Abstract
In order to fully understand the global tuberculosis (TB) epidemic it is important to investigate the population structure and dissemination of the causative agent that drives the epidemic. Mycobacterium tuberculosis strain family 11 (F11) genotype isolates (found in 21.4% of all infected patients) are at least as successful as the Beijing genotype family isolates (16.5%) in contributing to the TB problem in some Western Cape communities of South Africa. This study describes key molecular characteristics that define the F11 genotype. A data-mining approach coupled with additional molecular analysis showed that members of F11 can easily and uniquely be identified by PCR-based techniques such as spoligotyping and dot blot screening for a specific rrs491 polymorphism. Isolates of F11 not only are a major contributor to the TB epidemic in South Africa but also are present in four different continents and at least 25 other countries in the world. Careful study of dominant compared to rare strains should provide clues to their success and possibly provide new ideas for combating TB.
Prior to the last decade, the markers available to study the epidemiology of tuberculosis (TB) were drug susceptibility profiles and phage types (3, 9). These markers have limitations. In the last decade, a number of strain-specific genetic markers with different levels of discrimination, stability, and reproducibility to examine the molecular epidemiology and the spread of TB have been identified (12). These markers have been used in several different DNA fingerprinting methods to type stains and to study the molecular epidemiology of TB. The ability to accurately genotype clinical isolates of Mycobacterium tuberculosis in different settings has shown that the global epidemiology of TB is propagated by thousands of different genotypes of this organism (17, 19). These strains occur at different frequencies, and the relative frequencies in the different areas differ between districts, cities, countries, and continents (5, 16). The dynamics of the TB epidemic in a given area and time frame may therefore be a factor of the different strains circulating in that region.
Genotype data for M. tuberculosis isolates are usually generated only for a specific region, and this has resulted in the creation of numerous databases scattered around the world. Currently such information is not in the public domain, and comparative genotyping in a global context is therefore difficult. This has hampered progress in understanding the global population structure of M. tuberculosis. A concerted effort has been made to establish a spoligotype database which contains spoligotypes from clinical isolates originating from more than 90 countries (5, 6, 13, 14). Encoding of these data by data-mining approaches has already identified seven major strain groups (Beijing, LAM, EAI, Haarlem, CAS, X, and T) and several minor groupings (5, 6, 13, 14). Due to the worldwide occurrence and dominance of the Beijing genotype of M. tuberculosis in certain geographic regions, considerable effort was made to characterize and describe markers for this family of strains (2, 7). With the exception of spoligotypes (5, 6, 13, 14), limited information is available to describe comparable specific characteristics and markers for the other previously reported prominent strain groupings.
We have reported characteristics of three evolutionary lineages of M. tuberculosis, which dominate the TB epidemic in our study setting in the Western Cape of South Africa (19, 21). One of these families, designated F11, represents the largest proportion (21.4%) of all isolates from TB patients (n = 208 individual patients; July 1992 to December 1998) in this study community. The IS6110 restriction fragment length polymorphism (RFLP) banding pattern of F11 isolates varies from 11 to 19 (average, 14) bands, and the family consists of 29 clusters and 52 related unique strains, with an overall clustering of 68%. Representative RFLP banding patterns were given previously (18). All F11 isolates lack spoligotype spacers 9 to 11, 21 to 24, and 33 to 36, which appears to be a unique marker for this strain family (19, 21). F11 isolates have been classified into pathogenic group 2, according to single-nucleotide sequence polymorphisms (SNPs) of the katG463 and gyrA95 genes (20). SNP analysis of the 16S rRNA gene showed that all isolates in F11 have a unique C-T polymorphism in rrs491, which is absent in all the members of the other local strain families and unique strains (18). This SNP is therefore useful to uniquely identify members of F11 and may also be useful to further subclassify group 2 isolates.
The aim of this study was to assess whether F11 has a global distribution and, if so, how widespread it is. We show the value of comparative analysis of genotypic information in databases in Cape Town and in The Netherlands. The results also show how PCR-based techniques such as spoligotyping and analysis of a polymorphism at rrs491 can be used to rapidly and uniquely identify members of the F11 family.
MATERIALS AND METHODS
IS6110 RFLP comparison with the Dutch and international databases.
An IS6110 RFLP database for clinical isolates of M. tuberculosis (n = 1,772; July 1992 to December 1998), originating from high-incidence communities in the Western Cape of South Africa, was available for this study (1). From this database, 197 F11 IS6110 RFLP patterns were extracted. Using the BioNumerics 3.0 software (Applied Maths, Ghent, Belgium), these F11 patterns were compared to a matching list of the National Institute of Public Health and the Environment (RIVM) RFLP database containing a total of 9,998 IS6110 DNA fingerprints of isolates from patients in The Netherlands obtained in the period 1993 through 2001. The matching list (n = 7,011) contained at least one representative fingerprint of each cluster plus all unique DNA fingerprints in the database. Next, these patterns were matched with the RIVM international database, which contains 5,035 IS6110 DNA fingerprints from 43 different countries. The distribution of the number of isolates by country is reflected in Table 1. The similarity index of the IS6110 DNA fingerprints from different countries was calculated on the basis of the unweighted pair group method using arithmetic averages with the Dice coefficient and 1% position tolerance to identify groups of strains (clades). Strains that showed a high degree of similarity (>65%) with the IS6110 DNA fingerprints of the F11 genotype of the Western Cape were analyzed further to determine their origin.
TABLE 1.
Countries in which the Western Cape genotype F11 was found in the RFLP international database
| Continent (countries)a | No. of IS6110 DNA fingerprints available for comparison | No. (%) of isolates matching with F11 |
|---|---|---|
| Asia (China [300], India [154], Indonesia [93], Iran [98], Israel [14], Malaysia [2], Mongolia [23], Philippines [46], South Korea [14], Thailand [64], United Arab Emirates [8], Vietnam [424]), | 1,232 | 0 (0.0) |
| Europe (Austria [161, 1], Czech Republic [198, 1], Germany [348, 2], Denmark [729, 13], Italy [760, 17], The Netherlandsb [9998, 245], Russia [5], Spain [78, 10], Switzerland [17], United Kingdom [110, 2]) | 12,404 | 291 (2.3) |
| North Africa (Tunisia [228, 8], Ethiopia [184], Morocco [8]) | 420 | 8 (1.9) |
| Central Africa (Tanzania [131, 1], Burundi [1], Comoros [3], Guinea [5], Rwanda [58]) | 198 | 1 (0.5) |
| Southern Africa (Zambia [188, 8], South Africac [1,203, 229]) | 1,391 | 237 (17.0) |
| North America (United States [8, 1], Canada [40, 2], Honduras [11, 2], Cuba [185, 14]) | 244 | 19 (7.8) |
| South America (Chile [3, 2], Argentina [45, 3], Venuzuela [19, 3], Brazil [80, 4], Bolivia [55, 5], Colombia [1], Equador [3], Peru [1]) | 207 | 17 (8.2) |
| Total | 16,096 | 573 |
The number of IS6110 RFLP patterns available in the international database and the number of isolates with an F11 RFLP pattern found is indicated in brackets.
Patterns in the Dutch database.
Patterns in the South Africa and international database.
Polymorphism analysis.
Small aliquots of DNAs from isolates identified from the databases described above were used for rrs491 polymorphism analysis, using a dot blot hybridization strategy (18). To ensure accurate genotypic classification, amplified products of the reference strain H37Rv as well as 10 clinical isolates with known rrs491 sequences (two polymorphic variants) were included on each blot as negative and positive controls (15, 18). Automated sequence analysis with an ABI Prism (model 3100; Applied Biosystems) analyzer was done to confirm either a C or T at position 491 of the rrs gene in a subset of the isolates.
Spoligotyping and comparisons.
Spoligotyping was done according to the internationally standardized protocol (8, 11). In order to determine whether the spoligotypes identified in this study were found in other geographic regions, they were visually compared with the spoligotypes deposited in the worldwide spoligotype database (5, 6, 13, 14).
RESULTS
IS6110 RFLP patterns of isolates of the F11 genotype collected in South Africa (n = 197) were compared to the RFLP patterns from isolates collected from patients in The Netherlands. These F11 patterns matched with 245 isolates (2.5%) of the Dutch database (called clade F11). Twenty-two (9%) of the 245 isolates from The Netherlands in clade F11 matched with IS6110 RFLP patterns identical to those in the F11 genotype from South Africa, and 73 (33.8%) of the 245 Netherlands isolates shared a banding pattern with a similarity index of between 70 and 99%. Seventeen out of 22 RFLP patterns that showed a similarity index of 100% belong to one cluster in The Netherlands. Two isolates (chronologically at the onset of this cluster in 1993) originate from patients in The Netherlands who had migrated from South Africa. One of these patients is the index case for 11 other patients of this cluster.
Subsequent SNP analysis to identify the rrs491 polymorphism showed that all isolates from clade F11 in the RIVM database have the rrs491 polymorphism. Screening of representative isolates (n = 18) from five other predominant clades in the RIVM RFLP database did not identify additional isolates with this polymorphism. Spoligotyping showed that the Dutch isolates in clade F11 also have the characteristic spacers 9 to 11, 21 to 24, and 33 to 36 deleted, as described previously for members of the F11 genotype in South Africa (21). Together, these results confirm previous findings that the rrs491 polymorphism is a unique marker for members of F11 (18) and also strongly suggest that F11 and clade F11 isolates, which were isolated from two different continents, are indeed from the same lineage.
Comparison of the F11 RFLP patterns with the RIVM-based international database of 5,035 M. tuberculosis isolates identified 120 isolates (2.4%) showing a match with a similarity value of at least 65%. It should be noted that this collection is probably biased due to the nature of the input of samples into the database. Nevertheless, the distribution and number of isolates found per continent and country are shown in Table 1. In southern Africa, South America, and North America, 17.0, 8.2, and 7.8% of the isolates, respectively, showed a similarity value of at least 65% to the IS6110 RFLP patterns of the F11 genotype, whereas in Europe, North Africa, and Central Africa, fewer than 2.5% IS6110 RFLP patterns of the F11 genotype were found. Not one RFLP pattern of the F11 genotype was found among the 1,232 isolates originating from Asia. Eight (7%) of the 120 isolates match with IS6110 RFLP banding patterns identical to those of the F11 isolates from South Africa (these isolates originated from South Africa [n = 6]), Zambia, and Denmark), whereas 31 isolates (26%) shared a banding pattern with a similarity index between 70 and 99%. Collectively these 120 isolates originate from 43 different countries. DNA was available from isolates originating from Argentina (n = 3), Chile (n = 2), Canada (n = 1), and Zambia (n = 3), and SNP analysis positively identified the rrs491 polymorphism in all of these isolates. Subsequent spoligotyping on all isolates for which DNA was available showed that spacers 9 to 11, 21 to 24, and 33 to 36 were also deleted, as described preciously for F11 isolates in South Africa. A summary of the key molecular characteristics of F11 is given in Table 2.
TABLE 2.
Key molecular characteristics that define F11 isolates
| Characteristic | Reference |
|---|---|
| Principal genetic group 2 (katG463 CGG; gyrA95 ACC) | 20 |
| Polymorphism at rrs491 (C-T) | 16; this study |
| IS6110 banding pattern, from 11 to 19 inserts (average, 14) | 16; this study |
| Spoligotype 33 (spacers 9 to 11, 21 to 24, and 33 to 36 deleted) | 21; this study |
Based on the highly conserved spoligotype banding motif associated with the F11 evolutionary lineage, the international spoligotype database was searched for spoligotypes to determine the global distribution of F11. Spoligotype patterns (n = 13,008) of clinical isolates from more than 90 countries, primarily from Europe (73%) and the United States (39%), have been compiled in updated databases (5, 6, 13, 14). This collection may also be biased, similarly to the international RFLP database in The Netherlands. However, 108 isolates, called type 33, from The Netherlands, Chile, Honduras, Cuba, France, Italy, Austria, the United Kingdom, Spain, Brazil, Argentina, French Guiana, Sweden, Venezuela, and the United States that fully match the classic spoligotype pattern of F11 isolates (spacers 9 to 11, 21 to 24, and 33 to 36 deleted) were found in the spoligotype database, as were another 33 isolates from these countries that also belong to F11 (one extra spacer deleted in addition to spacers 9 to 11, 21 to 24, and 33 to 36). Again, no isolates from Asia with an F11 conserved spoligotype banding motif were identified in the international spoligotype database. It is interesting that spoligotype 33, which matches the spoligotype of the F11 genotype, was found to be one of the most ubiquitously distributed strain types among all isolates in the international spoligotype database. Figure 1 illustrates the global distribution of the Western Cape F11 genotype family.
FIG. 1.
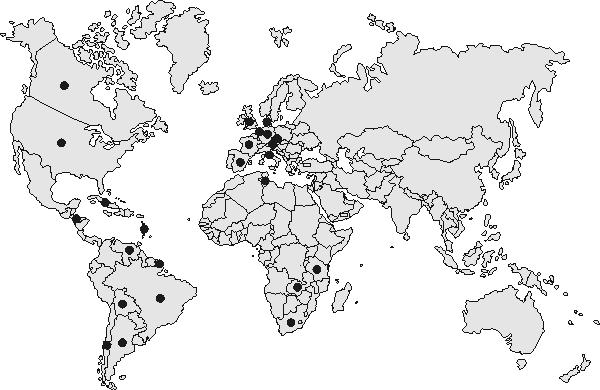
Worldwide distribution of the Western Cape F11 genotype family. Results are from the international RFLP database in The Netherlands (see countries in Table 1) and the spoligotype database in Gaudeloupe (The Netherlands, Chile, Honduras, Cuba, France, Italy, Austria, United Kingdom, Spain, Brazil, Argentina, French Guiana, Sweden, Venezuela, and United States).
DISCUSSION
Most outbreak- and population-based investigations of TB in the past have identified the clonal spread of an M. tuberculosis strain that has spread from person to person. Over time, evolution gives rise to strain variants that are detected on the basis of genetic changes. The identification of genetic markers common to members of a family, i.e., a clade of isolates, provides a second framework to classify these strains more broadly into a genotype family structure (2). The Beijing genotype of strains is the most well studied family and has led to the description of key molecular characteristics that defines this family of strains (2). The present study describes key molecular characteristics of the F11 genotype, which is the most successful strain of M. tuberculosis identified in the Western Cape communities in South Africa (19, 21). A data-mining approach coupled with additional molecular analysis collectively showed that members of F11 can easily be identified by PCR-based techniques such as the use of dot blot screening for the rrs491 polymorphism and spoligotyping to identify unique spacer deletions in members of this genotype. The results also showed that isolates of F11 not only are major contributors to the TB epidemic in South Africa but also are present in different continents and several other countries in the world. The precise contribution of this genotype in each country was difficult to assess in this study due to the nonrepresentative nature of isolates from different countries included in the databases. Interestingly, the F11 genotype was absent in Russia and countries in Asia in the international IS6110 RFLP database as well as in the spoligotype database. However, extended molecular epidemiology studies may identify F11 isolates in these regions in the future.
The factors that contribute to the success of any given M. tuberculosis genotype have not been unraveled yet. It must be cautioned that simple abundance is not necessarily an indicator of virulence (4). Conversely, molecular epidemiological data suggest that the Beijing genotype has a selective advantage over other clinical isolates to cause disease (2). This success could stem from increased transmissibility, stability, or an altered metabolism of as-yet-undefined virulence factors. There are several hypotheses to explain the large expansion of the Beijing genotype in Asia (2). These hypotheses can briefly be summarized as follows: (i) these strains spread as a result of their resistance to Mycobacterium bovis BCG-induced immunity, (ii) expansion of the Beijing genotype in the Asian continent resulted from the introduction of a new pathogen into a naive population, and (iii) the predominance of the Beijing genotype strains results from their reduced susceptibility to anti-TB drugs. However, less is known about the F11 genotype, which plays a major role in the TB epidemic in the Western Cape communities of South Africa. RFLP IS6110 banding patterns matching F11 have also been isolated in numerous other areas in South Africa (unpublished data). This suggests that F11 genotype strains (21.4% of the total number of isolates in our study community in the Western Cape of South Africa) are at least as successful as Beijing genotype strains (16.5%) in their ability to cause TB. It is unlikely that the predominance of F11 isolates results from reduced susceptibility to anti-TB drugs, since only 6% of the 197 patients from South Africa had F11 drug-resistant isolates. The markers described in this study can now be used to further investigate the prevalence and to study the relative importance of the F11 genotype in other settings.
Dominant strain groups other than the F11 and Beijing genotypes have been cited in the literature. Among these are major groups called LAM, EAI, Haarlem, CAS, X, and T (5, 6, 13). This is based on analysis of 13,008 spoligotype patterns from more than 90 countries. However, very few data are available to describe additional characteristics of isolates in these groupings. Spoligotype 33 in the SpolDB3 database is similar to the spoligotypes of F11 isolates described in this study. It is therefore likely that F11 (Western Cape, South Africa) is part of the LAM group, which includes isolates mostly from Latin American and the Mediterranean regions. It is likely that M. tuberculosis isolates will be categorized on the basis of synonymous SNPs in the future (10).
It is important that we add to the body of knowledge concerning the population structure of M. tuberculosis. This will allow us to monitor trends in the dynamics of this disease before, during, and after vaccination trials and to identify successful (fit) virulent strains, which in turn may lead to new treatment modalities.
Acknowledgments
We thank the IAEA (project SAF6003), the Harry Crossley Foundation, and GlaxoSmithKline for financial assistance.
We are grateful to H. Heersma for help with BioNummeris.
REFERENCES
- 1.Beyers, N., R. P. Gie, H. L. Zietsman, M. Kunneke, J. Hauman, M. Tatley, and P. R. Donald. 1996. The use of a geographical information system (GIS) to evaluate the distribution of tuberculosis in a high-incidence community. S. Afr. Med. J. 86:40-44. [PubMed] [Google Scholar]
- 2.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 3.Crawford, J. T., J. K. Fitzhugh, and J. H. Bates. 1981. Phage typing of the Mycobacterium avium-intracellulare-scrofulaceum complex. Am. Rev. Respir. Dis. 124:559-562. [DOI] [PubMed] [Google Scholar]
- 4.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 5.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, G. Kallenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. De Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. De Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 9.Gruft, H., R. Johnson, R. Claflin, and A. Loder. 1984. Phage-typing and drug-resistance patterns as tools in mycobacterial epidemiology. Am. Rev. Respir. Dis. 130:96-97. [DOI] [PubMed] [Google Scholar]
- 10.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms. Resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. Van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostrom, P., M. Gordon, C. Sola, M. Ridell, and N. Rastogi. 2002. Methods used in the molecular epidemiology of tuberculosis. Clin. Microbiol. Infect. 8:694-704. [DOI] [PubMed] [Google Scholar]
- 13.Sebban, M., I. Mokrousov, N. Rastogi, and C. Sola. 2002. A data-mining approach to spacer oligonucleotide typing of Mycobacterium tuberculosis. Bioinformatics 18:235-243. [DOI] [PubMed] [Google Scholar]
- 14.Sola, C., I. Filliol, M. C. Gutierrez, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Soolingen, D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]
- 17.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 18.Victor, T. C., A. van Rie, A. M. Jordaan, M. Richardson, G. D. Der Spuy, N. Beyers, P. D. van Helden, and R. Warren. 2001. Sequence polymorphism in the rrs gene of Mycobacterium tuberculosis is deeply rooted within an evolutionary clade and is not associated with streptomycin resistance. J. Clin. Microbiol. 39:4184-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren, R., M. Richardson, S. G. van der, T. Victor, S. Sampson, N. Beyers, and P. van Helden. 1999. DNA fingerprinting and molecular epidemiology of tuberculosis: use and interpretation in an epidemic setting. Electrophoresis 20:1807-1812. [DOI] [PubMed] [Google Scholar]
- 20.Warren, R. M., S. L. Sampson, M. Richardson, G. D. van der Spuy, C. J. Lombard, T. C. Victor, and P. D. van Helden. 2000. Mapping of IS6110 flanking regions in clinical isolates of M. tuberculosis demonstrates genome plasticity. Mol. Microbiol. 37:1405-1416. [DOI] [PubMed] [Google Scholar]
- 21.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]


