Abstract
The genetic basis for virulence in influenza virus is largely unknown. To explore the mutational basis for increased virulence in the lung, the H3N2 prototype clinical isolate, A/HK/1/68, was adapted to the mouse. Genomic sequencing provided the first demonstration, to our knowledge, that a group of 11 mutations can convert an avirulent virus to a virulent variant that can kill at a minimal dose. Thirteen of the 14 amino acid substitutions (93%) detected among clonal isolates were likely instrumental in adaptation because of their positive selection, location in functional regions, and/or independent occurrence in other virulent influenza viruses. Mutations in virulent variants repeatedly involved nuclear localization signals and sites of protein and RNA interaction, implicating them as novel modulators of virulence. Mouse-adapted variants with the same hemagglutinin mutations possessed different pH optima of fusion, indicating that fusion activity of hemagglutinin can be modulated by other viral genes. Experimental adaptation resulted in the selection of three mutations that were in common with the virulent human H5N1 isolate A/HK/156/97 and that may be instrumental in its extreme virulence. Analysis of viral adaptation by serial passage appears to provide the identification of biologically relevant mutations.
Virulence is the measure of the ability of a pathogen to damage its host. Human influenza A virus infection typically causes tracheobronchitis with a low incidence of fatal pneumonia. In 1918, a virulent influenza A virus variant arose, causing a devastating pandemic killing 50 million people (1). Although this virus was not isolated, it must have possessed mutations that increased its virulence. The genomic sequence of 1918 viruses are being determined from archival tissues and, whereas the sequence of the hemagglutinin (HA) and neuraminidase (NA) genes are now available (2), we do not yet have the understanding of the molecular basis for virulence needed to interpret this information. The difficulty of discerning mutations that control virulence among the background of unselected mutations has been exemplified by sequence analysis of the highly virulent A/Hong Kong/156/97-like (H5N1) virus that recently infected humans directly from birds in Hong Kong (HK) (3), where even the most closely related avian isolate, A/Ck/HK/220/97, differs from this virus at 28 amino acids (4). Subsequent human infections by related avian H9N2 viruses indicate a continued threat to the human population (1). There is thus a need to understand the genetic basis for virulence in influenza virus variants, with the hope that specific mutations will be indicators and thus predictors of virulence.
No clinical isolates of human influenza virus are known to differ in virulence (5), therefore necessitating the analysis of infection in animals. Influenza virus is partially host restricted, where virus from one host does not normally transmit or cause disease in other hosts. The A/HK/156/97 strain, however, is virulent for both chickens and mice (6), indicating that a shared genetic basis for disease production can exist among species. Adaptation of human influenza virus to mice by serial passage results in the selection of highly virulent variants that have acquired mutations in multiple genes (7–9). Analyses of the genetic basis for virulence by using reassortants that possess mixtures of genes from virulent and avirulent strains have identified various groupings of genes, which in aggregate implicate all eight genome segments (8). These data have led to the untested assumption that virulence cannot be genetically predicted, because there are too many degrees of freedom in the control of virulence.
A goal of the study of influenza pathogenesis is to define the roles of each viral gene in disease production. To begin to address this aim, a complete sequence comparison was done between the A/FM/1/47 (H1N1) parental strain (FM) and its mouse-adapted variant, FM-MA, which had increased 104.6-fold in virulence on the basis of LD50. This process identified single amino acid substitutions in 5 of its 10 genes (9). Reintroduction of each of these mutations into the parental FM strain confirmed their roles not only in increasing virulence but also in replicative fitness for the mouse (9). These findings are compelling, because they show a clear relationship between replicative fitness and the ability to damage the host. It was also surprising that the MA variant did not possess unselected mutations that typically accumulate in clinical isolates. This, however, would be predicted from studies of viral adaptation in cell culture, where genetic variation is a function of virus population size (10). Serial passage of large populations of virus under novel conditions permits competition among all possible mutants with the selection of optimal genotypes. In contrast, the transfer of small populations, typical of normal disease transmission, leads to the fixation of unselected and deleterious mutations because of stochastic effects, a process termed Muller's ratchet (10).
The primary feature of organisms with adaptive mutations is that they increase in prevalence in the population because of improved replicative fitness. A strong indicator of adaptive change at the molecular level is convergent evolution, characterized by the repeated and independent occurrence of common mutations in adapted variants. The occurrence of identical mutations is termed parallel evolution, and mutation at the same sites but with different amino acids is termed directional evolution, both of which are characteristic of instances of convergent evolution (11). This is the standard criterion for identifying mutations responsible for drug and inhibitor resistance, providing evidence for convergent evolution in many organisms, including influenza virus (12).
Because the parental A/FM/1/47 strain was subjected to mouse passage immediately on isolation in 1947 (13), it carries evidence of preexisting mouse adaptive mutations detectable on analysis by genetic reassortment (14, 15). In the present study, the prototype H3N2 clinical isolate, A/HK/1/68, without a prior history of mouse passage, was used to generate virulent variants by serial mouse-lung passage. The objective of this analysis was to identify and characterize the complexity and nature of the mutations that control virulence. Genomic sequencing of a highly virulent MA variant identified 11 mutations that were acquired on mouse adaptation. Sequencing of clonal variants showed that most of these mutations were positively selected in the population and affected specific regions of individual genes that identify functional themes for regulating virulence. Experimental evolution may have recapitulated natural evolution, at least in part, because the Hong Kong H5N1 lineage of viruses possessed several mutations in common with the MA strains, suggesting their instrumental operation in virulence.
Materials and Methods
Viruses.
The prototype human H3N2 isolate, A/HK/1/68, was obtained from the Laboratory Centre for Disease Control, Health Canada, Ottawa. Viruses were clonally purified by two plaque isolations in Madin–Darby canine kidney cell (MDCK) monolayer followed by stock preparation in the allantoic cavity of 9-day-old chicken embryos. Virus was passaged in mouse lungs by cycles of intranasal infection with 50 μl of 1/10 diluted lung extracts under halothane anesthesia for groups of 3 mice for 3 days, as described previously (14). MA viruses were clonally isolated and titrated by plaque assays on MDCK cell monolayer, as described (14)
Nucleotide Sequencing.
Viral RNA was purified by phenol extraction from stock virus, as previously described (15). Each genome segment was amplified by reverse transcription–PCR (RT-PCR) and purified in agarose gels or Sepahcryl S400 spin columns (Amersham Pharmacia) before direct dideoxy-terminated cycle sequencing by using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin–Elmer) and an Applied Biosystems automated sequencer, Model ABI 373. Genome segment-specific primers for RT-PCR were complementary to the first 16 nucleotides of each segment in combination with a primer complementary to the 12 nucleotides at the 3′ end of vRNA, and all possessed a 5′ terminal adapter sequence of CCGC. Sequencing primers (sequences available on request) were complementary to related H3N2 viruses (GenBank accession nos. J02135–40 and X59240, respectively).
SDS/PAGE.
MDCK cells were infected, [35S]Met-labeled, and analyzed, as described previously (14). Labeled virus was purified by adsorption to guinea pig erythrocytes. Trypsin was used at 1 μg/ml (15 min at 37°C) to cleave HA. Tunicamycin was used at 10 μg/ml from 5.5 h postinfection (pi) onward, with pulse labeling at 6.5 h pi for 1.5 h. Immunoprecipitations were performed as described previously, by using A/HK/1/68 specific rabbit immune serum (16).
Virulence Assay.
Virulence was measured as the lethal dose in CD1 strain Swiss–Webster mice. The median lethal dose (LD50) in plaque-forming units (pfu) was measured by intranasal infection of five groups of five mice each with serial 10-fold dilutions of virus, as described previously (14). Mortality from influenza virus infection occurs primarily before day 8 and was thus monitored for 10 days.
Statistical Analysis.
LD50 values were determined by using the Karber method (14). The significance of differences in virulence and growth values was determined by using the Z statistic for a standard normal distribution. The probability of multiple independent mutations in the same clonal isolate was predicted by the Poisson distribution by using the observed mutation frequency.
Growth in MDCK Cells, Chicken Allantoic Cavity, and Mouse Lung.
MDCK cells were infected at a multiplicity of infection of 5 pfu/cell and incubated at 34°C in the presence of 1 μg/ml trypsin (15). Supernatants were collected over a 48-h period, and plaque assays were performed in duplicate for each sample. Groups of CD-1 mice were infected intranasally with 5 × 103 pfu of each virus. Over a 10-day period, lungs were removed from groups of three mice, pooled, and sonicated for quantification by plaque assay on MDCK monolayers, as described above. The yield in chicken egg allantoic cavity was determined by infecting 9- to 10-day-old chicken embryos with 104 pfu and incubation at 48 h at 34°C. Pools of three eggs each (two to six pools/virus) were titrated by plaque assay on MDCK cells.
Hemolysis and Hemagglutination.
Hemolysis assays were performed by using human type O blood and citrate buffers in microtiter plates at 21°C, as described (17). Hemagglutination and hemagglutination inhibition assays were performed as described previously (18). α inhibitor was prepared by sonicating mouse lung (10% suspension) and centrifugation to prepare extracts, and mouse serum was used as a source of β inhibitor (17).
Results
MA variants of the human isolate, A/HK/1/68 (HK), were produced by serial lung-to-lung passage, beginning with intranasal inoculation of 106 pfu of virus per mouse. Subsequent passages involved the inoculation of diluted lung extracts, constituting dosages of 104–5 pfu/mouse (data not shown). Virulence was assayed by LD50 after 12 and 20 passages for both the total population of virus obtained directly from titrated lungs extracts without further culturing, as well as six clonal isolates from each of these passage levels obtained by plaque purification of 107-fold diluted lung extracts in MDCK cells (Table 1). The parental virus was totally avirulent for mice, LD50 >107.7 pfu; however, after 12 passages, the LD50 for the population of virus in lung extracts had decreased >104-fold and after 20 passages >105-fold, indicating that these virus populations had acquired mutations that profoundly affect virulence. Several HKMA clonal isolates from each passage level were similar in virulence to their respective populations and were thus representative of these populations; however, some were less virulent, indicating genetic heterogeneity within these populations. HKMA20C was the most virulent virus clone, LD50 = 102.5 pfu, similar to the passage 20 population (Table 1).
Table 1.
Virulence of mouse-adapted isolates of A/HK/1/68
| A/HK/1/68 viruses passaged in mouse lung | LD50, pfu | Change in LD50 (LOG10 decrease) |
|---|---|---|
| Unpassaged A/HK/1/68 parent clone HK | >107.7 | NA |
| Passage 12 population | 103.7 | >4.0 |
| Passage 12 clone HKMA12 | 104.0 | >3.7 |
| Passage 12 clone HKMA12A | 103.9 | >3.8 |
| Passage 12 clone HKMA12B | 103.6 | >4.1 |
| Passage 12 clone HKMA12C | 104.6 | >3.1 |
| Passage 12 clone HKMA12D | >104.7 | ND |
| Passage 12 clone HKMA12E | >104.9 | ND |
| Passage 20 population | 102.7* | >5.0 |
| Passage 20 clone HKMA20 | 104.2 | >3.5 |
| Passage 20 clone HKMA20A | 104.0 | >3.7 |
| Passage 20 clone HKMA20B | 103.0* | >4.7 |
| Passage 20 clone HKMA20C | 102.6±0.3 | >5.1 |
| Passage 20 clone HKMA20D | 103.5 | >4.2 |
| Passage 20 clone HKMA20E | 103.6 | >4.1 |
, viruses not significantly different in virulence relative to HKMA20C, (Z value, P ≤ 0.05); NA, not applicable; ND, not done.
Sequence Analysis.
To identify the mutations responsible for the increased ability to cause fatal lung infection, we initially sequenced the genome of the most virulent clonal isolate, HKMA20C, as well as the HK parent for comparison. A total of 11 amino acid substitutions involving 8 of the 10 viral proteins (Table 2) and 4 silent substitutions were detected, which produced 2 coding changes in each of PA, HA, and nucleoprotein (NP), as well as single amino acid substitutions in PB2, NA, M1, M2, and NS1, and no mutations in polymerase subunit PB1 and nuclear export protein NS2. The noncoding changes (data not shown) included an insertion of an extra A in the polyA coding region of the NA gene (Table 2).
Table 2.
The nature and location of predicted amino acid changes for HK and HKMA viruses (nucleotides in parentheses)
| PB2
|
PA
|
HA1
|
HA2
|
NP
|
NA
|
M1
|
M2
|
NS1
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 701 (2128) | 133 (422) | 556 (1691) | 218 (729) | 156 (1531) | 34 (145) | 480 (1483) | 468 (1422) | (1457)† | 167 (524) | 44 (155) | 103 (333) | |
| HK | Asp (G) | Glu (A) | Gln (A) | Gly (G) | Thr (C) | Asp (G) | Asp (G) | Pro (C) | − | Thr (A) | Asp (G) | Phe (T) |
| HKMA20C | Asn (A) | Gly (G) | Arg (G) | Trp (T) | Asn (A) | Asn (A) | Asn (A) | His (A) | (+Ade) | Ala (G) | Asn (A) | Leu (C) |
| HKMA12 | Asn (A)* | − | − | Trp (T) | Asn (A) | Asn (A) | − | His (A) | − | − | − | − |
| HKMA20 | Asn (A)* | − | Arg (G)* | Trp (T) | Asn (A) | Asn (A) | − | His (A) | − | − | − | − |
, from partial sequence; −, indicates wild-type sequence;
, indicates insertion of noncoding nucleotide.
Although it is clear that these mutations as a group must account for the difference in biology of the HKMA20C variant, it is not clear that they are all instrumental in adaptation to increased virulence. Because adaptive changes increase replicative fitness, viruses that possess these changes will be present at a greater frequency in the virus population than their rate of formation predicted from the mutation frequency. To detect mutations that were positively selected on mouse adaptation, we tested for the null hypothesis that the mutations in individual virus clones were randomly and thus independently generated. Given the observed frequency of coding mutations in HKMA20C of 2.2 × 10−3/amino acid, the probability of having two viruses with the same mutation because of independent mutational events is predicted by chance to be P = 5 × 10−6/n − 1, where n is the number of isolates tested. The occurrence of the same mutations in more than one isolate is thus significant evidence of positive selection rather than the occurrence of independent mutational events. The HKMA12 and HKMA20 variants were subjected to sequence analysis to determine whether they shared mutations with HKMA20C. Genome segments four to eight were completely sequenced, as well as the mutant loci detected in the PB2 and PA genes. The HKMA12 clone possessed 5 mutations in common with both of the passage 20 clones, and HKMA20 possessed 6 mutations in common with HKMA20C, indicating positive selection of 6 mutations (Table 2). The low probability of pairs of viruses with five or six mutations in common is also a clear indication of positive selection, as this is extremely unlikely to occur by chance (Poisson distribution P ≤ 1 × 10−13 and P ≤ 1.4 × 10−17).
Because two more mutations were detected in the HKMA20 clone (M1-Asp-232→Asn and NS1-Val-23→Ala), segments 7 and 8 (M1, M2, NS1, and NS2 genes) were sequenced for HKMA-12, −12A, −12B, and the 5 remaining HKMA20 series of clones (Table 1). This analysis identified a further mutation, NS2-Lys-88→Arg, and demonstrated positive selection for 4 of 6 mutations in the M and NS genes (Table 3).
Table 3.
Mutations in the M1, M2, NS1, and NS2 genes for clones of mouse-adapted A/HK/1/68
| Virus | Amino acid variation (nucleotide position)
|
|||||
|---|---|---|---|---|---|---|
| M1
|
M2
|
NS1
|
NS2
|
|||
| 167 (524) | 232 (617) | 44 (155) | 23 (94) | 103 (333) | 88 (289) | |
| HK | Thr (A) | Asp (G) | Asp (G) | Val (T) | Phe (T) | Lys (A) |
| HKMA12 | − | − | − | − | − | − |
| HKMA12A | − | − | − | − | − | − |
| HKMA12B | − | − | − | − | − | − |
| HKMA20 | − | Asn (A) | − | Ala (C) | − | − |
| HKMA20A | − | Asn (A) | − | Ala (C) | − | − |
| HKMA20B | − | − | Asn (A) | − | Leu (C) | Arg (G) |
| HKMA20C | Ala (G) | − | Asn (A) | − | Leu (C) | − |
| HKMA20D | − | − | − | − | Leu (C) | − |
| HKMA20E | − | − | Asn (A) | − | − | − |
−, indicates wild-type sequence.
Protein Changes.
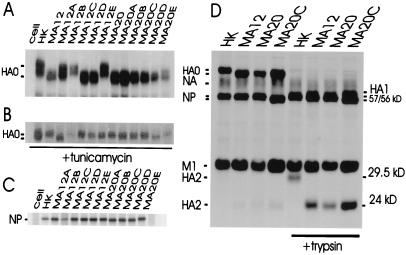
The HA2-Thr-156→Asn substitution is expected to result in the loss of a glycosylation site that would be detectable by increased mobility on SDS/PAGE. Electrophoresis of infected MDCK cell proteins showed that the uncleaved form of HA protein (HA0) possessed higher electrophoretic mobility than HK for 3 of 6 passage 12 clones and 6 of 6 passage 20 clones (Fig. 1A). The increased electrophoretic mobility was caused by decreased glycosylation, because the unglycosylated forms of their HA0 proteins resulting from tunicamycin treatment were indistinguishable in electrophoretic mobility from the parental virus (Fig. 1B), and the HA of HKMA12A was distinct in its properties. The mutations affecting glycosylation were confirmed to map to the HA2 subunit by trypsin cleavage of purified virions, because this subunit was shifted to higher relative electrophoretic mobility, corresponding to a 5.5-kDa decrease in apparent size relative to HK HA2 (Fig. 1D).
Figure 1.
SDS/PAGE of proteins of parental and MA clones of A/HK/1/68. (A) 35[S]met-labeled infected MDCK cell proteins were immunoprecipitated to show the position of the HA0 precursor. (B) The unglycosylated form of HA0 protein synthesized in the presence of tunicamycin. (C) The position of the NP proteins is shifted for HKMA20B, -C, and -D. (D) The shift in mobility of the HA2 subunit generated by trypsin in variants.
On SDS/PAGE analysis, it was observed that the NP proteins of HKMA20-B, -C, and -D possessed increased mobility that corresponded to a 1-kDa decrease in apparent size (Fig. 1C). This correlated with the presence of the mutation at amino acid 480 of HKMA20C relative to HKMA20 and HKMA12 and indicated positive selection of this mutation.
Changes in Biology of HA.
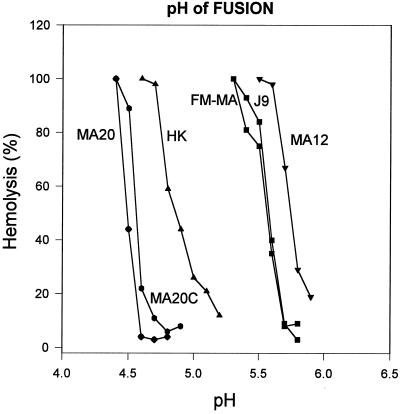
The Gly-218→Trp mutation site in HA1 has also been shown to mutate in antibody escape mutants that become altered in inhibitor sensitivity as well as pH of fusion (19). We assessed the pH optimum of fusion of HK, HKMA-12, −20, and −20C (Fig. 2) and, as expected, the three MA variants possessed different pH optima of fusion relative to the HK parent, but surprisingly they differed from each other as well. The HK parent possessed a median pH of fusion (±1 SD) of 4.9 ± .04 relative to 5.7 ± .02, 4.5 ± .02, and 4.6 ± .02 for HKMA-12, −20, and −20C, respectively. This was not expected, because each of the MA viruses had the same HA gene sequence. Consistent with the conformational change triggered by low pH, the pH sensitivity of hemagglutination activity was found to reflect the relationship shown for pH of fusion (data not shown).
Figure 2.
The pH dependence of hemolysis by A/HK/1/68 and the HKMA12, HKMA20, HKMA20C virulent MA variants. The relative extent of hemolysis was measured in duplicate for each of two experiments at 0.1 pH increments. The hemolysis profiles of FM-MA and the J9 reassortant that possesses the same HA gene (17) are shown as controls.
The susceptibility of MA variants to nonspecific inhibitors was assessed by hemagglutination inhibition assay with mouse serum or lung extract as a source of β and α inhibitor, respectively. The three MA strains had all increased in resistance to β inhibitor, seen as a 4-fold reduction in inhibition of hemagglutination (reduction in titer from 1,280 to 320). Only the HKMA20 clone had become resistant to α inhibitor (reduction in titer from 8 to <2).
Growth in Different Hosts.
The difference in virulence of the MA variants was not caused by an increased ability to infect mice, because the HK parent was already highly infectious (median infectious dose of 13 pfu, as assessed by serological conversion). The relative ability of HK and the HKMA-12, −20, and −20C clones to replicate was assessed in mouse lung, MDCK cells, and chicken allantoic cavity. All of the MA clones grew faster and to higher titer than HK in mouse lung (Fig. 3A). Comparison of the yield 1 day postinfection as well as the maximum yields of infectious virus indicated that the HKMA20C virus had the greatest increase in replicative fitness relative to HK in the mouse, which was consistent with its relative virulence (Table 4). The increases in fitness 1 day postinfection were dramatic, demonstrating the replicative basis for selection of MA variants. All of the MA clones tested also had an increased ability to replicate in MDCK cells (Fig. 3B); however, both of the passage 20 MA clones yielded less infectious virus in chicken allantoic cavity than the HK parent or the less passaged HKMA12 clone (P ≤ 0.05) (Fig. 3C). Thus mouse adaptation selected variants with increased replicative fitness in the mouse, but some of the mutations conferring these properties were host specific.
Figure 3.
Yield of HK and HKMA variants in different hosts. (A) Mouse lung; each point represents the average of three mice; values were not obtained for HKMA20C after 4 days because of lethality. (B) Yield in MDCK cells. (C) The yield of infectious virus in chicken allantoic cavity; values are the average plus standard deviation of two to six pools of three eggs.
Table 4.
Relative ability of HK and HKMA variants to replicate in mouse lung
| Virus | Yield in mouse lung 1 day
postinfection
|
Maximum yield in mouse lung
|
||
|---|---|---|---|---|
| Titer, pfu/ml | Relative fitness | Titer, pfu/ml | Relative fitness | |
| HK | 1.1e5 | 1 | 4.8e6 | 1 |
| HKMA12 | 2.0e7 | 180 | 3.6e7 | 7.5 |
| HKMA20 | 5.6e7 | 510 | 5.6e7 | 12 |
| HKMA20C | 1.3e8 | 1,180 | 1.3e8 | 27 |
Evidence of Parallel Evolution on Mouse Adaptation.
To assess whether the MA variants of HK had selected any mutations in common with FM-MA, their genome sequences were compared. The PA-556, NP-34, and M1–232 mutations occurred at sites that differed between HK and FM-MA virus. The probability of each of these sites being mutated purely by chance was extremely unlikely (P ≤ 0.0002), given the low number of “target” amino acid substitutions in the FM-MA genes relative to HK (30 of 716, 24 of 498, and 4 of 252, respectively). These mutations existed in the prototype A/FM/1/47 and may have been selected on mouse adaptation of this virus in 1947. These sites are positioned on the genetic map of influenza A virus by blue downward arrows, relative to the unique mutations in FM-MA (black downward arrows) and the mutations observed among HKMA variants (black upward arrows) (Fig. 4).
Figure 4.
Location of mutations selected on mouse adaptation on the functional genetic map of Influenza A virus. The location of mutations seen in FM-MA that have been confirmed to control virulence (9) are shown with black downward arrows, and those preexisting in FM virus are indicated in blue. Mutations observed in HKMA clones are shown with upward arrows. The functional regions and binding sites are described elsewhere (12). NLS, nuclear localization signal; POL, polymerase; NTP, nucleotide triphosphate; PKR, dsRNA-dependent protein kinase; effector indicates a multiple protein-binding region; NES, nuclear export signal; regions labeled with influenza virus protein names indicate sites of interaction.
Discussion
Adaptation is believed to be the driving force in evolution, where organisms are selected in nature because of increased fitness conferred by beneficial mutations. This paper extends the analysis of the mutational basis for virulence by using mouse adaptation, because this system provides the identification of mutations that generate evolutionarily relevant phenotypic variation. The pattern of mutations associated with HK-MA variants was consistent with multiple events in the virus population followed by assembly of mutations into genomes by reassortment and competition among viruses with different mutations. Thus the passage 12 virus had already acquired 5 mutations that were predominant in the population at passage 20. Between passages 12 and 20, subsequent mutations or combinations of mutations in individual genome segments resulted in subpopulations as shown for segment 7 (M1 and M2 genes) and 8 (NS1 and NS2 genes). Such cycles of competition among mutant viruses drive rapid evolution in influenza virus to result in progressive selection of variants. The reason for the replacement of the parental virus population by the adapted variants is a function of increased replication rate (180- to 1,180-fold) as well as peak yield (7.5- to 27-fold) that produced dominant viruses that outcompete less fit viruses.
Positive Selection.
A total of 14 coding mutations and one insertion mutation were observed on mouse adaptation of HK. The positive selection of 11 of 14 mutations indicates they confer a replicative advantage; however, there is also reason to believe that the remaining three mutations observed in individual clones from the passage 20 population were also adaptive. The Glu-133→Gly mutation in the PA protein occurred in the middle of the first nuclear localizations signal at amino acids 124–139 (Fig. 4; ref. 12) and may affect this activity. The M1-Thr-167→Ala mutation was independently selected in 1972 to become fixed in the H3N2 lineage (data not shown), indicating that this mutation has been selected on human passage. The NS2-Lys-88→Arg mutation was found in several virulent viruses, including the virulent Hong Kong H5N1 lineage of viruses, suggesting that this is an adaptive mutation that plays a role in modulating virulence (data not shown). Thus 13 of 14 mutations detected on mouse passage of HK virus were deemed to be instrumental in adaptation to increased virulence, because they either were being positively selected, involved functionally important regions of proteins (discussed below) and/or were found to have occurred independently in other virulent influenza viruses (data not shown) (Figs. 2–4). From previous genetic analysis of the A/FM/1/47-MA variant, all of the HKMA mutations would be expected to be adaptive (9, 15).
Parallel Evolution Associated with Virulence.
Three mutations observed on mouse adaptation of HK occurred at sites of preexisting variation in FM, which may represent adaptive changes selected on mouse adaptation in 1947 and is consistent with the presence of virulence determinants in this virus (9, 14, 15). Ten of 14 sites that mutated in HKMA variants were also found to have mutated in other naturally virulent or laboratory-adapted strains (data not shown). With respect to the virulent A/HK/156/97 strain, it is interesting that this virus possessed three amino acid mutations, PB2-Asp-701→Asn, NS1-Phe-103→Leu, and NS2-Lys-88→Arg, in parallel with HK MA variants, and suggests they may operate in this virus to mediate its virulence. The PB2-Asp-701→Asn mutation is located between the second nuclear localization signal (NLS2) at 736–739 and the cap-binding motif at 634–650 (Fig. 4; ref. 12), both of which could be modified in activity. The NS1 gene is required for counteracting several antiviral effects of IFN (20), and so the Phe-103→Leu mutation may be affecting this function, because mice lacking an IFN response can develop a systemic infection with neurovirulent influenza virus (20) that is similar to A/HK/156/97 infection of wild-type mice (6). The remaining mutation, NS2-Lys-88→Arg, may be affecting the function of this protein in nuclear export of viral nucleocapsids (12). Sequence analysis of A/HK/156/97 clones that are attenuated in virulence for mice was associated with the loss of the PB2-Asp-701→Asn mutation as well as two mutations, NS1-Asp-101→Asn and HA1-Pro-211→Thr, which are near the NS1–103 and HA1–218 mutations observed on mouse adaptation (21), further supporting the relevance of these mutations as determinants of virulence.
Parallel Mutation with Human Lineages of Virus.
Two of the MA mutations had also been independently selected in humans. The NS1-Val-23→Ala mutation is convergent with most influenza viruses, including human H1N1 and H2N2 strains (data not shown). The M1-Thr-167→Ala mutation was also established in the human H3N2 lineage in 1972 (data not shown). Thus, experimental evolution appears to recapitulate the selective processes of natural evolution of influenza viruses.
MA Variants Possess Mutations in Specific Functional Regions.
It is apparent that increased virulence is because of mutations that increase the ability of influenza virus to exploit the host environment. Given the location of mutations selected on mouse adaptation in FM-MA and HKMA variants (Fig. 4), they appear to implicate the central processes of viral replication and sites of interaction with host and viral components. Specific types of functional regions that are repeatedly mutated include RNA-binding sites, as seen for NS1–23, M1–139, and possibly NP-34; nuclear localization signals of polymerase subunits, PB2–482, and PA-133, as well as mutations at subunit interfaces of HA, HA1–218 in HKMA variants, and HA2–47 in FM-MA (Table 4; ref. 14). The NP-480 mutation is adjacent to amino acid 479, which affects NP oligomerization (12), and the PB2-binding site of the PB1 polymerase subunit is mutated in FM-MA (9, 12).
The Thr-156→Asn mutation of HA2 results in the loss of a glycosylation site adjacent to the cleavage site of HA0 and may be relevant to cleavage activation, because there is detectable cleavage of HA without trypsin for the HKMA-12, −20, and −20C strains produced in MDCK cells (Fig. 1D). This is analogous to the loss of a glycosylation site near the cleavage site of the HA of A/CK/Pennsylvania/1370/83 (H5N2), which resulted in increased cleavability and high virulence (22). The other HA mutation, Gly-218→W of HA1, occurs at the trimer interface adjacent to the receptor-binding site. Mutations at this site in monoclonal antibody escape mutants have been shown not only to affect HA structure such that antibody binding is maintained without neutralization but concomitantly to result in alteration in receptor specificity and the pH threshold for membrane fusion (19). Consistent with these data, the HKMA-12, -20, and -20C variants were altered in pH of fusion, resistance to β-inhibitor (mouse serum lectin), and receptor binding (data not shown). A common feature of other MA variants is mutations in HA associated with resistance to inhibitors and in some instances altered pH of fusion, as seen for FM-MA and A/Philippines/2/82/BS/ML10 (8, 17).
Extragenic Modulation of HA Function.
The observation that the HKMA-12, −20, and −20C strains had the same HA amino acid sequence but were heterogeneous in their pH of fusion as well as resistance to α inhibitor indicates that other mutations in the genome presumably present on an interacting protein were responsible for modulating the properties of HA (Fig. 4). The M1 protein is a strong candidate for this novel role, because it differs in both HKMA20 and HKMA20C relative to HKMA12 and has been implicated in the modulation of HA function. Strains of human influenza H1N1 viruses with identical HA molecules differ in their ability to agglutinate chick erythrocytes because of the nature of their M1 proteins (23). Interestingly, fixation of the M1–167 mutation in the human lineage in 1972 coincided with a change in receptor specificity (24) that did not correlate with changes in HA sequence, again suggesting the involvement of another protein in HA function. Because there is direct evidence that the M1 protein interacts with the cytoplasmic projections of HA and NA (25), it is possible that the nature of this interaction affects HA biology.
The M2–44 site, at the cytoplasmic border of the transmembrane domain (Fig. 4), is also mutated in two highly virulent avian strains, A/Ck/FPV/Rostock/34 (FPV) (H7N7) and A/CK/Penn/1370/83 (H5N2). Previous studies have shown that the FPV M2 protein is required to prevent premature acid-induced conformational change of its HA in endosomes (26). Reassortants of A/CK/Penn/1370/83 that possess its HA gene invariably possess its M1 and M2 genes (27), suggesting a functional requirement for another gene interaction.
Host-Restrictive Mutation.
The nature of the polymerase, NP, and NS proteins has been implicated in host restriction (12). The HKMA20C variant has become host restricted in its growth characteristics, possibly because of the PA-133 mutation in the first nuclear localization signal (NLS1). The FM-MA variant has the same growth properties because of a combination of mutations in the PB2 (NLS1) and PB1 (PB2-binding site) subunits (9). The PA-556 mutation may also be relevant to host specificity, because this site has mutated only in two other viruses, both with a history of mouse adaptation (A/PR/8/34 and A/WS/33). The NP gene has evolved into host-specific lineages (12), where Asp-34 is found in all human clinical isolates but has mutated in HKMA variants.
In conclusion, mouse adaptation is caused by selection of host-dependent as well as host-independent mutations that result in heightened exploitation of the host, seen clinically as increased disease. The means of achieving optimal competitive advantage appears to be controlled by acquisition of a relatively discrete subset of mutations that involve specific types of functional regions of the influenza virus genome. The location of these mutations identifies novel modulators of virulence. The adaptive approach to analyzing virulence should have general applicability to other viruses (11).
Acknowledgments
Sequence analysis was initiated in R. Levandowski's laboratory, Office of Vaccines Research and Review, Center for Biologics Evaluation and Research, Federal Drug Administration. Helpful criticism was provided by E. D. Kilbourne and K. W. Wright. This work was funded by the Natural Sciences and Engineering Research Council of Canada, Grant 0GP004177. L.C.K. and M.N. were funded by Natural Sciences and Engineering Research Council (Canada) Undergraduate Research Awards and H.L. by the Chinese Scholarship Council, People's Republic of China.
Abbreviations
- MDCK
Madin–Darby canine kidney
- MA
mouse adapted
- PB1
PB2, and PA, RNA polymerase subunits
- HA
hemagglutinin
- NP
nucleoprotein
- NA
neuraminidase
- NS1
nonstructural protein
- HK
Hong Kong
- pfu
plaque-forming unit
Footnotes
References
- 1.Lin Y P, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, et al. Proc Natl Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. . (First Published August 1, 2000, 10.1073/pnas.160270697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 4.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilbourne E D. J Clin Invest. 1959;38:266–274. doi: 10.1172/JCI103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley A J, Kawaoka Y. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rott R. Arch Virol. 1979;59:285–298. doi: 10.1007/BF01317469. [DOI] [PubMed] [Google Scholar]
- 8.Ward A C. Virus Genes. 1997;14:187–194. doi: 10.1023/a:1007979709403. [DOI] [PubMed] [Google Scholar]
- 9.Brown E G, Bailly J E. Virus Res. 1999;61:63–76. doi: 10.1016/s0168-1702(99)00027-1. [DOI] [PubMed] [Google Scholar]
- 10.Domingo E, Menendez-Arias L, Holland J J. Rev Med Virol. 1997;7:87–96. doi: 10.1002/(sici)1099-1654(199707)7:2<87::aid-rmv188>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 11.Bull J J, Badgett M R, Wichman H A, Huelsenbeck J P, Hillis D M, Gulati A, Ho C, Molineux I J. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown E G. Biomed Pharmacother. 2000;54:196–209. doi: 10.1016/S0753-3322(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen A F, Jr, Stokes J C, Smadel J E. Am J Hygiene. 1948;47:142–149. doi: 10.1093/oxfordjournals.aje.a119191. [DOI] [PubMed] [Google Scholar]
- 14.Brown E G. J Virol. 1990;64:4523. doi: 10.1128/jvi.64.9.4523-4533.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smeenk C, Brown E G. J Virol. 1994;68:530–534. doi: 10.1128/jvi.68.1.530-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou S, Brown E G. Virus Res. 1996;40:75–89. doi: 10.1016/0168-1702(95)01261-3. [DOI] [PubMed] [Google Scholar]
- 17.Smeenk C A, Wright K E, Burns B F, Thaker A J, Brown E G. Virus Res. 1996;44:79–95. doi: 10.1016/0168-1702(96)01329-9. [DOI] [PubMed] [Google Scholar]
- 18.Brown E G. J Clin Microsc. 1988;26:313–318. doi: 10.1128/jcm.26.2.313-318.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniels P S, Jeffries S, Yates P, Schild G C, Rogers G N, Paulson J C, Wharton S A, Douglas A R, Skehel J J, Wiley D C. EMBO J. 1987;6:1459–1465. doi: 10.1002/j.1460-2075.1987.tb02387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Sastre A, Durbin R K, Zheng H, Palese P, Gertner R, Levy D E, Durbin J E. J Virol. 1998;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiromoto Y, Saito T, Lindstrom S, Nerome K. Virology. 2000;272:429–437. doi: 10.1006/viro.2000.0371. [DOI] [PubMed] [Google Scholar]
- 22.Deshpande K L, Fried V A, Ando M, Webster R G. Proc Natl Acad Sci USA. 1987;84:36–40. doi: 10.1073/pnas.84.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong N, Nobusawa E, Morishita M, Nakajima S, Nakajima K. J Gen Virol. 1998;79:2425–2434. doi: 10.1099/0022-1317-79-10-2425. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Virology. 1997;227:493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- 25.Ali A, Avalos R T, Ponimaskin E, Nayak D P. J Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinhauer D A, Wharton S A, Skehel J J, Wiley D C, Hay A J. Proc Natl Acad Sci USA. 1991;88:11525–11529. doi: 10.1073/pnas.88.24.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster R G, Kawaoka Y, Bean W J. Virology. 1989;171:484–492. doi: 10.1016/0042-6822(89)90618-1. [DOI] [PubMed] [Google Scholar]