The Yorkie/YAP pathway is a major determinant of organ size in the fly and in mammalian systems and has been linked to many forms of cancer. Nagaraj et al. identify a new target of this pathway as the mitochondrion and show that Yorkie up-regulates the genes that control mitochondrial fusion, resulting in reduced reactive oxygen species levels. These results demonstrate the conserved regulation of mitochondrial function by the Yorkie pathway, how genetic alteration of mitochondrial fusion can block cell overgrowth, and how an oncogene can manipulate the cell biology of the mitochondria.
Keywords: Hippo pathway, ROS, Yap, Yorkie, marf, mitochondrial fusion, opa
Abstract
Mitochondrial structure and function are highly dynamic, but the potential roles for cell signaling pathways in influencing these properties are not fully understood. Reduced mitochondrial function has been shown to cause cell cycle arrest, and a direct role of signaling pathways in controlling mitochondrial function during development and disease is an active area of investigation. Here, we show that the conserved Yorkie/YAP signaling pathway implicated in the control of organ size also functions in the regulation of mitochondria in Drosophila as well as human cells. In Drosophila, activation of Yorkie causes direct transcriptional up-regulation of genes that regulate mitochondrial fusion, such as opa1-like (opa1) and mitochondria assembly regulatory factor (Marf), and results in fused mitochondria with dramatic reduction in reactive oxygen species (ROS) levels. When mitochondrial fusion is genetically attenuated, the Yorkie-induced cell proliferation and tissue overgrowth are significantly suppressed. The function of Yorkie is conserved across evolution, as activation of YAP2 in human cell lines causes increased mitochondrial fusion. Thus, mitochondrial fusion is an essential and direct target of the Yorkie/YAP pathway in the regulation of organ size control during development and could play a similar role in the genesis of cancer.
The Mst/Lats/YAP pathway functions in the process of contact inhibition to regulate organ size in mammals, and loss in this pathway's function is implicated in multiple cancer types (Zhao et al. 2008). In Drosophila, where this pathway, commonly referred to as the “Hippo pathway,” was first identified and best understood, the corresponding proteins are Hippo/Warts/Yorkie (Hariharan 2006; Pan 2007; Zhang et al. 2009). Loss-of-function mutations in any upstream component of the Hippo pathway will cause Yorkie (Yki) to remain unphosphorylated and actively signal within the nucleus, causing an overgrowth phenotype (Dong et al. 2007; Oh and Irvine 2008). YAP/Yki functions as a transcriptional coactivator, interacting with a DNA-binding partner such as mammalian TEAD proteins/Drosophila Scalloped (Sd) (Huang et al. 2005; Goulev et al. 2008; Wu et al. 2008; Zhang et al. 2008). In Drosophila, the Hippo pathway promotes proliferation by positively regulating cyclin E and the microRNA bantam (Harvey et al. 2003; Wu et al. 2003; Huang et al. 2005; Nolo et al. 2006; Thompson and Cohen 2006). This pathway also inhibits apoptosis by controlling DIAP1 expression (Harvey et al. 2003; Huang et al. 2005; Dong et al. 2007; Oh and Irvine 2008). Overexpression of Yki in the late larval and pupal eye disc cells causes an increased adult eye size (Huang et al. 2005; Dong et al. 2007). This phenotype is further enhanced by coexpression of Sd (Goulev et al. 2008; Wu et al. 2008; Zhang et al. 2008). The extreme effect of this pathway on growth and its prominent role in cancer progression prompted us to investigate a possible link to cellular metabolism.
Results
Mitochondrial phenotype upon Yki pathway activation
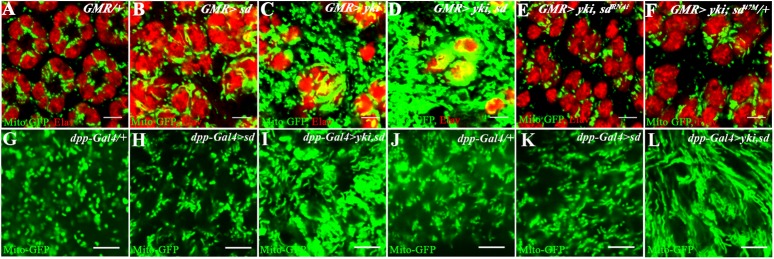
GFP targeted to the mitochondrial matrix (mitoGFP) is trapped and stabilized and allows visualization of the mitochondrial network (Clark et al. 2006; Goyal et al. 2007; Deng et al. 2008). This reagent was used to assess the effects of activating the yki/sd pathway on mitochondria. In the wild-type pupal eye disc, the majority of mitochondria are oval in shape and scattered around the cell with no visible signs of an interconnected network (Fig. 1A; Supplemental Fig. S1). Upon overexpression of Sd, the DNA-binding partner of Yki, only a modest increase in mitochondrial size seems evident (Fig. 1B). However, overexpression of Yki (GMR-Gal4, UAS-yki) causes a dramatic increase in mitochondria (Fig. 1C; Supplemental Fig. S1). This phenotype is further enhanced upon a simultaneous coexpression of Yki and Sd (Fig. 1D). Inactivation of sd in a GMR-Gal4, UAS-yki mutant background by either a single-copy loss of sd or a coexpression of sdRNAi suppresses the Yki-induced mitochondrial expansion (Fig. 1E,F). In contrast, knocking down Homothorax (hth), another known transcriptional partner of Yorkie (Peng et al. 2009), in a Yorkie overexpression background did not modify the mitochondrial phenotypes of Yorkie (data not shown), suggesting that the control of mitochondria by this pathway involves the Yki/Sd and not the Yki/Hth complex. The phenotype of an enhanced mitochondrial network upon Yki/Sd expression is not specific to the eye, as it is also seen in the wing and leg imaginal discs (Fig. 1G–L; additional controls in Supplemental Figs. S2, S3).
Figure 1.
Yki activation causes mitochondrial phenotypes. (A–F) Mid-pupal eye discs. mitoGFP (green) marks the mitochondrial matrix. ELAV (red) marks photoreceptor nuclei. Bar, 5 μm. (A) Control (GMR-Gal4, UAS-mitoGFP) mitochondria (green) are seen as individual dots. (B) Overexpression of Scalloped (GMR-Gal4, UAS-mitoGFP UAS-sd) causes a modest increase in mitochondria. (C) Overexpression of Yki (GMR-Gal4, UAS-mitoGFP UAS-yki) results in extensive mitochondrial (green) expansion. (D) Combined overexpression of Yki and Sd (GMR-Gal4, UAS-mitoGFP UAS-yki, UAS-sd) causes further enhancement of the mitochondrial expansion phenotype. (E) In the background of Yki activation, a reduction in sd function (GMR-Gal4, UAS-mitoGFP, UAS-yki, UAS-sdRNAi) suppresses the Yki-mediated expansion of mitochondria. (F) Single-copy loss of sd in a Yki-activated background (GMR-Gal4, UAS-mitoGFP, UAS-yki, sd47M/+) causes a suppression in mitochondrial expansion compared with that seen upon Yki overexpression (cf. C). (G–L) Third instar wing (G–I) and leg (J–L) imaginal discs. mitoGFP (green) marks the mitochondria. (G) Control (dpp-Gal4, UAS-mitoGFP) wing disc; the mitochondria (green) are seen as individual dots. (H) Overexpression of Sd (dpp-Gal4, UAS-mitoGFP, UAS-sd) does not significantly affect mitochondria. (I) A combined overexpression of Yki and Sd in the wing disc (dpp-Gal4, UAS-mitoGFP, UAS-yki, UAS-sd) results in extensive increase in mitochondria. (J) Control (dpp-Gal4, UAS-mitoGFP) leg disc. (K) Overexpression of Sd (dpp-Gal4, UAS-mitoGFP, UAS-sd) is similar to control. (L) Combined overexpression of Yki and Sd (dpp-Gal4, UAS-mitoGFP, UAS-yki, UAS-sd) causes extensive expansion of mitochondria.
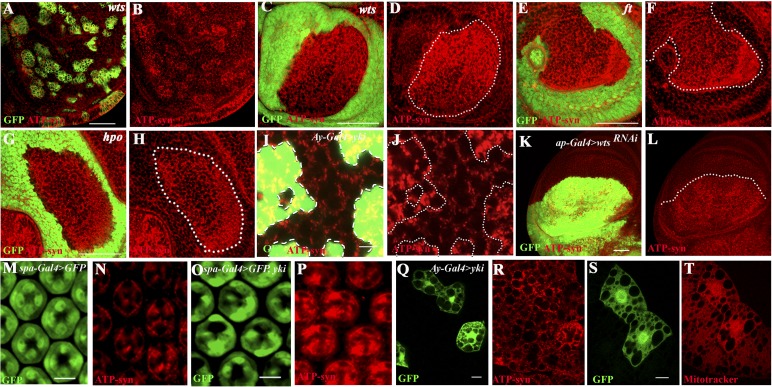
To determine whether Yki functions autonomously in mitochondrial control, we generated mutant clones of the canonical Hippo pathway components: hippo, which encodes an upstream Ste-20 class kinase (Harvey et al. 2003; Wu et al. 2003); warts (wts), which encodes the terminal kinase that phosphorylates Yki (Justice et al. 1995; Harvey et al. 2003); and fat (ft), which encodes a receptor for the Hippo pathway (Bennett and Harvey 2006; Silva et al. 2006). We also generated clones that overexpress Yki. In all cases, mutant cells show an autonomous increase in the expression of mitochondrial markers (Fig. 2A–J). Furthermore, down-regulation of wts and ft using RNAi in the dorsal compartment of the wing disc also causes an increase in mitochondrial staining within the mutant tissue (Fig. 2K,L; Supplemental Fig. S4). Reduction of Yki by RNAi suppresses the increased mitochondrial staining observed in wts mutant cells, showing that yki functions downstream from wts to regulate mitochondrial expansion (Supplemental Fig. S5). The microRNA bantam is a transcriptional target of Yki, and its overexpression causes increased cell proliferation and organ size overgrowth (Nolo et al. 2006; Thompson and Cohen 2006). However, overexpression of bantam does not result in a mitochondrial phenotype (Supplemental Fig. S6A–D). Furthermore, although bantam overexpression enhances tissue overgrowth due to Yki, no enhancement of the Yki mitochondrial phenotype by bantam overexpression is apparent in the eye disc (Supplemental Fig. S6E–H).
Figure 2.
Mitochondrial phenotypes in Hippo pathway mutant backgrounds. (A–L) Third instar wing discs. (A,B) Isolated mutant clones of wts (green) generated using MARCM and stained for ATP-syn (red in B) show increased staining in the mutant (green) cells. (C–H) Third instar wing discs containing mutant clones (nongreen) for warts (wts) (C,D), fat (ft) (E,F), and hippo (hpo) (G,H) show increased mitochondrial staining using ATP synthase-α antibody (red) in the mutant cells (nongreen) compared with adjacent wild-type tissue (green). (B,D,F,H) Red channel alone is shown for clarity. Bar, 50 μm. (I,J) Clones of cells in a wing disc expressing Yki (green) using the Ay-Gal4 system (see the Materials and Methods). Increased expression of the mitochondrial marker ATP synthase-α (red) is seen in Yki overexpressing cells (green) as compared with the adjacent wild-type cells (nongreen). (J) Red channel alone is shown for clarity. Bar, 5 μm. (K,L) Knockdown of wts function in the dorsal compartment of the wing disc (K; green, ap-Gal4 UAS-GFP, UAS-wtsRNAi) causes an increase in mitochondrial marker ATP synthase-α staining (L; red). The white dotted line marks the dorsal–ventral boundary. Bar, 50 μm. (M–P) Pupal eye discs. (M,N) Control, pupal eye disc from spa-Gal4, UAS-GFP background. The cone cells are marked with GFP (M; green) and stained for mitochondrial marker ATP-syn (N; red). (O,P) Overexpression of Yki in cone cells (spa-Gal4, UAS-GFP, UAS-yki) (O; green) causes a significant increase in mitochondrial marker ATP-syn staining (P; red). Bar, 5 μm. (Q–T) Clones of cells in the fat body expressing Yki (shown in Q and S in green) using the Ay-Gal4 system (see the Materials and Methods). Increased staining with ATP synthase-α antibody (R; red) and increased uptake of the mitochondrial membrane potential-sensitive dye MitoTracker Red (S; red) can be seen in Yki overexpressing cells (S; green) at the perinuclear regions when compared with adjacent wild-type cells (S; nongreen). Bars, 5 μm.
The Yki mitochondrial phenotype is independent of cell proliferation
The use of GMR-Gal4 as a driver to express Yki causes cells to proliferate, making it difficult to assess whether the observed change in mitochondrial morphology is a primary result of the activation of this pathway or a secondary consequence of increased proliferation. We therefore used spa-Gal4 as a driver to activate Yki in post-mitotic cone cells (green in Fig. 2M,O) in the eye. The spa-Gal4, UAS-yki combination does not cause overgrowth, and yet a robust increase in mitochondrial staining is readily apparent (Fig. 2M–P), suggesting that even in the absence of cell proliferation, this pathway is capable of promoting mitochondrial biogenesis. This phenotype is specific to Yki, since overexpression of other growth-promoting factors such as activation of the EGFR or the Wingless pathway using the spa-Gal4 driver does not cause a similar increase in mitochondrial biogenesis (Supplemental Fig. S7). Furthermore, loss of two other tumor suppressor genes (scribble and avalanche), each of which shows an extensive growth phenotype similar to that seen upon Yki activation, also does not result in increased mitochondrial staining (Supplemental Fig. S8).
Finally, we asked whether the increased mitochondrial staining seen with mitoGFP and ATP-synthase is also reflected as higher mitochondrial membrane potential by staining with the dye MitoTracker (Baltzer et al. 2009). As evidenced by clones generated in the post-mitotic cells of the fat body, Yki activation causes an increase in mitochondrial markers and in the intensity of MitoTracker staining (Fig. 2Q–T).
The mitochondrial phenotype of Yki is conserved in humans
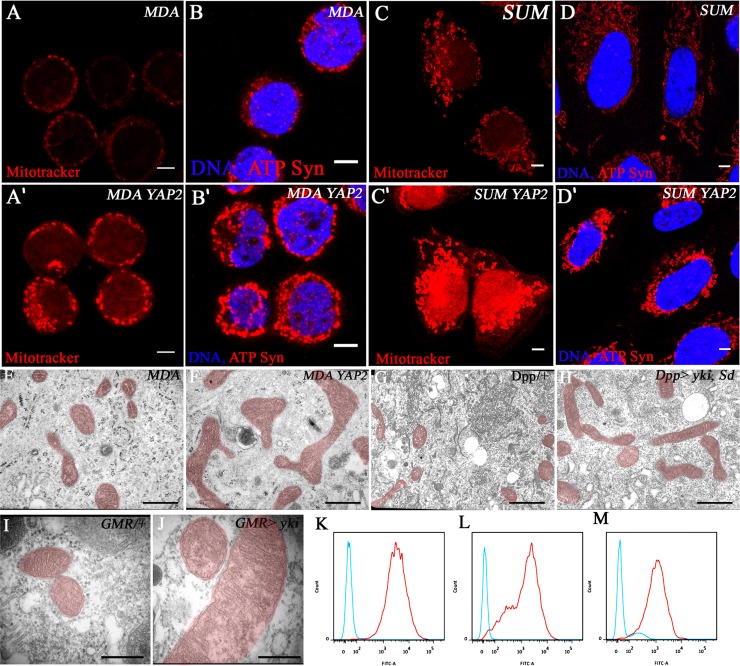
To determine whether the role of the Hippo pathway in the control of mitochondrial network formation is conserved across species, we stably overexpressed a Flag-tagged form of YAP2 (one of the human homologs of Yorkie) in several human cancer cell lines, which endogenously express YAP2, as assessed by protein blots (data not shown), and thus might have functional cofactors. Mitochondria were analyzed with MitoTracker Red staining and an antibody against the mitochondrial protein ATP synthase-α. In control cells from the breast cancer cell lines MDAMB453 and SUM159PT, we observed a small number of mitochondria predominantly localized in perinuclear regions (Fig. 3A–D). Overexpression of YAP2 causes an increase in MitoTracker and mitochondrial ATP synthase-α staining (Fig. 3A′–D′). We also examined three additional cell lines derived from different human cancers that express varying levels of endogenous YAP2. In two cases, we observed a correlation between increased YAP2 expression and enhanced mitochondrial mass, but this correlation was not seen in the third cell line (data not shown). Thus, YAP expression may be correlative but is not the sole determinant of mitochondrial morphology in human cancer cells. However, the phenotypes that we observed upon YAP2 overexpression are remarkably similar to those seen in Drosophila and suggest that regulation of mitochondria by the Yki pathway is conserved in humans.
Figure 3.
Mitochondrial phenotypes of Yki/YAP2-activated human and Drosophila cells. (A–D) Controls. Mitochondrial staining in MDAMB453 and SUM159PT cells expressing empty vector using MitoTracker Red (A,C) and ATP-synthase-α antibody (B,D). Stable expression of Flag-tagged YAP2 in MDAMB453 and SUM159PT cells causes an increase in mitochondria monitored using MitoTracker Red (A′,C′) and ATP-synthase-α antibody (B′,D′). Bar, 5 μm. (E–J) Ultrastructural analysis of mitochondria (highlighted in red) upon Yki/YAP2 overexpression. (E,F) Electron micrographs of mitochondria from human cells. YAP2 overexpressing MDAMT453 cells (F) have elongated and enlarged mitochondria compared with their vector-transformed controls (E). Bar, 1 μm. (G,H) Electron micrographs of mitochondria from Drosophila salivary glands. The mitochondria are elongated upon combined overexpression of Yki and Sd (H) compared with wild-type control (G). Bar, 1 μm. (I,J) Electron micrographs of mitochondria from Drosophila pupal eye discs. High-magnification view of mitochondria from control (I) and Yki-activated (J) pupal eye tissue. Bar, 0.5 μm. (K–M) Reduction of ROS in YAP2 overexpressing cells. Flow cytometric analysis of ROS-labeled control cells (red trace) and YAP2-overexpressing cells (blue trace) in HS578T (K), SUM159PT (L), and MDAMB231 (M) cell lines. The ROS levels show a dramatic decrease of ∼100-fold in all three cell lines upon YAP2 expression.
Ultrastructural and metabolic analysis of mitochondria upon Yki activation
To characterize the mitochondrial phenotype seen above at the ultrastructural level, we carried out an electron microscopic (EM) analysis in cultured human cells overexpressing YAP2 (Fig. 3E,F) as well as in Drosophila tissues in which Yki is activated (Fig. 3G–J). Consistent with the light microscopic immunohistochemical results, at the EM level, the mitochondria are elongated and enlarged (Fig. 3E–J), suggesting that the observed phenotypes are due to mitochondrial fusion. The elongated mitochondria continue to maintain normally structured cristae. Quantitation of the fusion phenotype revealed an average twofold increase in the length of mitochondria upon Yki/Sd activation (P = 0.0009) (Supplemental Fig S9A). Mitochondrial numbers are moderately increased (52%) (Supplemental Fig. S9B), which could be due to either increased biogenesis or decreased turnover, which is reported to result from increased fusion (Twig et al. 2008).
To assess the functional effects of Yki/YAP2 on mitochondria, we measured several metabolic outputs in human and Drosophila cells. ATP levels are not significantly altered upon Yki/YAP2 overexpression in either Drosophila or human cells, and consumption of glucose, oxygen, and glutamine and production of lactate and glutamate were also unaffected in several human cell lines (Supplemental Fig. S10). However, levels of reactive oxygen species (ROS) show a dramatic, two orders of magnitude decrease when YAP2 is overexpressed in the three independent cell lines tested (Fig. 3K–M). These results demonstrate that one consequence of the mammalian YAP2 pathway activation is a reduction of ROS, which is likely to increase cellular resistance to oxidative stress.
Mitochondrial targets of Yki activation
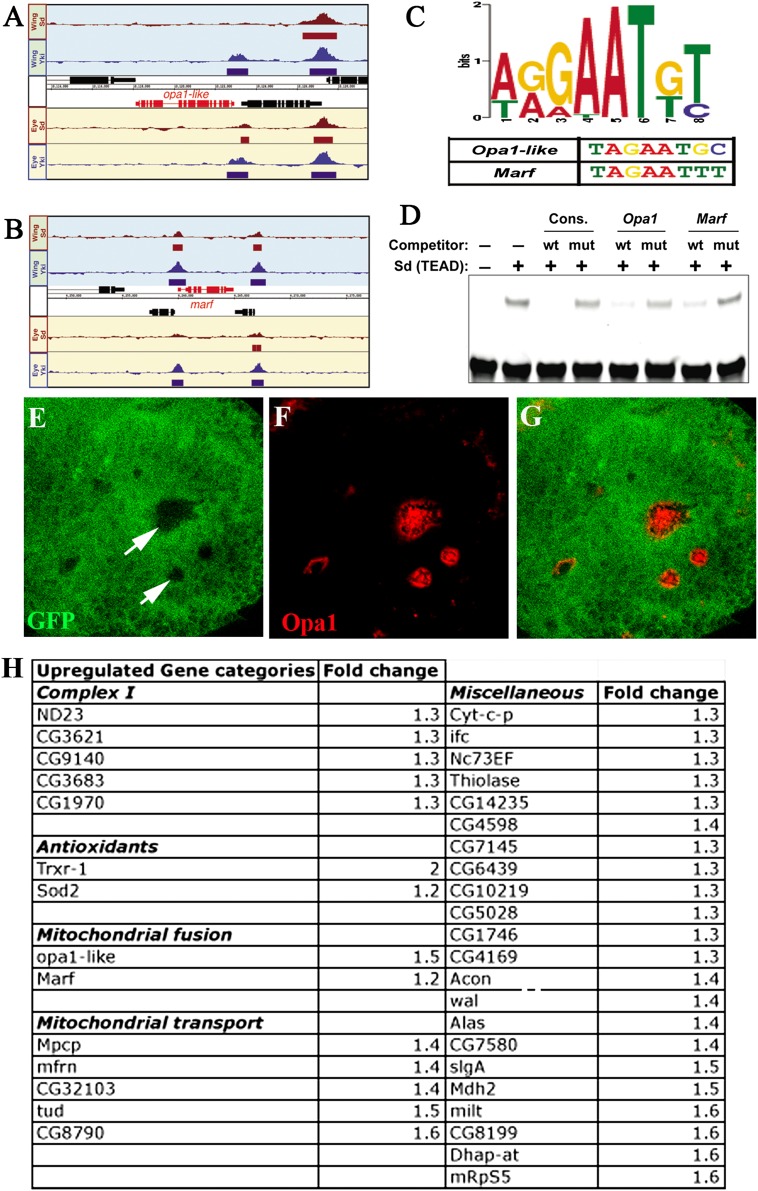
To probe mechanistic determinants of Yki function in mitochondrial expansion, we conducted a genome-wide microarray experiment and specifically compared expression patterns of mitochondria-related transcripts from control (GMR-Gal4) and Yorkie overexpressing (GMR-Gal4; UAS-yki) pupal eye discs. Genes that have previously been implicated in basal mitochondrial biogenesis, such as Spargel (PGC-1α) and DELG (NRF2) (Baltzer et al. 2009), are not altered upon overexpression of Yki. However, a number of other genes associated with mitochondrial function are up-regulated by Yki, including the two mitochondrial fusion genes opa1-like (opa1) and mitochondria assembly regulatory factor (Marf) (Supplemental Fig S11A; Chan 2006). These could potentially provide a link between Yki activation and enhanced mitochondrial fusion evident from the ultrastructural analysis.
To determine whether Yki and Sd are directly bound to DNA corresponding to mitochondrial genes, we used chromatin immunoprecipitation (ChIP) and whole-genome tiling arrays (ChIP–chip) to identify regions bound by these factors in eye–antennal and wing imaginal discs. We found that Yki and Sd together bind to enhancer elements corresponding to several genes related to mitochondrial fusion, including opa1 and Marf (Fig. 4A,B; Supplemental S11B). These genes include a consensus motif (AGGAATGT) in their upstream sequence that matches with the published Sd-binding consensus site. Importantly, most of the Sd-bound regions around these genes also contain strong matches to the Sd consensus site that we defined using in vitro selection followed by next-generation sequencing (Fig. 4C). Gel shift assays established that the Sd consensus sites within opa1 and Marf are indeed bound by Sd (Fig. 4D). Luciferase reporter assays in S2 cells using the Sd-binding domain containing enhancer regions of opa1 and Marf showed a 15-fold and fivefold induction of opa1 and Marf, respectively, upon activation with Yki/Sd (Supplemental Fig. S12). In situ hybridization studies further show that in wts mutant clones, opa1 expression is up-regulated (Fig. 4E–G). Based on the genetic, microarray, and ChIP data and reporter assays, we conclude that the Yki/Sd complex controls expression of the mitochondrial fusion genes opa1 and Marf that lead to the formation of the extensively fused mitochondria observed upon Yki activation.
Figure 4.
Direct regulation of mitochondria-related genes by the Yorkie/Scalloped complex. (A,B) Sd and Yki are bound near the loci of mitochondrial morphogenesis genes. Sd and Yki ChIP–chip binding profiles at opa1 (A) and Marf (B). Regions designated as bound by a given factor (5% FDR [false discovery rate]) are indicated by solid rectangles below a given peak. (C) Sd consensus motif identified by in vitro selection and next-generation sequencing in mitochondrial fusion genes. (D) Gel mobility shift assay. Purified TEA domain of Sd protein used for mobility shift assay shows binding to wild-type but not mutated consensus Sd-binding sites as indicated. opa1- and Marf-specific DNA sequences containing wild-type but not mutated putative Sd-binding sites can compete and block Sd binding to the probe. (E–G) Fluorescence in situ hybridization to visualize opa1 transcripts in wts mutant clones. wts mutant clones (nongreen, marked by arrows in E) were generated in third instar eye discs using the Flp/FRT system. In situ hybridization for GFP (green) marks the wild-type tissue. (F) opa1 expression (red) is up-regulated in the mutant (nongreen) tissue. The merged image is shown in G. Note that the level of expression of opa1 in the wild-type eye disc is below the levels of detection. However, in these experiments, we were able to detect wild-type opa1 expression in the wing disc pouch, ring glands, and eye disc upon overexpression of opa1 using the combination GMR-Gal4, UAS-opa1. (H) List of categories of mitochondrial genes that are up-regulated with their fold change (judged by microarray analysis) that are also bound by both Yki and Sd (based on ChIP–chip data set generated for Yki and Sd).
The ChIP–chip array showed that 261 mitochondrial genes were bound by both Sd and Yki, of which 36 are also up-regulated at least 1.2-fold in the microarray analysis (Fig. 4H). We confirmed the up-regulation of opa1 and Marf by quantitative PCR (Supplemental Fig. S13). The 36 genes include various mitochondrial enzymes and transporters, including complex I genes, antioxidants, and the two mitochondrial fusion genes opa1 and Marf, which implies that the Yki/Sd complex directly binds to the upstream regions of these genes to regulate their transcription. In addition to these fusion proteins, the two major classes of up-regulated proteins whose relevance seems evident from our genetic analysis include the antioxidant proteins and members of complex I of the electron transport chain. Both of these protein classes have demonstrated functions related to ROS levels (Finkel 2003; Owusu-Ansah et al. 2008), and their up-regulation by the Yki/YAP pathway helps explain the dramatic reduction in ROS levels upon YAP2 activation (Fig. 3K–M) and suggests a conserved function for Yki/YAP2 signaling in the modulation of oxidative stress. The best-characterized target of the Yki/Sd pathway, Diap1, is also up-regulated at levels similar to the mitochondrial genes (1.2-fold) in our transcriptome analysis.
opa1 and Marf suppress Yki growth and mitochondrial phenotypes
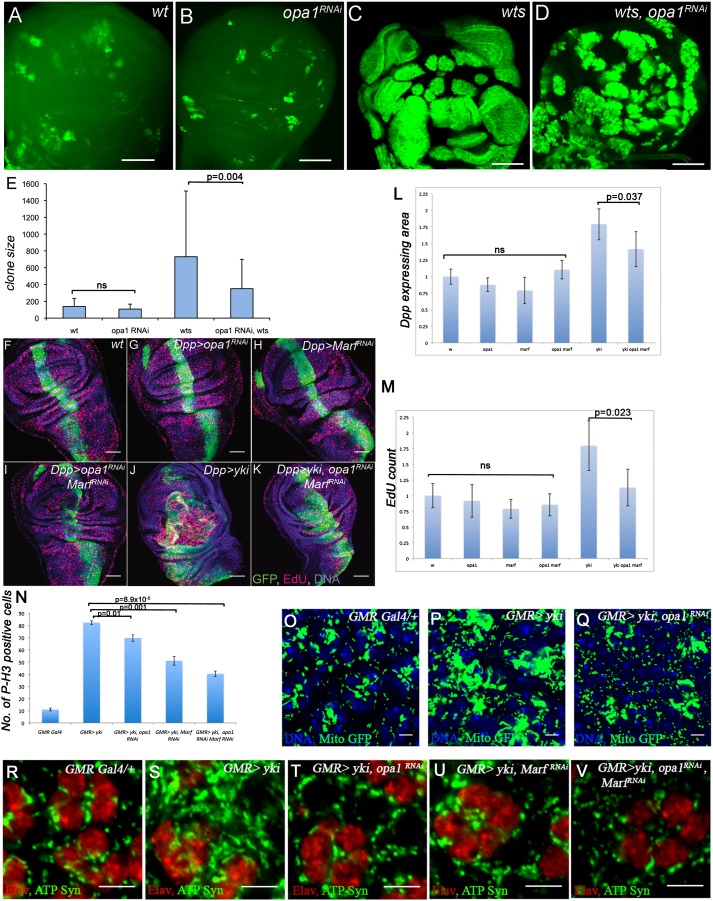
The role of opa1 and Marf as downstream targets of Yki was further dissected through genetic analysis. MARCM clones of wts in wing discs in which opa1 function is attenuated using UAS- opa1RNAi causes significant suppression in the growth of mutant clones (Fig. 5A–E). Using the dpp-Gal4 driver, we found that combined inactivation of opa1 and Marf significantly suppresses the increased tissue growth observed upon overexpression of Yki (Fig. 5F–L). This effect is Yki-specific, as overgrowth due to loss of other tumor suppressor backgrounds is not suppressed by opa1 (Supplemental Fig. S14).
Figure 5.
Mutations in mitochondrial fusion genes suppress phenotypes caused by Yki activation. (A–E) MARCM clones of wts in the wing imaginal discs. (A) Mock clones (green) generated with wild-type FRT82B chromosome. (B) Mock clones (green) expressing opa1RNAi. (C) wts clones (green) showing an extensive overgrowth phenotype. (D) wts clones (green) expressing opa1RNAi suppress the overgrowth phenotype seen upon loss of wts function (cf. C and D). (E) A quantitative analysis of clone sizes in different genotypes as indicated at the bottom of the graph. The clone sizes in the wild type and opa1RNAi are similar with no significant differences, while expression of opa1RNAi in wts clones results in a significant (P = 0.004) suppression of clone size. Bars, 50 μm. (F–K) Representative images of third instar wing discs labeled with EdU to mark S-phase cells. dpp-Gal4, UAS-GFP is used to drive appropriate RNAi/ UAS-Yki overexpression. Genotypes are indicated (RNAi constructs used are opa1KK105706 and MarfKK105261). Bars, 50 μm. (L) Quantification of the area of the dpp-Gal4, UAS-GFP-expressing tissue. RNAi targeting opa1, Marf, or both does not significantly alter growth in the wing disc, as demonstrated by measuring the area of dpp-expressing tissue normalized to control (dpp-Gal4 UAS-GFP crossed to w1118, opa1 RNAi, Marf RNAi, or opa1 RNAi/Marf RNAi). Overexpression of yorkie (dpp-Gal4 UAS-GFP, UAS-yki) causes a robust increase in tissue size (P = 1.2 × 10−4), and this is significantly reduced by combined inactivation of opa1 and Marf (P = 0.037). Average sizes relative to control and error bars indicating standard deviation are shown. (M) Quantification of EdU-positive cells within the dpp-Gal4, UAS-GFP-expressing tissue. Genotypes are as indicated. Compared with control (dpp-Gal4, UAS-GFP), the reduction of opa1 or Marf function by RNAi, separately or in combination, does not significantly reduce the number of EdU-labeled cells. Overexpression of yorkie (dpp-Gal4, UAS-GFP, UAS-yki) causes a dramatic increase in EdU-positive cells (P = 0.008), and coexpression of opa1 and Marf RNAis with yorkie (dpp-Gal4, UAS-GFP, UAS-yki, UAS-opa1 RNAi, UAS-Marf RNAi) leads to a significant suppression of EdU-positive cell number (P = 0.023). Average number of EdU spots in the dpp(+) area is plotted relative to control, and error bars indicate standard deviation. (N) Quantitative analysis of phospho-Histone H3 (P-H3)-positive cells upon Yki activation and in combination with a reduction of opa1 and Marf function. Genotypes are as indicated. Overexpression of Yki in the third instar eye discs causes a significant increase in the number of P-H3-positive cells (P = 2.7 × 10−6). Knockdown of opa1 alone, Marf alone, or both in a Yki overexpression background significantly suppresses the number of P-H3-positive cells (P = 0.01, 0.001, and 8.9 × 10−5, respectively). (O–Q) Suppression of Yki-induced mitochondrial phenotype upon loss of opa1 function. Mitochondrial morphology is monitored using mitoGFP (green). Cell nuclei are marked with Topro (blue). Bar, 5 μm. (R–V) Suppression of Yki-induced mitochondrial phenotype upon loss of opa1 and Marf function. Mitochondrial morphology is monitored using ATP synthase-α staining (green). Photoreceptor nuclei are marked with ELAV (red). Bar, 5 μm. Combined reduction of opa1 and Marf in a Yki-activated background causes strong suppression of mitochondrial expansion.
The attenuation in Yki-mediated tissue growth can be traced to a reduction in cell proliferation. In the larval wing disc, knockdown of opa1 and Marf using RNAi suppresses the enhanced EdU incorporation observed upon Yki overexpression (Fig. 5F–K,M). Likewise, overexpression of Yki causes increased proliferation of cells in the third instar eye imaginal disc, and this phenotype is significantly suppressed upon reduction of either opa1 or Marf as measured by phospho-Histone H3 staining (Fig. 5N; additional controls in Supplemental Figs. S15, S16) in the eye. We analyzed Caspase-3 staining in multiple RNAi backgrounds for both opa1 and Marf used in this study and did not detect an increase in cleaved Caspase-3 (Supplemental Fig. S17). The suppression of Yki-mediated proliferation by the loss of fusion genes correlates with the suppression in Yki-induced mitochondrial fusion observed in the pupal eye disc (Fig. 5O–V; further controls in Supplemental Fig. S18). The relevance of opa1-mediated mitochondrial fusion to the Yki-mediated mitochondrial phenotype is underscored by the dosage-dependent interaction between a single-copy loss of opa1 and Yki overexpression (Supplemental Fig. S19).
Discussion
Previous studies have shown that the Hippo pathway functions in flies as well as vertebrates to control organ size (Dong et al. 2007). Furthermore, mutations in components of this pathway have been implicated in multiple forms of cancer (Edgar 2006). This pathway has been shown to directly promote cell proliferation and repress apoptosis. Here, we show that the mitochondrion is an important additional target of the Hippo pathway. An increase in Yki activity causes an increase in mitochondrial fusion due to direct transcriptional activation of major mitochondrial fusion genes. Increased mitochondrial fusion has been linked to regulation of the G1–S checkpoint and Cyclin E activity (Mandel et al. 2005; Mitra et al. 2009), and cells harboring fused mitochondria are resistant to stress-induced apoptosis (Tondera et al. 2009; van der Bliek 2009). Interestingly, these are precisely the phenotypes seen upon activation of the Hpo/Yki pathway. It seems likely that this pathway independently activates the cell cycle, represses apoptosis (Harvey et al. 2003; Hay and Guo 2003; Wu et al. 2003), and promotes mitochondrial and metabolic changes described in this study that together cause tumor growth. However, since mitochondrial fusion plays a role in S-phase entry and stress resistance (Mitra et al. 2009), it is attractive to speculate that the mitochondrial effects could indirectly affect Yki's up-regulation of Cyclin E and DIAP. This is supported by our observation that in hpo, opa1 double-mutant clones, Cyclin E expression is suppressed when compared with that seen in hpo mutant clones (Supplemental Fig. S21). The increase in fusion gene levels upon Yki activation is modest. This may be important in producing the observed phenotype of fused but functional mitochondria. Gross overexpression of fusion genes leads to abnormal and globular mitochondria (Supplemental Fig. S20). Moreover, the modification of mitochondrial structure by Yki is accompanied by an up-regulation of antioxidant enzymes and subunits of complex I of the electron transport chain and a dramatic reduction in intracellular ROS. It has recently been demonstrated that other oncogenes (Ras, Raf, and Myc) also reduce ROS, but in our system, these oncogenes do not up-regulate mitochondrial fusion as seen with activation of the Yki pathway, suggesting that different oncogenic pathways could alter ROS by distinct mechanisms (DeNicola et al. 2011). The importance of mitochondrial fusion in Yki signaling is further highlighted by our observation that a reduction in mitochondrial fusion suppresses Yki-mediated growth phenotypes. This observed link between the Hpo/Yki pathway and mitochondrial fusion is of significance to both normal development and cancer biology. Our study reveals that increased mitochondrial fusion and expansion by Yki is evolutionarily conserved and that this function of the Hippo pathway is relevant for both normal and patho–physiological situations.
Materials and methods
Fly stocks
The following stocks were used: sd47M; UAS-sd; UAS-mitoGFP; spa-Gal4; GMR-Gal4; ap-Gal4; Ay-Gal4; dpp-Gal4 (Bloomington Stock Center); UAS-yki (D. Pan); Sd-GFP (A. Spradling); UAS-opa1RNAi (KK105706 and HMS000349); UAS-MarfRNAi (GD40478, KK105261, and JF01650) (multiple opa1 and Marf RNAis were tested for adult eye size suppression and in other assays described in Fig. 5); UAS-wtsRNAi (KK106174 and HMS00026); UAS-ft RNAi (KK108863 and HMS00932); UAS-ykiRNAi (KK104523); UAS-sdRNAi (KK108264); UAS-avlRNAi (KK107264); UAS-scribRNAi (KK106065); FRT hpo 42-47 (I. Hariharan); UAS-ban (S. Cohen); UAS-opa1 (J. Chung); and MARCM stocks (J. Martinez).
Immunohistochemistry
We used the following antibodies: anti-mouse ATP synthase-α (1:100; Mitosciences), anti-rabbit Mn-SODII (1:100; Stressgen), MitoTracker Red (Molecular Probes), anti-rat anti-phospho-histone 3 (1:300), and anti-Yki (1:500; D. Pan). BrdU labeling was done by incubating freshly dissected eye discs in 75 μg/mL BrdU for 30 min, followed by fixation, washing in PBS, and mounting. Antibody staining was performed as described previously (Rogge et al. 1991).
Ay-Gal4 flip-out clones
Flip-out clones were generated in the eye and in the fat body. hs-flp was used to flip out the stuffer cassette from act-FRTyFRT-Gal4 to generate act Gal4, which expresses Gal4 in all cells of the eye imaginal disc and fat body (Ito et al. 1997). The respective crosses were maintained at 18°C. At mid-second instar, a 10-min heat shock at 37°C was applied (Ito et al. 1997). Tissues were dissected in the third instar, fixed, and stained with appropriate antibodies or MitoTracker Red (Invitrogen).
EdU labeling and tissue growth measurements
Appropriate genetic crosses were allowed to lay eggs on grape juice/agar plates, and larvae hatched over a 3-h period were collected and transferred to normal food plates maintained at 29°C. Upon reaching the third instar, larvae were dissected in room temperature Schneider's Drosophila medium, imaginal wing discs were incubated for 30 min in 10 μM EdU, and then discs were fixed and stained according to standard protocol (Click-iT EdU Alexa Fluor 555 imaging kit, Life Technologies). For visualization of GFP, tissue was stained with a monoclonal GFP antibody (Millipore) at a dilution of 1:100. To quantify the number of EdU-positive cells, images were analyzed with ImageJ software. Briefly, automatic thresholds were applied to green and red channels, and the green channel was inverted and subtracted from the red channel to select only EdU spots within the dpp-expressing area. Watershed segmentation was then performed to separate overlapping EdU spots, and particles 10 μm2 in size or greater were counted. To measure tissue sizes, ImageJ was used to automatically threshold the fluorescent channel, and area was measured.
MARCM clones
MARCM clones for wts (third chromosome) and its suppression by opa1RNAi (second chromosome) were done using egg collection in vials for 3 h. Larvae from the crosses were heat-shocked for 45 min at 37°C at 48 h after egg laying. Mid-third instar larvae were collected for dissection, and the discs were fixed and mounted in VectaShield for confocal microscopy. The area under the clone was measured using ImageJ software, and statistical data analysis was performed in Excel.
In situ hybridization
wts mutant clones were generated using the Flp/FRT system by a 30-min heat-shock pulse at 37°C during the mid-second instar. Plasmids for the generation of GFP and Opa1 probes were obtained from Kathy Ngo and the Drosophila Genomics Resource Center (DGRC), respectively. The opa1 probe was labeled with digoxygenin-tagged alkaline phosphatase, and the GFP probe was labeled with fluorescein-tagged HA. For tissue in situ hybridization experiments, third instar larvae from the cross were washed in DEPC-treated water, dissected in Schneider's medium, fixed in 3.7% paraformaldehyde in 100 mM PIPES buffer for 45 min on ice, and subsequently dehydrated in ethanol for 12 h. The following day, discs were subjected to Proteinase K treatment (10 μg/mL) for 4 min, followed by washes in glycine buffer (20 mg/mL). The discs were then incubated in prehybridization buffer (2 h) and then incubated in hybridization buffer for 2 h at 55°C. The probe was prepared in 200 μL of hybridization buffer and heated at 90°C, followed by cooling for 5 min on ice for denaturation. The discs were incubated with probe in hybridization buffer for 18–22 h at 55°C. The following day, the discs were washed multiple times in PBT at 55°C. The signal was amplified with fluorescent-labeled antibodies (fl-HA for GFP and Dig-AP for opa1), and the discs were then mounted in VectaShield. Images were captured using confocal microscopy.
Imaging
Samples were imaged using a Bio-Rad Radiance 2000 confocal with LaserSharp 2000 acquisition software and a Zeiss LSM700 confocal with Zen 2009 acquisition software. Fluorescent intensity quantifications were analyzed by ImageJ software.
Microarray experiments
Eye discs (100 for each genotype) were dissected from GMR-Gal4 (control) and GMR-Gal4, UAS-yki genotypes ∼40 h after pupariation. Total RNA was isolated from the dissected discs (Trizol method and Qiagen RNAeasy kit) and used to generate microarray probes that were hybridized to Drosophila genome 2 arrays (Affymetrix). The GeneChip Operating system (Affymetrix) and dCHIP program (Harvard University) were used to generate pairwise comparisons between the transcription profiles of control and Yorkie overexpressing discs.
ChIP, ChIP–chip, and motif analysis
Wandering third instar larvae were dissected and fixed. Imaginal discs were collected in PBS on ice. Discs were fixed with 1.8% formaldehyde, and cross-linked chromatin was sonicated to an average size of 500 base pairs (bp). Chromatin preparation and ChIPs were performed as described (Estella et al. 2008). Both rabbit anti-Yki (D. Pan) and rabbit anti-GFP (Abcam, ab290) were used at final dilutions of 1:300. Immunoprecipitated DNA and input DNA were amplified for array hybridization using the GenomePlex WGA4 whole-genome amplification kit (Sigma). The samples were then labeled according to Affymetrix protocols and hybridized on Affymetrix GeneChip Drosophila Tiling 2.0R arrays. ChIP–chip for each factor and tissue was performed in biological triplicate. All raw and processed ChIP–chip data are publicly available through the Gene Expression Omnibus (GEO; accession no. GSE26678).
Tiling array data were processed with MAT (model-based analysis of tiling arrays), and peaks were called at a 5% FDR (false discovery rate) using MAT. For a given tissue, peaks were called as shared if the Yki and Sd peaks overlapped by 1 bp or more. Target genes were called based on the transcription start site nearest a given peak. At the 5% FDR threshold, 6713 Sd+Yki peaks were called in the wing disc and 5170 Sd+Yki peaks were called in the eye–antenna disc. Altogether, 5203 genes are called as Sd+Yki targets in either the wing or eye–antenna. Sd+Yki target gene GO (gene ontology) overlap probabilities were calculated from the hypergeometric distribution based on the number of FlyBase GO annotated Sd+Yki target genes in the given category and the total number of Drosophila melanogaster genes in the annotation category.
In vitro selection (or SELEX) was performed with GST-Sd immobilized on glutathione agarose. The input SELEX library sequence was 5′-GTTCAGAGTTCTACAGTCCGACGATCTGGNNNNNNNNNNNNNNNNCCAGCTGTCGTATGCCGTCTTCTGCTTG-3′, where “N” represents A/G/T/C. Eluted DNA was PCR-amplified and prepared for Illumina sequencing by three rounds of PCR with the following primers: 5′-CAAGCAGAAGACGGCATACGA-3′ and 5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA-3′. Illumina sequencing (once for 36 cycles) was performed according to the manufacturer's protocol for small RNA sequencing, as this is optimized for sequencing short DNA fragments. The Sd-binding motif was constructed from the top 300 16-mers selected after two rounds of in vitro selection with full-length GST-tagged Sd. A position weight matrix (PWM) was generated using MEME (Bailey and Elkan 1994) and the following parameters: minimum motif width of 6, maximum motif width of 8, one per sequence.
Gel shift assays were performed essentially as described previously (Estella et al. 2008), except the oligonucleotide probe was dual-labeled with 5′ IRDye 700 (Integrated DNA Technologies), and gels were scanned using the Odyssey infrared imaging system (Li-Cor Biosciences). The TEA domain of Sd, described previously (Halder and Carroll 2001), was expressed in BL21 and purified using Ni-NTA agarose (Qiagen). The following probes were used: Consensus, 5′-TTCGATACACTTGTGGAATGTGTTTGATTTGTTAGCCCCG-3′; Consensus-mutated, 5′-TTCGATACACTTGcatAATaaGTTTGATTTGTTAGCCCCG-3′; Opa1, 5′-GTTTCTTTTTAATATTTAGAATGCACTTTGCAAAGAAGTA-3′; Opa1-mutated, 5′-GTTTCTTTTTAATATTcatAATaaACTTTGCAAAGAAGTA-3′; Marf, 5′-AATCATTGTCAGGGTAAATTCTATTTTTAAATTAGACAAA-3′; and Marf-mutated, 5′- AATCATTGTCAGGGTttATTatgTTTTTAAATTAGACAAA-3′.
MitoTracker and antibody staining of human cells
Flag-tagged YAP2 was cloned into the pLJM1 lentiviral vector, and breast cancer cells stably expressing Flag-YAP2 were selected with puromycin. SUM159PT and MDAMB453 cells as well as culture conditions are described previously (Neve et al. 2006). For MitoTracker staining, human cells plated on coverslips were treated with 250 nm of MitoTracker Red (Invitrogen) in culture medium for 30 min at 37°C. The cells were fixed, washed, and mounted in VectaShield (Vector Laboratories) for taking images.
For antibody staining, cells were fixed in 4% paraformaldehyde for 5 min, washed for 2 min (PBS + 0.05% Triton X-100), permeabilized with PBS + 0.1% TX-100 PBS for 5 min, and washed twice for 5 min each. The cells were blocked with 10% normal goat serum for 30 min, followed by two washes for 5 min each. Cells were then incubated with primary antibody for 1 h at room temperature, followed by two 5-min washes and incubation with the secondary antibody for 1 h at room temperature. Finally, the cells were washed three times for 5 min each and mounted on glass slides with VectaShield (Vector Laboratories). TO-PRO-3 (Invitrogen) was applied in the second wash.
ATP measurements
Drosophila imaginal discs from 50 larvae expressing Yorkie with a T80-Gal4 driver were dissected and collected in lysis buffer, and ATP was extracted according to the manufacturer's instructions using an ATP Bioluminescence assay kit HS II (Roche). ATP was measured by using a luminometer, and the values were normalized with protein amounts measured from the same lysed extracts using a Pierce BCA protein assay kit (Thermo Scientific). ATP measurements from human cells were carried out using the same procedure.
Metabolite measurements
Cells (106) of empty vector or YAP2 transfected cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin, and the medium was replaced 24 h prior to analysis. Cell numbers were counted using a Beckman Coulter Z1 particle counter, and glucose, lactate, glutamine, and glutamate concentrations were simultaneously analyzed using a NOVA Basic 4 Bioanalyzer. Metabolite consumption/production was expressed as moles per 106 cells per hour.
Electron microscopy
Drosophila tissues were dissected in PBS and fixed in 2% glutaraldehyde and 2.5% formaldehyde in PBS, and human cells were fixed in 2% glutaraldehyde in PBS. Embedding, thin sectioning, and staining were carried out according to standard protocol (Afzelius and Maunsbach 2004). The sections were stained with uranyl acetate and lead citrate and examined on a JEOL 100CX electron microscope at 80 kV.
ROS measurements
The ROS dye (CM-H2DCFDA, Invitrogen) was dissolved in DMSO and added to serum-containing medium to a final concentration of 5 μM. Cells were incubated with the dye-containing medium for 20 min at 37°C. Cells were rinsed twice with PBS, trypsinized, and recovered in full medium followed by a 5-min spin down at 200g. Cells were resuspended in 400 μL of PBS and used for flow cytometric analysis using the FITC channel.
Reporter assays
Putative enhancers upstream of opa1-like (chromosome 2R, 10123270–10123692) and Marf (chromosome X, 6259520–6260412) were inserted between the MluI and XhoI sites of the pGL2 vector to generate luciferase reporter constructs. The expanded and 3xSd reporter constructs, also pGL-based, and pIE7-Flag-Sd and pAc-HA-Yki expression constructs were described previously (Nicolay et al. 2011). S2 cells were transfected with an individual reporter plasmid, Sd and/or Yki expression plasmids, and a Renilla luciferase plasmid (pAC-Rluc) to control for transfection efficiency. Transfections were performed with Effectene transfection reagent (Qiagen), and reporter assays were carried out in triplicate 48 h post-transfection. Reporter assays were performed using the Dual-Glo Luciferase assay system (Promega).
Acknowledgments
We thank K. Ngo, D. Pan, I. Hariharan, A. Spradling, J. Chung, S. Cohen, and the stock centers of Bloomington, the National Institute of Genetics (Tokyo), and the Vienna Drosophila RNAi Center (Vienna) for providing Drosophila stocks and reagents. We thank H. Richardson, H. McNeill, and T. Orr-Weaver for Cyclin E antibody. We gratefully acknowledge Xiaoyue Wang's help in the luciferase reporter assays. We thank and acknowledge Derek Cheung's contribution in analyzing data. We thank W. Freije for help with microarray analysis, and Genaro Villa for help with creating YAP2 constructs. We acknowledge the help from the microarray core facility in the Department of Pathology at the University of California at Los Angeles. We also thank Sirus Kohan and the Brain Research Institute EM facility for assistance with electron microscopy. K.T.J. is supported by a post-doctoral fellowship (#PF-10-130-01-DDC) from the American Cancer Society. This work is supported by National Institutes of Health Grant RO1EY008152 to U.B.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.183061.111.
References
- Afzelius BA, Maunsbach AB 2004. Biological ultrastructure research; the first 50 years. Tissue Cell 36: 83–94 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Baltzer C, Tiefenbock SK, Marti M, Frei C 2009. Nutrition controls mitochondrial biogenesis in the Drosophila adipose tissue through Delg and cyclin D/Cdk4. PLoS ONE 4: e6935 doi: 10.1371/journal.pone.0006935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF 2006. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol 16: 2101–2110 [DOI] [PubMed] [Google Scholar]
- Chan DC 2006. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol 22: 79–99 [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M 2006. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M 2008. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci 105: 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. 2011. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA 2006. From cell structure to transcription: Hippo forges a new path. Cell 124: 267–273 [DOI] [PubMed] [Google Scholar]
- Estella C, McKay DJ, Mann RS 2008. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell 14: 86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T 2003. Oxidant signals and oxidative stress. Curr Opin Cell Biol 15: 247–254 [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A 2008. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol 18: 435–441 [DOI] [PubMed] [Google Scholar]
- Goyal G, Fell B, Sarin A, Youle RJ, Sriram V 2007. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell 12: 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Carroll SB 2001. Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development 128: 3295–3305 [DOI] [PubMed] [Google Scholar]
- Hariharan IK 2006. Growth regulation: A beginning for the hippo pathway. Curr Biol 16: R1037–R1039 doi: 10.1016/j.cub.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114: 457–467 [DOI] [PubMed] [Google Scholar]
- Hay BA, Guo M 2003. Coupling cell growth, proliferation, and death. Hippo weighs in. Dev Cell 5: 361–363 [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122: 421–434 [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124: 761–771 [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9: 534–546 [DOI] [PubMed] [Google Scholar]
- Mandal S, Guptan P, Owusu-Ansah E, Banerjee U 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J 2009. A hyperfused mitochondrial state achieved at G1–S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci 106: 11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV 2011. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev 25: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G 2006. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol 16: 1895–1904 [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD 2008. In vivo regulation of Yorkie phosphorylation and localization. Development 135: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Mandal S, Banerjee U 2008. Distinct mitochondrial retrograde signals control the G1–S cell cycle checkpoint. Nat Genet 40: 356–361 [DOI] [PubMed] [Google Scholar]
- Pan D 2007. Hippo signaling in organ size control. Genes Dev 21: 886–897 [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS 2009. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev 23: 2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge RD, Karlovich CA, Banerjee U 1991. Genetic dissection of a neurodevelopmental pathway: Son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell 64: 39–48 [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H 2006. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol 16: 2081–2089 [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM 2006. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126: 767–774 [DOI] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, et al. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 28: 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM 2009. Fussy mitochondria fuse in response to stress. EMBO J 28: 1533–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445–456 [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D 2008. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 14: 388–398 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J 2008. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell 14: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yue T, Jiang J 2009. Hippo signaling pathway and organ size control. Fly (Austin) 3: 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lei QY, Guan KL 2008. The Hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr Opin Cell Biol 20: 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]