin, unpublished observations). Several structurally diverse endogenous compounds are also known to inhibit human DDAH-1: S-nitroso-l-homocysteine, 4-hydroxnonenal and zinc (II).22–27 However, these endogenous compounds are not easily converted into drug-like inhibitors. Therefore, we decided to pursue a more conservative approach for discovery of DDAH-1 inhibitors. Herein, we describe HTS of a library of fragment-sized molecules for inhibitors of DDAH-1. The goal of screening these low molecular weight compounds (≤ 300 Da) is not to immediately discover high potency inhibitors, but rather to discover novel DDAH-1 inhibitor scaffolds with promise for future development. Screening is conducted with subsaturating substrate concentrations ([S] = KM) to increase the probability of detecting competitive inhibitors. Additionally, a set of validation steps were devised and implemented to eliminate false positives and reactive electrophiles. These procedures resulted in the successful identification of structurally-diverse scaffolds capable of inhibiting human DDAH-1.
2. Results and Discussion
Towards the goal of developing inhibitors of human DDAH-1 that are structurally dissimilar to substrate, we developed a robust HTS assay capable of querying large and diverse compound libraries. The HTS assay performance has been documented elsewhere.19, 20 To discover novel scaffolds suitable for inhibitor development, we chose a more conservative fragment-based approach in which smaller, less potent compounds are identified as starting points for drug development. The benefits of this approach are reviewed elsewhere, and include the ability to identify compact, easily modified core structures in which most of the atoms contribute directly to binding affinity.28, 29 The potency of these core structures can then be optimized incrementally by linking to other fragments (a “fragment-linking” approach) or by stepwise modification (a “fragment-growing” approach). The economy of functional structure found in the initial hits is typically gauged by Ligand Efficiency values that are ≥ 0.3 kcal / mol / non-hydrogen atom (L.E. = ΔGbinding / number of non-hydrogen atoms), as benchmarked by a typical drug-like compound of 500 Da with 38 non-hydrogen atoms and a Kd value of 10 nM.30 Although fragment-sized inhibitors usually have much weaker potencies (Kd = 0.1 to > 10 mM) than drug-like compounds, fragments with satisfactory L.E. values are considered to be valuable starting points for inhibitor development.
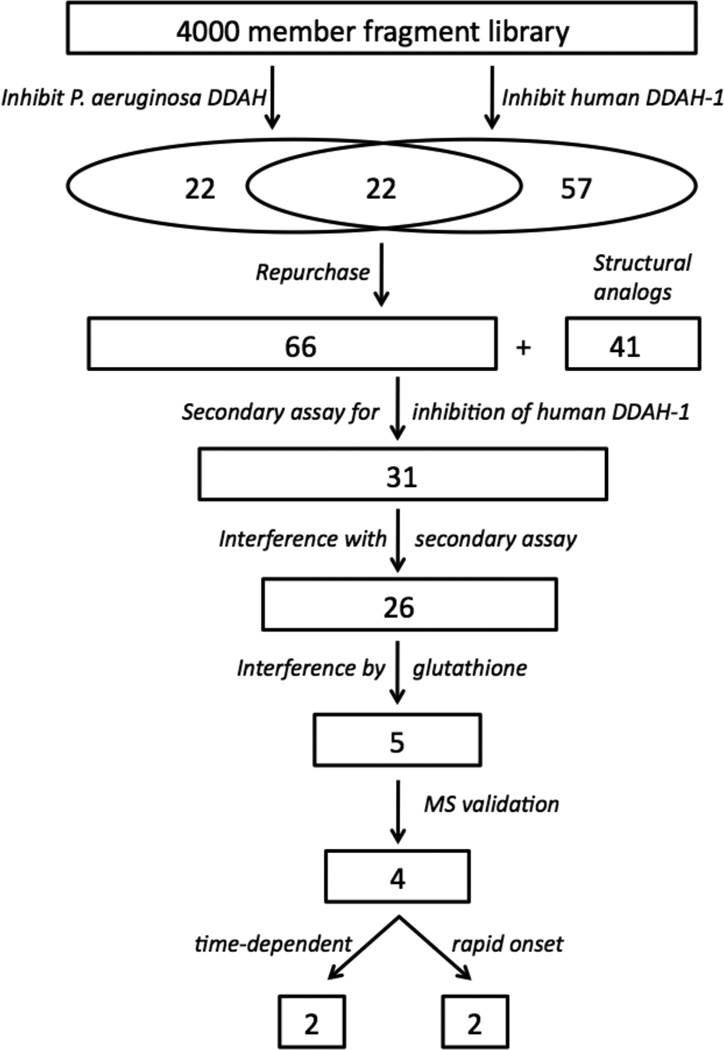
For HTS, we selected a commercially-available library of compounds that conform to the “rule of three”: Molecular weight < 300 Da; Clog P < 3; H-bond donors ≤ 3; H-bond acceptors ≤ 3; rotatable bonds ≤ 3.31 We previously screened this library for inhibitors of human DDAH-1 and reported only on the assay’s performance.19 The primary screening hits from that assay are now reported here, and are combined with additional primary screening hits from a second HTS assay of the same library completed by using the P. aeruginosa DDAH isoform. We then designed a rigorous series of validation tests that were applied to these pooled primary hits. We reasoned that including both isoforms in the primary screening step would enhance the probability of finding DDAH inhibitors because the structural and kinetic differences between isoforms and the methodological differences between their HTS assays might enhance the diversity of primary screening hits. The overall workflow for hit discovery and validation is given in Figure 2.
Figure 2.
Diagram of the workflow for inhibitor discovery and validation. The numbers indicate how many compounds progressed to each step. See Results and Discussion for details.
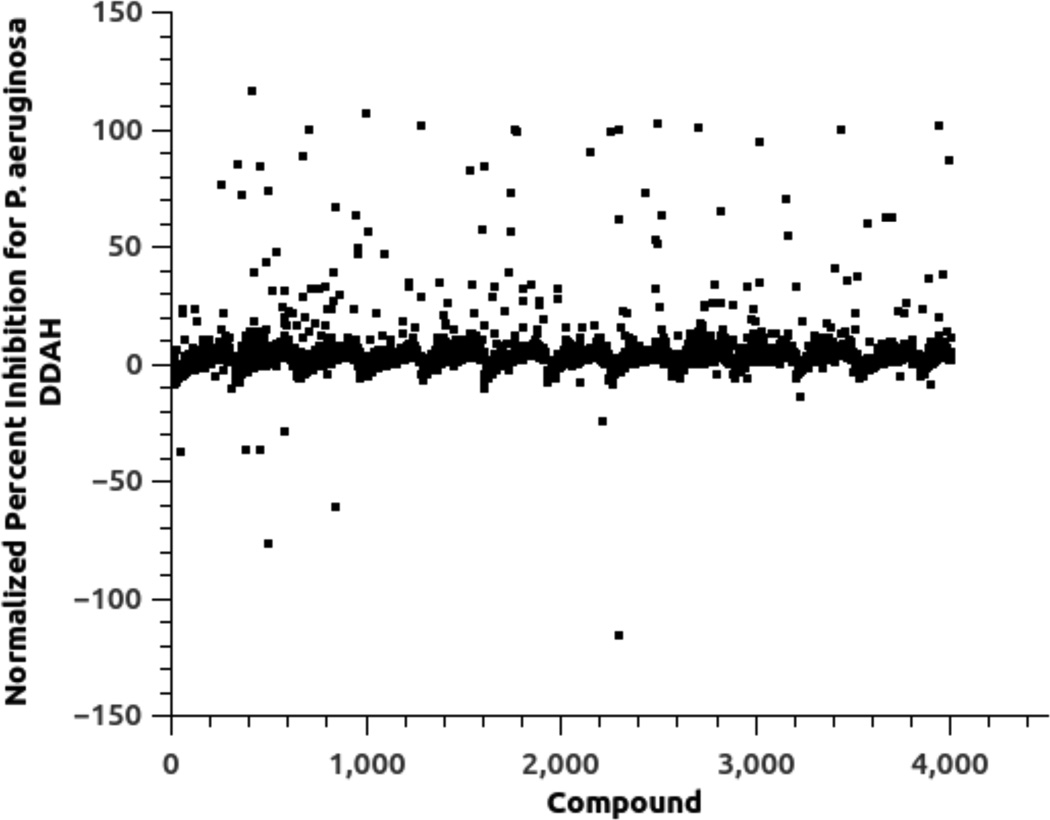
In brief, the HTS assay for each isoform relies on enzyme-catalyzed hydrolysis of an alternative substrate, S-methyl-l-thiocitrulline, to produce methanethiol as an alternative product.32 This thiol is then detected by a chromogenic or fluorogenic reagent and the resulting increase in signal is compared to control wells to observe any apparent inhibition by library compounds.19, 20 Each assay is done in the presence of a non-ionic detergent to reduce non-selective inhibition by “aggregators.”33, 34 Primary screens of the 4000-member fragment library in duplicate at 100 µM of each compound for each DDAH isoform identified 44 compounds as possible inhibitors of P. aeruginosa DDAH and 79 compounds as possible inhibitors of human DDAH-1, reflecting a 1 % and 2 % primary hit rate, respectively (Figure 3). This primary hit rate is much higher than is typically seen when screening diverse libraries of drug-like molecules, but is typical for libraries of fragment-sized molecules.28 A subset of these hits (22 compounds) was identified in both screens, resulting in a total of 101 unique molecules identified by the primary screens. These compounds were manually categorized into groups of similar structure, and representative compounds from each group were repurchased for validation tests. Only one representative was chosen from structurally similar groups containing moieties that were likely to be thiol-reactive. Other groups of compounds were supplemented by the purchase of additional compounds with related structures. For example, a number of the primary hits contained a 2-substituted benimidazole moiety. So, other 2-substituted benzimidazole derivatives were purchased to more fully explore related chemical space during the secondary screen (vide infra). Compounds that were not readily available for repurchase were abandoned. This process resulted in selection of 66 compounds from the primary hits and an additional 41 supplemental compounds, to result in a total of 107 compounds that progressed to further study.
Figure 3.
Primary HTS results for inhibition of the P. aeruginosa DDAH isoforms by a 4000-member library of fragment-sized compounds. Primary HTS identified 44 compounds as potential inhibitors. A comparable plot for primary screening of the human DDAH-1 isoform with the same library is found in reference (19). See Experimental Procedures for details.
A series of validation tests to eliminate false positives were designed and performed. All of the enzyme assays subsequent to the primary screen were completed using human DDAH-1 (unless otherwise indicated) because this particular isoform is the desired target. First, false positives due to interference with the primary HTS assay were considered. These hits could be the result of fluorescence quenching, scavenging of the methanethiol reaction product, direct reaction with the thiol-reactive reporter molecules, or oxidation effects. To eliminate some of these possibilities, the 107 compounds were screened using a secondary assay that uses a different detection method than used in the primary assay. Instead of an artificial substrate, the native substrate Nω, Nω-dimethyl-l-arginine (1) is used (at 300 µM, which is 2.5-fold higher than the KM value determined using the same assay format), and formation of the reaction product l-citrulline is detected via a discontinuous derivatization assay.35 Using this orthogonal secondary assay, 31 compounds (including 22 of the 66 compounds directly identified by the primary screen) showed ≥ 20 % inhibition at a concentration of 400 µM and were chosen to progress through further validation tests. This cutoff value is relaxed in the secondary assay to enable enhanced detection of inhibition by compounds in this group. These results suggest that a significant fraction (68 %) of the original primary hits were false positives due to assay interference. This percentage of reactive and otherwise undesirable compounds is similar to other CPM-based HTS assays.36 The high ligand concentrations used in the primary fragment screen may also contribute to the frequency of false positives. Additional experiments suggested that a number of the false positives identified here may be non-selective thiol-reactive compounds or may quench the fluorescence of the reporter molecule after its reaction with methanethiol (data not shown).
Next, we considered the possibility that some of the 31 hits identified using the secondary orthogonal assay were false positives due to interference with the secondary assay, or due to non-selective modification of the catalytic Cys residue in DDAH-1. So, two additional validation tests were used to eliminate these possibilities. First, assay interference was tested. The entire 107 panel of primary hits and structurally-related analogs, in the absence of enzyme, were incubated both with and without l-citrulline and derivatized as before. An observed decrease (≥ 20 %) in l-citrulline (product) detection indicates that a library compound interferes with the secondary assay. These observed decreases were detected for 9 compounds, 5 of which are present in the panel of 31 hits that showed apparent DDAH-1 inhibition by the orthogonal assay. These compounds were removed from further consideration. Second, library compounds that display non-selective thiol-reactivity or that cause thiol oxidation were identified. The remaining 26 hits were preincubated with or without an excess of reduced glutathione and then assayed for DDAH-1 inhibition using Nω, Nω-dimethyl-l-arginine (1) and the secondary assay. Compounds were eliminated from consideration if the preincubation with glutathione reduced the apparent enzyme inhibition by ≥ 50 %. Based on this assay, 21 of the 26 non-interfering compounds were eliminated as likely to be oxidizing or thiol-reactive species.
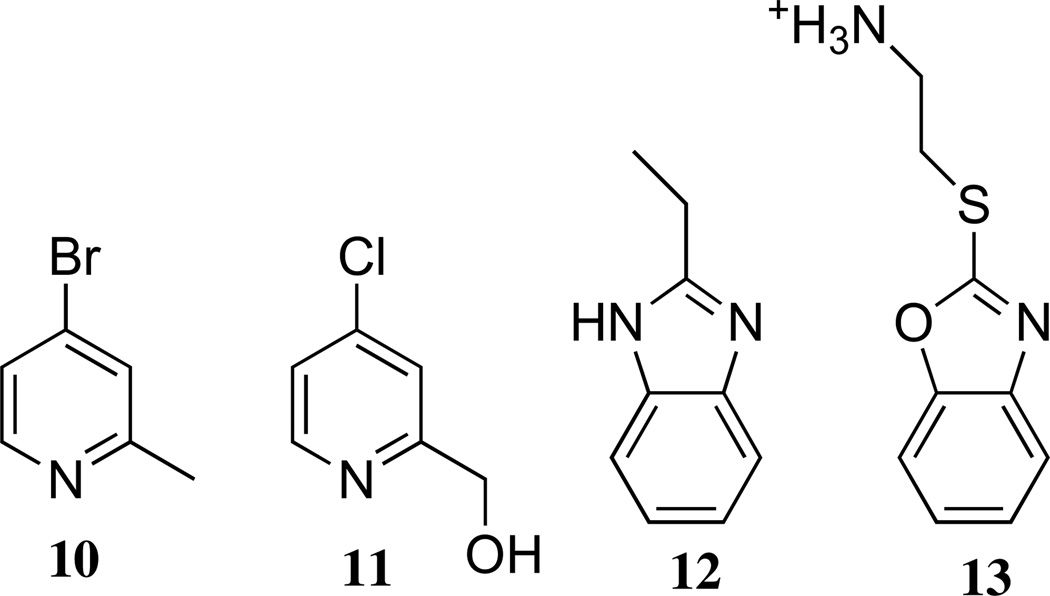
The stock solutions of the 5 remaining hits in DMSO were subjected to further validation by mass spectrometry to verify the identity assigned to each chemical. Four compounds, 2-methyl-4-bromopyridine (10)37, 2-hydroxymethyl-4-chloropyridine (11)38, 2-ethyl-benzimidazole (12; (m/z) M + H+ calc’d for 147.09, found 147.00) and 2-(1,3-benzoxazol-2-ylthio)ethanamine (13; (m/z) M + H+ calc’d for 195.09, found 195.08) showed their expected masses (Figure 4). Results for the fifth compound, 3-amino-5-fluoro-1,3-dihydro-2H-indol-2-one (MWcalcd = 166.15 Da; MWobsd = 179.08 Da), did not match the expected mass, so this compound was discarded from consideration. The remaining four compounds, 10–13, passed all of the validation controls (summarized in Table 1) and were considered to be genuine inhibitors of DDAH-1. These four structures can be sorted into two categories, 4-halopyridines (10, 11) and benzimidazole-like compounds (12, 13), of which each group was subsequently shown to have functional differences in their modes of inhibition.
Figure 4.
Four validated inhibitors of human DDAH-1.
Table 1.
Inhibition Results for Compounds 10 – 13
| Compound | Inhibition in Primary Screen (Pa; Hu)a (%) |
Inhibition in Secondary Screen (%) |
Interference With Secondary Screenb (%) |
Inhibition in Secondary Screen + GSHc (%) |
kinact/KI (M−1s−1) |
IC50 (mM) |
Hill Coefficient |
Ki (mM) |
|---|---|---|---|---|---|---|---|---|
| 10 | 89; 55 | 74 ± 10 | − 5 ± 4 | 95 ± 4 | 4.8 ± 0.3d | N.A.e | N.A. | N.A. |
| 11 | 50; 17 | 56 ± 7 | 4 ± 4 | 46 ± 5 | 0.65 ± 0.07 f | N.A. | N.A. | N.A. |
| 12 | 1; 4 | 36 ± 5 | − 11 ± 6 | 25 ± 4 | N.A. | 1.4 ± 0.2 | 1.5 ± 0.2 | 0.8 ± 0.2 |
| 13 | − 60; Dg | 21 ± 4 | 0 ± 4 | 23 ± 5 | N.A. | 2.7 ± 0.5 | 1.0 ± 0.3 | 1.7 ± 0.4 |
Percent inhibition by each compound (100 µM) as measured in the primary HTS assay for P. aeruginosa DDAH (Pa) and human DDAH-1 (Hu) using [S] = KM conditions
Extent to which the library compound interferes with detection of a l-citrulline standard in the absence of enzyme
Extent to which human DDAH-1 is inhibited by the library compound in the presence of excess glutathione.
Value from ref (37)
Not Applicable
Value from ref (38)
Negative value indicates apparent increase in product. D indicates the sample was “discarded” due to high background fluorescence values.
Compounds 10 and 11, which comprise the 4-halopyridine group, were found to be time-dependent inactivators of P. aeruginosa DDAH with kinact / KI values of 4.8 ± 0.3 and 0.65 ± 0.07 M−1 s−1, respectively.37, 38 Although a full characterization will be presented elsewhere, the same compounds are also time-dependent inactivators of human DDAH-1. Using an inactivation time course at one inhibitor concentration, the kinact / KI values were estimated to be 0.05 and 0.04 M−1 s−1 for 10 and 11, respectively (Gayle Burstein, W.F. ; University of Texas, Austin; data not shown). Since these compounds were not deemed to be reactive electrophiles or tight-binding inhibitors, their time-dependent inhibition mechanisms were investigated in detail and described elsewhere.37, 38 Briefly, both of these 4-halopyridines act as quiescent affinity-labels that are relatively unreactive toward glutathione in solution. However, when bound to P. aeruginosa DDAH, the protonated pyridinium form of 10 and 11 is stabilized by Asp66, which greatly enhances the reactivity of each compound. A subsequent attack by Cys249 results in displacement of approximately one equivalent of halide and results in an irreversible covalent inactivation. To our knowledge, 4-halopyridines had not previously been shown to be capable of modifying proteins. Therefore, they represent a significant discovery by our HTS: a novel “warhead” useful for inhibitor design in which pairs of residues, rather than a single reactive nucleophile, are targeted when arrayed in the proper conformation around a binding site large enough to fit the pyridine ring.
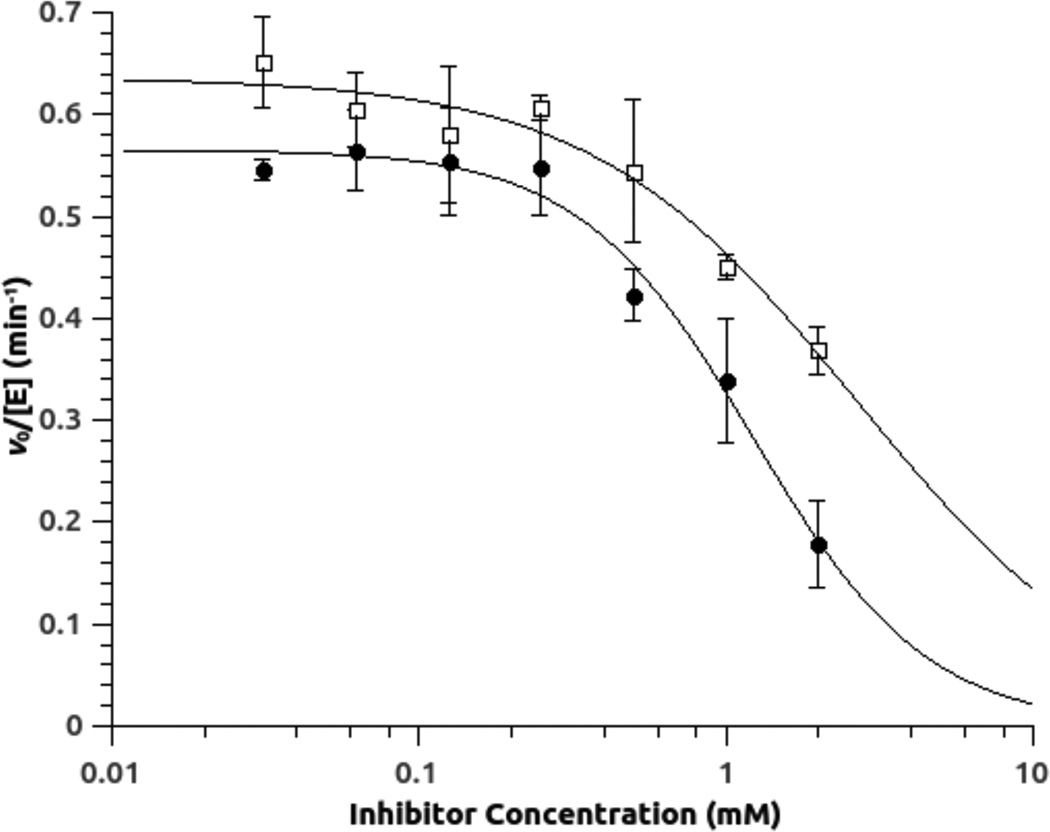
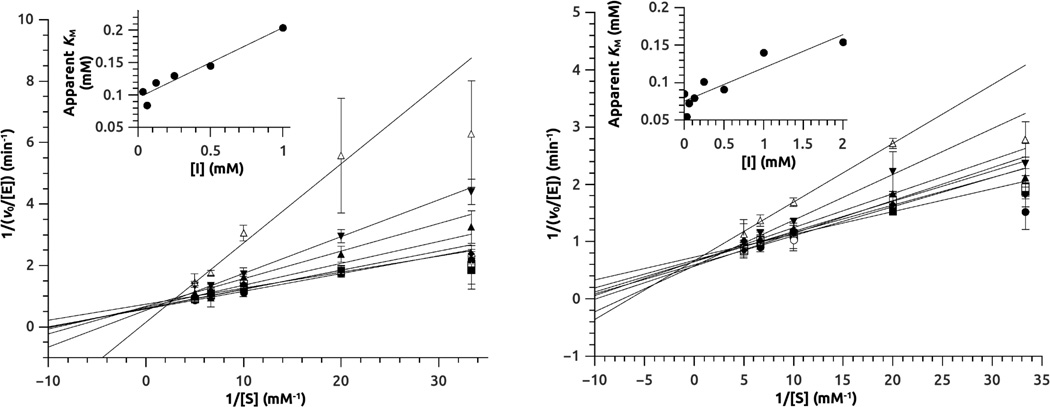
In contrast to the 4-halopyridines, the benzimidazole-like group of compounds showed rapid onset of inhibition, with no lag period observable during the experimental timeframe. Mixtures of 12 and 13 with human DDAH-1 were diluted into excess substrate and full activity was rapidly regained, indicating that inhibition was also rapidly reversible (Supplementary Data Figure S1). The potency of reversible inhibitors 12 and 13 for human DDAH-1 was initially assessed using concentration-response plots (Figure 5, Table 1). The potency for each is weak, but well within the expected range for fragment-sized inhibitors. Notably, the Hill coefficients are approximately 1, consistent with a 1:1 binding stoichiometry to the enzyme. In contrast, the P. aeruginosa isoform of DDAH did not show any inhibition by 12 at concentrations up to 1 mM, and 13 interfered with the assay (vide infra). To determine the mode of inhibition with human DDAH-1, enzyme activity was measured in the presence of varying amounts of substrate and inhibitor. Addition of either 12 or 13 had a large effect on the KM value, but not on the kcat value, consistent with assigning a competitive mode of inhibition to both of these compounds (Figure 6). Non-linear fits of the data to a competitive inhibition model determined Ki values for inhibitor 12 (0.8 ± 0.2 mM) and 13 (1.7 ± 0.4 mM), both within the expected range of potency for fragment-sized inhibitors. More pertinent are the Ligand Efficiences of inhibitors 12 and 13, which are 0.4 and 0.3 kcal / mol / heavy (non-hydrogen) atom, respectively. These values are at, or above, the typical 0.3 cutoff used in fragment optimization and indicate that these scaffolds are suitable starting points for inhibitor development.30
Figure 5.
Concentration-response plots for inhibition of human DDAH-1 by 12 (●) and 13 (□). Experiments are done in the presence of competing substrate Nω, Nω-dimethyl-l-arginine (50 µM; 0.53 × the KM value determined using the same assay format) and results are fitted to inhibition parameters for compound 12 (IC50 = 1.4 ± 0.2 mM; Hill = 1.5 ± 0.2) and 13 (IC50 = 2.7 ± 0.5 mM; Hill = 1.0 ± 0.3).
Figure 6.
Mode-of-inhibition studies for inhibitors 12 (Left panel) and 13 (Right panel). Human DDAH-1 activity is measured at different substrate concentrations in the presence of increasing concentrations of inhibitor. Non-linear curve fitting is used to quantify the effects of inhibitor on kcat and KM values (not shown). Double reciprocal plots with linear fits are shown here for visual display with the insets showing variation in apparent KM values with inhibitor concentration. The uninhibited KM for the substrate ADMA is 94 ± 7 µM when determined using the same assay format. E stands for enzyme. S stands for substrate. I stands for inhibitor, which is included at the following concentrations: 0 (●), 0.3 (■), 0.6 (○), 0.12 (□), 0.25 (♦), 0.5 (▲), 1 (▼), 2 (△) mM. The Ki values for competitive inhibitors 12 and 13 are determined to be 0.8 ± 0.2 mM and 1.7 ± 0.4 mM, respectively.
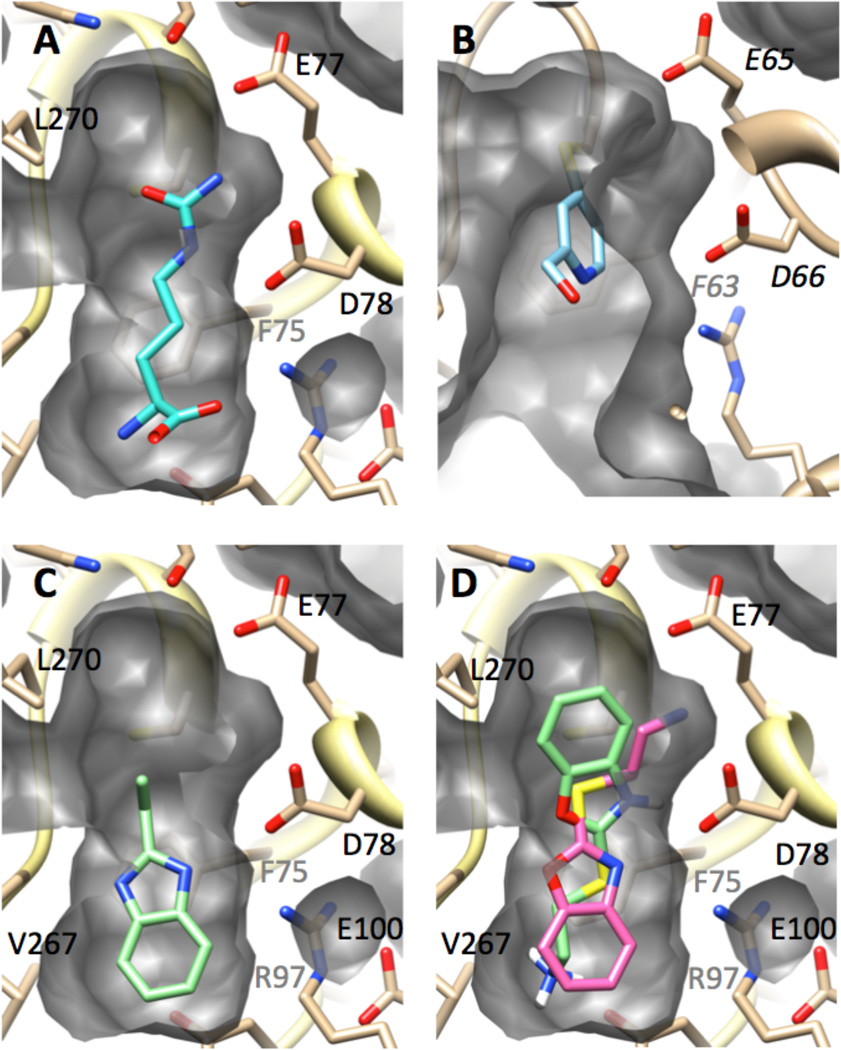
Another consideration in selecting fragment-sized inhibitors for further development is the availability of sites for derivatization to enhance binding affinity. Experimental and computational methods were used to explore possible binding conformations of these fragments (Figure 7). As a reference structure, the product l-citrulline is observed by X-ray crystallography to make a number of hydrophobic and H-bonding interactions at the active site of human DDAH-1.10 Specifically, the urea group in l-citrulline occupies the same site that binds the substrate’s guanidinium, and the sidechain and amino acid moieties of both the substrate and product make the same interactions with the enzyme. In this product-bound structure, a small vacant pocket remains near residue Leu270. This pocket likely accommodates the Nω-methyl groups of the substrate. The structures of inhibited DDAH can be compared to the product-bound form (Figure 7). An X-ray structure of P. aeruginosa DDAH, after inactivation by 11, reflects the slightly larger active-site pocket of this isoform.38 The pyridine ring of the inactivator, which is covalently attached to the active-site cysteine, occupies only the space near that of the guanidinium binding site, leaving both the Nω-methyl- and the amino acid-binding pockets vacant. The pendant alcohol of 11 (and likely the methyl group of 10) point toward the vacant amino acid binding pocket and is suggested as a promising site to target for derivatization in a “fragment-growing” approach to increase the potency of this inhibitor scaffold.
Figure 7.
Observed and modeled structures of inhibitors bound at the DDAH active site. Each panel is shown from a similar vantage point and shows a cut-away view of the active-site cavity’s surface (grey). The enzyme backbone is shown in ribbons (tan), with selected residues depicted in stick form and labeled in black or grey (if their view is obscured). Ligands are shown in stick form. Nitrogens are shown in blue, oxygens in red, sulfur in yellow. A) X-ray structure of l-citrulline (cyan) bound to human DDAH-1.10 B) X-ray structure of P. aeruginosa DDAH after inactivation by 11 (light blue).38 C) Modeled structure of human DDAH-1 bound to 12 (light green). D) Modeled structure of human DDAH-1 bound to 13 in two poses (light green, pink). See Experimental Procedures and Results and Discussion for details.
The modeled conformations of the reversible competitive inhibitors 12 and 13 occupy a different part of the active site than the 4-halopyridines (Figure 7). Both inhibitors are predicted to occupy the pocket that usually accommodates the side chain of the substrate by stacking over the side chain of Phe75 and by making additional hydrophobic interactions with the side chain of Leu171 (not shown), thereby interacting with two residues that help form a hydrophobic portion of the active site. Specifically, ten poses were generated for inhibitor 12. These grouped into only one cluster with the lowest computed binding energy (−5.50 kcal / mol) being close to the average for all ten poses (−5.49 kcal / mol), and a RMSD < 0.06 Å for all poses. The modeled conformation of inhibitor 12 predicts H-bonding interactions with the side chain of Asp78 and the backbone carbonyl of Val267 (not shown). The ethyl group in 12 projects towards the guanidinium and Nω-methyl-binding pockets, and immediately suggests a site to derivatize in a “fragment-growing” approach, or a point of possible attachment to a halopyridine fragment in a “fragment-linking” approach, for future attempts to enhance the potency of each inhibitor.29 The benzyl ring appears to be tightly packed in the hydrophobic part of the active site. This result is consistent with the observation that a number of benzimidazole-like structures that contain substitutions in this ring did not show inhibition during primary HTS. However, since inhibition by 12 was not first detected during the primary screen (see below), caution should be used when drawing conclusions based on these negative results.
Ten poses were also generated for compound 13, and were grouped into two low energy clusters with similar values for the lowest calculated binding energy in each group (−7.74 and −7.68 kcal / mol). The first group, comprised of six poses, had RMSD values < 1.23 Å, and the second group of four poses had RMSD values < 1.38 Å, with only one pose in both groups having an RMSD > 1.0 Å. The first group of poses for 13 (Figure 7D, shown in light green) has an orientation “flipped” from that of 12, with the heterocycle of 13 occupying the guanidinium-binding pocket and the pendant amine occupying the same site as the substrate’s α-amine. In contrast, the second group of poses (Figure 7D, shown in pink) recapitulates many of the same interactions seen with compound 12, but, notably, the longer alkyl group of 13 extends a pendant amine into the guanidinium-binding site, making H-bonding interactions with another oxygen in the carboxylate sidechain of Asp78. Despite the potential to make this additional interaction, this compound has a weaker binding affinity, perhaps reflecting the inability of the benzoxazole oxygen to H-bond with the backbone carbonyl of Val267 or reflecting an unoptimized linker length between the bicyclic ring system and the pendant amine. Changes in active-site dynamics and conformation upon inhibitor binding might also play a role in binding affinities and are not reflected in these models. Nevertheless, the conformations of inhibitor 13 illustrates the possibility of making additional interactions at the guanidinium-binding site and reinforces the suggestion of selecting the ethyl substituent of inhibitor 12 as a site for future derivatization.
A retrospective analysis of where these four inhibitors were discovered during the HTS process suggests how future screens might be improved (Table 1, Figure 2). As expected, inhibitors 10 and 11 were initially detected in the primary screen of P. aeruginosa DDAH, but only compound 10 was a “hit” in the primary human DDAH-1 screen. This result is rooted in the experimental methodology of each screen. The duration of the enzyme and inhibitor preincubation step in the P. aeruginosa DDAH screen (≤ 1 h) was longer than in the human DDAH-1 screen (10 min).19, 20 Since inhibitors 10 and 11 are time-dependent inactivators, an increase in the duration of this step results in a lower apparent IC50 value and, therefore, a more easily detectable “hit.” Future HTS protocols that lengthen the duration of this step would have an enhanced ability to detect other time-dependent inactivators.
The origins of hits 12 and 13 were more surprising. Neither compound was found as a hit in the primary assay, but these compounds were instead purchased as part of the set of 41 structural analogs of primary hits that were eventually eliminated during the secondary assay or verification tests. The weak inhibition by 12 did not rise above the cutoff value during the primary screen, but was detected once the inhibition threshold was relaxed during secondary screening. Compound 13 showed apparent negative inhibition (activation) in the primary screen of P. aeruginosa DDAH and was discarded from the primary screen of human DDAH-1 due to high background fluorescence. The effects observed for compound 13 were likely due to degradation of the library stock solution. Degradation would liberate the thiol cysteamine, which could react directly with the detection reagents. Subsequent repurchase of compound 13 allowed measurement of its inhibition of human DDAH-1 before degradation. The suspected instability of 13 highlights another benefit of starting instead with compound 12 for future inhibitor development. These results suggest that a more lenient inhibition cutoff threshold of the primary screen in future HTS of fragment libraries may allow for increased detection of less potent fragment-sized inhibitors that still maintain suitable Ligand Efficiency values. Supplementing the primary hits with structural analogs also proved to be a useful step in hit discovery.
3. Summary and Conclusion
Development of a robust and sensitive high-throughput screening assay for two isoforms of DDAH allowed the primary screening of 4000 fragment-sized molecules. Screening in the presence of subsaturating substrate concentrations ([S] = KM) allowed the discovery of four previously unknown inhibitors of human DDAH-1 that use different modes of inhibition. A set of validation tests also allowed elimination of a number of false positives and reactive electrophiles that would lack suitable selectivity for optimization as biologically useful inhibitors. These filtering steps are valuable, in part, because reactive electrophiles have dominated the primary hits in some previous screens.20, 21 Two of the hits resulting from our screen are 4-halopyridines and are classified as quiescent affinity labels. Covalent inactivators have an advantage over reversible competitive inhibitors when the desired endpoint is accumulation of the enzyme’s substrate, as is the case with human DDAH-1. Additionally, these newly identified 4-halopyridines will likely have a much wider application as a novel covalent “warhead” for use with other protein targets. The remaining two hits are benzimidazole-like and are determined to be reversible competitive inhibitors. In general, benzimidazoles are considered to be “privileged scaffolds” in drug discovery (ligands capable of binding diverse receptors).39 The Ligand Efficiency values of these particular hits are 0.4 (12) and 0.3 (13) kcal / mol / heavy (non-hydrogen) atom, which indicates they are suitable for further development. Both inhibitor scaffold types have obvious sites to derivatize when using either “fragment-linking” or “fragment-growing” approaches to improve potency. Therefore, these findings contribute substantially toward the goal of developing inhibitors of human DDAH-1 with structures dissimilar to substrate and provide two new scaffolds for inhibitor development.
4. Experimental Procedures
4.1 Materials
All reagents were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise specified. Black 384-well polypropylene plates (Catalog #264576) were from Nalge Nunc (Rochester, NY). Microcentrifuge tubes (1.5 mL) were purchased from Fisher Scientific (Pittsburgh, PA). Trifluoroacetic acid, HEPES, K2HPO4, and KH2PO4 were purchased from Thermo Fisher Scientific (Waltham, MA, USA). All buffer solutions were filtered by 0.22 µm Express PLUS filters (Millipore, Billerica, MA, USA). A 4000 member Fragment Library (ChemBridge Corporation, San Diego, CA, USA) was obtained from the inventory of the Automation and High Throughput Screening Facility at the Texas Institute for Drug and Diagnostics Development (TI-3D, The University of Texas at Austin).
Stock solutions of the thiol-reactive fluorogenic reagent 7-diethylamino-3-(4’-maleimidylphenyl)-4-methylcoumarin (CPM) were prepared by dissolving CPM in dimethyl sulfoxide (≥ 99 %, Fisher Scientific, Pittsburgh, PA) to a final concentration of 2 mM and stored in 1 mL aliquots in opaque amber microcentrifuge tubes (Fisher Scientific) at −20 °C. Stock solutions of the thiol-reactive chromogenic reagent 5,5’-dithiobis-(2-nitrobenzoic acid) (DTNB) were prepared by dissolving DTNB to a final concentration of 5 mM in HEPES buffer (10 mM), KCl (100 mM), with a final pH of 6.5.
4.2 Expression and purification of DDAH isoforms
The DDAH isoform from Pseudomonas aeruginosa was heterologously over expressed in Echerichia coli BL21 (DE3) cells and purified as previously described.40 To remove inhibitory metal ions, the resulting protein was dialyzed overnight at 4 °C against 4 L of 1,10-phenanthroline (2 mM), HEPES buffer (10 mM) and 100 mM KCl (100 mM) at pH 7.3. This was followed by two consecutive dialysis steps of 4 – 16 h each at 4 °C against 4 L of Chelex-100-treated (BioRad Laboratories, Hercules, CA) HEPES buffer (10 mM), KCl (100 mM) at pH 7.3. The homogeneity of purified protein was determined to be > 97 % by coomassie-blue stained SDS-PAGE, and the expected mass of the protein (30,503 KDa for a His6-tagged P. aeuroginosa DDAH lacking the N-terminal methionine) was confirmed by electrospray ionization mass spectrometry (ESI-MS) to be 30,498 ± 10 KDa (Analytical Core Facility, College of Pharmacy, University of Texas at Austin, USA. To determine protein concentration, purified P. aeruginosa DDAH was added to Denaturing Buffer (guanidinium hydrochloride (6 M final concentration), sodium phosphate (20 mM) at pH 6.6). The absorbance of the sample at 280 nm was determined using a Cary 50 UV-vis spectrophotometer (Varian, Walnut Creek, CA, USA). The P. aeruginosa DDAH extinction coefficient (17,210 M−1cm−1) was calculated (http://workbench.sdsc.edu) based on amino acid sequence and its final concentration was calculated using Beer's Law. Protein was then aliquotted, flash frozen in liquid nitrogen, stored at −80 °C, and thawed on ice immediately before use.
The human DDAH-1 isoform was heterologously over expressed in E. coli BL21 (DE3) cells and purified as previously described.12 To remove inhibitory metal ions, the protein was dialyzed overnight at 4 °C against 1 L of 1,10-phenanthroline (2 mM), KH2PO4 (10 mM), and KH2PO4 (100 mM) at pH 7.3. The chelator was then removed by three additional dialyses for at least 4 h each at 4 °C against 1 L of KH2PO4 (10 mM), KCl (100 mM), and glycerol (10% (v/v)) at pH 7.3, which had been previously treated by passage through a Chelex-100 column. The homogeneity of purified protein was determined to be > 95 % by Coomassie-blue stained SDS-PAGE, and the expected mass of the protein (33,304 KDa) was confirmed by electrospray ionization mass spectrometry (ESI-MS) to be 33,305 ± 10 KDa, To determine protein concentration, purified human DDAH-1 was assayed and stored as described above for P. aeruginosa DDAH except that a different published extinction coefficient (7680 M−1cm−1) was used.12
4.3 Primary high-throughput screen for inhibitors of DDAH
Primary screening of the 4000-member library of fragment-sized compounds for inhibitors of P. aeruginosa DDAH was completed using a previously published methodology.20 Here, the final assay mixtures contained library compounds at 100 µM in K2HPO4 (172 mM), KCl (172 mM), EDTA (2 mM), Tween-20 (0.02 %), DMSO (1 % v/v), DTNB (0.5 mM), S-methyl-l-thiocitrulline (SMTC) substrate (20 µM), and P. aeruginosa DDAH (20 nM) at pH 7.3. Primary hits were identified as compounds with inhibition of ≥ 3σ from the sample mean (≥ 48 % inhibition).
The primary screening of the same library for inhibitors of the human DDAH-1 isoform was previously completed, and the hits are now disclosed here.19 For the purposes of easy comparison, the final assay components are given: library compound (100 µM), K2HPO4 (172 mM), KCl (172 mM), EDTA (2mM), Tween-20 (0.02% v/v), DMSO (1 %), CPM (3.5 µM), S-methyl-l-thiocitrulline substrate (0.4 µM), and purified human DDAH-1 (20 nM) at pH 7.3. Primary hits were identified as compounds with a normalized percent inhibition ≥ 50 %.
4.4 Secondary assay for rapid-onset, time-dependent, and reversibility studies of human DDAH-1 inhibitors
To evaluate inhibition that occurs upon mixing (rapid-onset) by selected library compounds, the substrate Nω, Nω-dimethyl-l-arginine (300 µM) was mixed with library compound (400 µM) in Reaction Buffer [Na2HPO4 (172 mM), KCl (172 mM), Tween-20 (0.02 % v/v), EDTA (2 mM) at pH 7.3]. Reactions were initiated by adding enzyme (1 µM final concentration) to a final volume of 60 µL, incubated for 1 – 2 h, and quenched with HCl (1 M final concentration). The reactions were then assayed for uriedo groups using a published colorimetric ureido derivatization assay.35 Due to the constraint of keeping product formation rates linear and detectable over the reaction time, the substrate concentration was kept in 2.5-fold excess of the KM value (120 µM when determined using the same assay format). To compensate for potential substrate competition, the concentrations of the library compounds were also raised 4-fold from those used in the primary screen. All compounds showing ≥ 20 % inhibition were considered to be “hits.”
To evaluate time-dependent inhibition by selected library compounds, each of a selected set of library compounds (400 µM) was added to Reaction Buffer containing human DDAH-1 (1 µM) and incubated at room temperature for 10 – 20 min. The reaction was then initiated by adding Nω, Nω-dimethyl-l-arginine (300 µM), incubated at room temperature for 1 – 2 h, quenched with HCl (1 M final concentration), and assayed as described above. Results were compared with parallel experiments that omit the preincubation step.
To evaluate whether inhibition could be rapidly reversed upon dilution into excess substrate, human DDAH-1 (30 µM) was incubated at room temperature in Na2HPO4 (100 mM), NaCl (100 mM), EDTA (2 mM), Tween-20 (0.02 % v/v) at pH 7.25. Some samples also included reduced glutathione (2 mM). Each of a selected set of library compounds (0 – 2 mM) was added to start the incubation. After various incubation times (0 – 120 min), 1.5 µL of the mixture was diluted 40-fold into excess substrate Nω, Nω-dimethyl-l-arginine (1 mM), Na2HPO4 (100 mM), NaCl (100 mM), EDTA (2 mM), Tween-20 (0.02 %) at pH 7.25 to a final volume of 60 µL. The resulting mixtures were incubated for an additional 120 min, quenched with HCl (300 mM final concentration), and assayed for the product l-citrulline as described above.
4.5 Concentration-response assays for IC50 and Ki determinations
In a clear, polystyrene, 96-well microtiter plate (Nalge Nunc), Nω, Nω-dimethyl-l-arginine (30 – 200 µM final concentration) was mixed with 2.4 µL of DMSO stock solutions for each of a selected set of library compounds (0 – 2 mM final concentration). Reactions were initiated by adding human DDAH-1 (0.3 µM) in Reaction Buffer to a final volume of 60 µL and incubated at room temperature for 80 min. Reactions were quenched with HCl (1 M final concentration) and assayed for the product l-citrulline using the secondary assay described above. The exact concentrations and fitting methods used to determine IC50 and Ki values are described in more detail in Figures 5 and 6.
4.6 Tests for assay interference
Library compounds might be falsely identified as DDAH inhibitors if they are electrophilic compounds that react directly with free thiols like methanethiol (the product of the enzymatic reaction in the primary screen) or non-selectively with the active-site cysteine of DDAH. To filter out such compounds, the secondary assay described above was repeated in the presence of reduced glutathione to observe any decrease in observed inhibition. Briefly, each of a selected set of library compounds (400 µM) was added to Reaction Buffer containing reduced glutathione (2 mM) and human DDAH-1 (1 µM). The mixtures were incubated at room temperature for 10 – 20 min. Subsequently, the enzyme-catalyzed reaction was initiated by adding Nω, Nω-dimethyl-l-arginine (300 µM), incubated at room temperature for 1 – 2 h, quenched with HCl (1 M final concentration), and assayed as described above.
Compounds that interfere with the orthogonal secondary assay might also result in false identification as a DDAH inhibitor. Such compounds were filtered out by mixing each of a set of selected library compounds (400 µM), with and without l-citrulline (100 µM) in Reaction Buffer in the absence of enzyme. Samples were derivatized using the colorimetric uriedo derivatization assay.35 A value for the “Interference with Secondary Screen” (Table 1) was defined as 100 × (1 – (Abs540 nm in the presence of the library compound ÷ Abs540 nm in the absence of the library compound).
4.7 Computational docking of inhibitors to human DDAH-1
Docking was completed using the program Autodock 4.2, starting with the 2.3 Å resolution structure of human DDAH-1 in complex with l-citrulline (PDB accession code 2JAI) after l-citrulline was manually deleted from the PDB file.10, 41 Three-dimentional coordinates for a selected set of library compounds were generated from their corresponding SMILES strings using the Openbabel software suite (http://openbabel.org/). The docking site was specified as a box extending 8 Å from the original placement of the product l-citrulline. The energy grid was prepared with spacing of 0.1 Å, and a Lamarckian genetic algorithm was used with a population size of 300, mutation rate of 0.02, and crossover rate of 0.8. Simulations were run for a maximum of 25,000,000 energy evaluations. Each ligand was docked 10 times, and only the lowest energy conformations of each cluster are shown. Docking was performed on a dual-processor computer with two dual-core AMD Opteron 2218 chips, running Ubuntu Linux 11.10.
4.8 Calculation of Ligand Efficiency
Ligand Efficiency values for compounds 12 and 13 were calculated using the method described in reference (30), using the Ki values determined experimentally for each compound (Table 1, Figure 6).
4.9 General data analysis
Linear fits for absorbance versus time plots were calculated using the SciPy python module (http://www.scipy.org). OpenOffice Calc (http://www.openoffice.org/) was used for additional calculations in spreadsheets. Nonlinear regression was performed using the open source suite QtiPlot (http://soft.proindependent.com/).
Supplementary Material
Acknowledgements
For her assistance with the high-throughput screening, we thank Dr. Eun Jeong Cho and the Automation and High Throughput Screening Facility at the Texas Institute for Drug and Diagnostic Development (TI-3D) at the University of Texas, Austin. This work was supported in part by grants from the National Institutes of Health (GM69754), the Robert A. Welch Foundation (F-1572) and a seed grant from the CTT/TI-3D Chemistry and Molecularly-Targeted Therapeutic Development grant program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Data
Supplementary Data is included in the online version of this article, at doi:.
References
- 1.Fukumura D, Kashiwagi S, Jain RK. Nat Rev Cancer. 2006;6:521. doi: 10.1038/nrc1910. [DOI] [PubMed] [Google Scholar]
- 2.Leiper J, Nandi M. Nat Rev Drug Discov. 2011;10:277. doi: 10.1038/nrd3358. [DOI] [PubMed] [Google Scholar]
- 3.Bakker J, Grover R, McLuckie A, Holzapfel L, Andersson J, Lodato R, Watson D, Grossman S, Donaldson J, Takala J. Crit Care Med. 2004;32:1. doi: 10.1097/01.CCM.0000105118.66983.19. [DOI] [PubMed] [Google Scholar]
- 4.Watson D, Grover R, Anzueto A, Lorente J, Smithies M, Bellomo R, Guntupalli K, Grossman S, Donaldson J, Le Gall JR. Crit Care Med. 2004;32:13. doi: 10.1097/01.CCM.0000104209.07273.FC. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa T, Kimoto M, Sasaoka K. J Biol Chem. 1989;264:10205. [PubMed] [Google Scholar]
- 6.Knipp M. Chembiochem. 2006;7:879. doi: 10.1002/cbic.200500527. [DOI] [PubMed] [Google Scholar]
- 7.Leiper JM, Santa Maria J, Chubb A, MacAllister RJ, Charles IG, Whitley GS, Vallance P. Biochem J. 1999;343(Pt 1):209. [PMC free article] [PubMed] [Google Scholar]
- 8.Tran CT, Fox MF, Vallance P, Leiper JM. Genomics. 2000;68:101. doi: 10.1006/geno.2000.6262. [DOI] [PubMed] [Google Scholar]
- 9.Rossiter S, Smith CL, Malaki M, Nandi M, Gill H, Leiper JM, Vallance P, Selwood DL. J Med Chem. 2005;48:4670. doi: 10.1021/jm050187a. [DOI] [PubMed] [Google Scholar]
- 10.Leiper J, Nandi M, Torondel B, Murray-Rust J, Malaki M, O'Hara B, Rossiter S, Anthony S, Madhani M, Selwood D, Smith C, Wojciak-Stothard B, Rudiger A, Stidwill R, McDonald NQ, Vallance P. Nat Med. 2007;13:198. doi: 10.1038/nm1543. [DOI] [PubMed] [Google Scholar]
- 11.Kotthaus J, Schade D, Muschick N, Beitz E, Clement B. Bioorg Med Chem. 2008;16:10205. doi: 10.1016/j.bmc.2008.10.058. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Monzingo AF, Hu S, Schaller TH, Robertus JD, Fast W. Biochemistry. 2009;48:8624. doi: 10.1021/bi9007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Hu S, Fast W. J Am Chem Soc. 2009;131:15096. doi: 10.1021/ja906432e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lluis M, Wang Y, Monzingo AF, Fast W, Robertus JD. ChemMedChem. 2011;6:81. doi: 10.1002/cmdc.201000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Closs EI, Basha FZ, Habermeier A, Forstermann U. Nitric Oxide. 1997;1:65. doi: 10.1006/niox.1996.0106. [DOI] [PubMed] [Google Scholar]
- 16.Kotthaus J, Schade D, Clement B. J Enzyme Inhib Med Chem. 2011;27:24. doi: 10.3109/14756366.2011.573480. [DOI] [PubMed] [Google Scholar]
- 17.Hartzoulakis B, Rossiter S, Gill H, O'Hara B, Steinke E, Gane PJ, Hurtado-Guerrero R, Leiper JM, Vallance P, Rust JM, Selwood DL. Bioorg Med Chem Lett. 2007;17:3953. doi: 10.1016/j.bmcl.2007.04.095. [DOI] [PubMed] [Google Scholar]
- 18.Vallance P, Bush HD, Mok BJ, Hurtado-Guerrero R, Gill H, Rossiter S, Wilden JD, Caddick S. Chem Commun (Camb) 2005:5563. doi: 10.1039/b510709a. [DOI] [PubMed] [Google Scholar]
- 19.Linsky T, Fast W. J Biomol Screen. 2011;16:1089. doi: 10.1177/1087057111417712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linsky T, Wang Y, Fast W. ACS Med Chem Lett. 2011;2:592. doi: 10.1021/ml2000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghebremariam YT, Erlanson DA, Yamada K, Cooke JP. J Biomol Screen. 2012;17:651. doi: 10.1177/1087057112441521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knipp M, Braun O, Vasak M. J Am Chem Soc. 2005;127:2372. doi: 10.1021/ja0430200. [DOI] [PubMed] [Google Scholar]
- 23.Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure. 2006;14:901. doi: 10.1016/j.str.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Braun O, Knipp M, Chesnov S, Vasak M. Protein Sci. 2007;16:1522. doi: 10.1110/ps.062718507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong L, Fast W. J. Biol. Chem. 2007;282:34684. doi: 10.1074/jbc.M707231200. [DOI] [PubMed] [Google Scholar]
- 26.Forbes SP, Druhan LJ, Guzman JE, Parinandi N, Zhang L, Green-Church KB, Cardounel AJ. Biochemistry. 2008;47:1819. doi: 10.1021/bi701659n. [DOI] [PubMed] [Google Scholar]
- 27.Knipp M, Charnock JM, Garner CD, Vasak M. J Biol Chem. 2001;276:40449. doi: 10.1074/jbc.M104056200. [DOI] [PubMed] [Google Scholar]
- 28.Carr RA, Congreve M, Murray CW, Rees DC. Drug Discov Today. 2005;10:987. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 29.Erlanson DA. Top Curr Chem. 2012;317:1. doi: 10.1007/128_2011_180. [DOI] [PubMed] [Google Scholar]
- 30.Hopkins AL, Groom CR, Alex A. Drug Discov Today. 2004;9:430. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 31.Congreve M, Carr R, Murray C, Jhoti H. Drug Discov Today. 2003;8:876. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 32.Stone EM, Fast W. Anal Biochem. 2005;343:335. doi: 10.1016/j.ab.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Ryan AJ, Gray NM, Lowe PN, Chung CW. J Med Chem. 2003;46:3448. doi: 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- 34.Coan KE, Shoichet BK. J Am Chem Soc. 2008;130:9606. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knipp M, Vasak M. Anal Biochem. 2000;286:257. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 36.Chung CC, Ohwaki K, Schneeweis JE, Stec E, Varnerin JP, Goudreau PN, Chang A, Cassaday J, Yang L, Yamakawa T, Kornienko O, Hodder P, Inglese J, Ferrer M, Strulovici B, Kusunoki J, Tota MR, Takagi T. Assay Drug Dev Technol. 2008;6:361. doi: 10.1089/adt.2007.105. [DOI] [PubMed] [Google Scholar]
- 37.Johnson CM, Linsky TW, Yoon DW, Person MD, Fast W. J Am Chem Soc. 2011;133:1553. doi: 10.1021/ja109207m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson CM, Monzingo AF, Ke Z, Yoon DW, Linsky TW, Guo H, Robertus JD, Fast W. J Am Chem Soc. 2011;133:10951. doi: 10.1021/ja2033684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welsch ME, Snyder SA, Stockwell BR. Curr Opin Chem Biol. 2010;14:347. doi: 10.1016/j.cbpa.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone EM, Person MD, Costello NJ, Fast W. Biochemistry. 2005;44:7069. doi: 10.1021/bi047407r. [DOI] [PubMed] [Google Scholar]
- 41.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Comput Chem. 2009;30:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.