In loss-of-function Blimp1/Prdm1-null mouse embryos, spiral artery-associated trophoblast giant cells (SpA-TGCs) are disrupted and the spongiotrophoblast layer is compromised, resulting in mid-gestation lethality. Here, the authors investigate the cause of these tissue disturbances. They identify a Blimp1/Prdm1-positive multipotent progenitor cell in the spongiotrophoblast that sustains the growth of the placenta and find that Blimp1/Prdm1 is genetically required for the formation of SpA-TGcs.
Keywords: Blimp1/Prdm1, placenta, spiral artery, endovascular trophoblast giant cells, trophoblast stem cells
Abstract
Developmental arrest of Blimp1/Prdm1 mutant embryos at around embryonic day 10.5 (E10.5) has been attributed to placental disturbances. Here we investigate Blimp1/Prdm1 requirements in the trophoblast cell lineage. Loss of function disrupts specification of the invasive spiral artery-associated trophoblast giant cells (SpA-TGCs) surrounding maternal blood vessels and severely compromises the ability of the spongiotrophoblast layer to expand appropriately, secondarily causing collapse of the underlying labyrinth layer. Additionally, we identify a population of proliferating Blimp1+ diploid cells present within the spongiotrophoblast layer. Lineage tracing experiments exploiting a novel Prdm1.Cre-LacZ allele demonstrate that these Blimp1+ cells give rise to the mature SpA-TGCs, canal TGCs, and glycogen trophoblasts. In sum, the transcriptional repressor Blimp1/Prdm1 is required for terminal differentiation of SpA-TGCs and defines a lineage-restricted progenitor cell population contributing to placental growth and morphogenesis.
Growth and survival of the mammalian embryo within the maternal uterine environment depends on development of the placenta. Reciprocal signaling cues between closely interacting maternal, extraembryonic, and embryonic tissues govern the balance between cell proliferation and terminal differentiation of trophoblast progenitors during formation of this specialized organ necessary to promote maternal–fetal exchange, provide an immunological barrier, and produce hormones that systemically influence maternal physiology during pregnancy (Watson and Cross 2005; Moffett and Loke 2006).
Shortly after implantation of the blastocyst, mural trophodermal cells differentiate into highly polyploid migratory primary parietal trophoblast giant cells (P-TGCs). In contrast, the polar trophectoderm immediately overlying the inner cell mass continues to proliferate, giving rise to the extraembryonic ectoderm (ExE) and its derivatives, the so-termed ectoplacental cone (EPC) and the chorionic ectoderm. Diverse trophoblast cell subtypes identified on the basis of restricted gene expression profiles, cell morphology, and localization become allocated in an orderly sequence (Simmons and Cross 2005; Simmons et al. 2007; Hu and Cross 2010). The EPC generates secondary P-TGCs and the highly invasive spiral artery-associated TGCs (SpA-TGCs) that migrate into the deciduum to surround, invade, and remodel the incoming maternal vasculature (Adamson et al. 2002; Hemberger et al. 2003; Simmons and Cross 2005). Glycogen trophoblasts (Gly-Ts) are specified within the early spongiotrophoblast cell population (Bouillot et al. 2006; Tesser et al. 2010) and at later stages invade into the maternal tissue. The canal-associated TGCs (C-TGCs) differentiate at later developmental stages to line the blood canals of the spongiotrophoblast layer (Adamson et al. 2002). Derivatives of the chorionic ectoderm closely interact with fetal endothelial cells during formation of the labyrinth layer. The chorionic trophoblast progenitors differentiate into polyploid mononuclear sinusoidal cells (S-TGCs) and multinucleated syncitiotrophoblasts (Syn-Ts) lining maternal blood sinuses (Watson and Cross 2005; Simmons et al. 2008a). Continuous growth of the labyrinth layer is necessary for efficient maternal–fetal exchange to support the dramatic growth of the embryo during the second half of gestation.
Gene targeting experiments have identified transcriptional regulators and signaling pathways required for development of the trophoblast cell lineage. The homeodomain protein Cdx2 and its downstream target, the T-box family member Eomesodermin (Eomes), are essential to promote trophectoderm development shortly after implantation (Russ et al. 2000; Niwa et al. 2005; Strumpf et al. 2005). At slightly later stages, Eomes expression marks the distal ExE (Arnold et al. 2009). Self-renewing stem cells within the ExE are maintained in the undifferentiated state by the combined activities of Fgf4 and localized Nodal signaling from the underlying epiblast (Tanaka et al. 1998; Guzman-Ayala et al. 2004). Trophoblast stem (TS) cell cultures deprived of Fgf and Activin/Nodal spontaneously differentiate to become mature TGCs (Tanaka et al. 1998; Simmons et al. 2007). Thus, terminal differentiation may represent the default pathway, whereas continuous exposure to growth factor signaling is required to maintain stem cell capabilities.
Placental morphogenesis also crucially depends on regulatory networks governing specification of mature post-mitotic TGC subtypes at the correct time in the appropriate placental location. Formation of polyploid TGCs at the periphery of the EPC is controlled by the antagonistic actions of the basic helix–loop–helix (bHLH) family members Hand1 and Ascl2. Hand1 promotes TGC formation (Riley et al. 1998), whereas Ascl2, coexpressed with Hand1 in the EPC, suppresses TGC terminal differentiation (Guillemot et al. 1994; Tanaka et al. 1997; Scott et al. 2000). A key feature of terminal differentiation is that TGCs exit from the cell cycle and undergo multiple rounds of DNA endoreduplication. Cyclin E is required to promote endoreduplication within TGCs (Geng et al. 2003; Parisi et al. 2003). Endoreduplication has also been causally linked to the cell cycle regulator Geminin (Gonzalez et al. 2006). Loss of Geminin function causes the totipotent cells present at the morula stage to undergo endoreduplication and acquire a TGC phenotype.
The transcriptional repressor Blimp1 (encoded by the Prdm1 gene), originally identified as a master regulator of plasma cell terminal differentiation, also controls gene expression profiles in T-cell subsets, macrophages, the sebaceous gland, and skin epidermis (Horsley et al. 2006; Magnusdottir et al. 2007). Within lineage-restricted T cells, Blimp1 plays multiple roles controlling the balance between TH1 and TH2 subsets, memory and effector CD8 T cells, and maturation of CD4 follicular helper T cells (Kallies and Nutt et al. 2007; Nutt 2007; Martins and Calame 2008; Welch 2009; Crotty et al. 2010). In the early mouse embryo, Blimp1 governs primordial germ cell (PGC) specification (Ohinata et al. 2005; Vincent et al. 2005) and regulates development of the forelimb and caudal pharyngeal arches (Robertson et al. 2007). Recent studies demonstrate that Blimp1 plays an essential role in reprogramming of the intestinal epithelium during the suckling-to-weaning transition (Harper et al. 2011).
Blimp1 contains an N-terminal PR/SET domain and five C-terminal C2H2 zinc fingers that mediate nuclear import and DNA binding. Its ability to mediate gene silencing and reorganize chromatin architecture at specific target sites depends on recruitment of epigenetic partners. Associations with histone deactylases (HDACs), the G9a methyl transferase, and the lysine-specific demethylase LSD1 have been shown to regulate plasma cell maturation (Bikoff et al. 2009). Complexes with the arginine methyltransferase Prmt5 govern epigenetic changes in the germ cell lineage (Ancelin et al. 2006). Considerable evidence suggests that Blimp1 transcriptional targets are cell type-specific. Thus, in B cells, macrophages, and sebaceous gland progenitors, Blimp1 directly represses c-Myc expression to arrest cell cycle progression (Horsley et al. 2006; Martins and Calame 2008). However, c-Myc is not a transcriptional target in T lymphocytes. Rather, Blimp1 blocks proliferation by direct repression of the T-cell cytokine IL2. Blimp1 regulates the key targets Ifnγ, Tbx21, and Bcl6 to shift the balance between TH1/TH2 lineages (Martins and Calame 2008). Its ability to silence Bcl6 expression is also crucial during terminal differentiation of plasma cells and CD4+ follicular helper T lymphocytes (Crotty et al. 2010). In contrast, Bcl6 is not a transcriptional target in mature natural killer (NK) effector cells (Kallies et al. 2011). Genome-wide chromatin immunoprecipitation (ChIP) experiments demonstrate widespread promoter occupancy at hundreds of candidate target genes (Doody et al. 2010). Context-dependent local chromatin structure probably also has an impact on target site selection and recruitment of corepressors.
Blimp1/Prdm1 mutant embryos arrest at around embryonic day 10.5 (E10.5) due to placental defects (Vincent et al. 2005; Robertson et al. 2007). The present experiments investigate the underlying causes of these tissue disturbances. We demonstrate that the invading SpA-TGCs display robust Blimp1 expression and that Blimp1 functional loss selectively disrupts specification of this discrete TGC subtype. Transcriptional profiling experiments identified additional SpA-TGC lineage-restricted marker genes that potentially regulate placental morphogenesis. Blimp1 expression also delineates a discrete population of diploid cells present within the spongiotrophoblast layer at all stages examined. We engineered a novel Prdm1.Cre-IRES-LacZ reporter allele for fate-mapping studies. The Blimp1+ subset of spongiotrophoblasts gives rise exclusively not only to the invading SpA-TGC, but also the Gly-T cell population and terminally differentiated C-TGCs. Blimp1 functional activities in diverse trophoblast cell types regulate their crucial contributions to placental growth and development.
Results
Blimp1 is expressed by multiple trophoblast cell subtypes in the developing placenta
Several trophoblast cell subtypes have been defined on the basis of cell morphology, gene expression profiles, and tissue localization (Simmons and Cross 2005; Simmons et al. 2007). To examine temporally and spatially restricted Blimp1 expression patterns by trophoblast cell subpopulations in the developing placenta, we performed in situ hybridization and immunostaining experiments. Recent evidence suggests that Blimp1 regulates NK cell maturation and effector functions (Smith et al. 2010; Kallies et al. 2011). Additionally, uterine NK cells may be required for remodeling of maternal blood vessels during placentation (Adamson et al. 2002; Moffett and Loke 2006). To exclude possible maternal contributions, we analyzed expression of a paternally inherited hypomorphic Blimp1.gfp reporter allele (Kallies et al. 2004; Robertson et al. 2007).
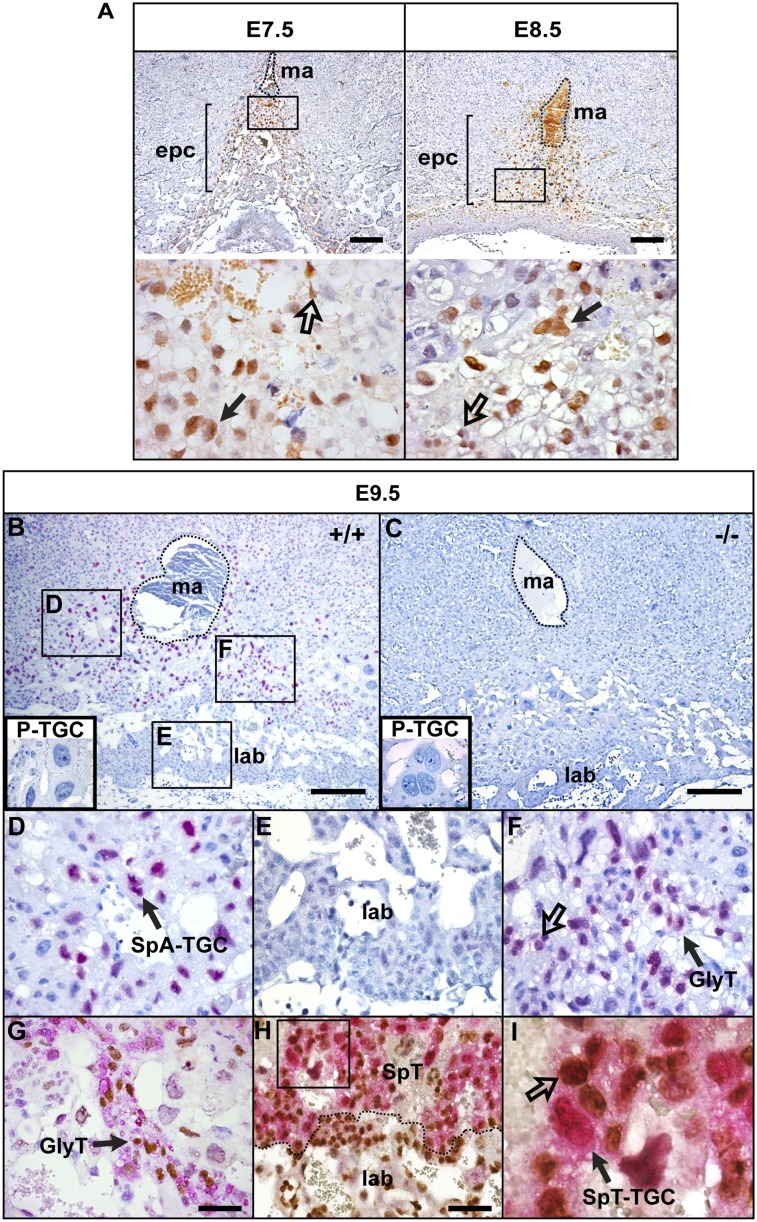
Gfp transcripts were detectable at E7.5 in a subset of cells within the EPC (Supplemental Fig. 1A). Slightly later (E8.5), we observed expression within a subset of cells lying subjacent to the P-TGC layer as well as the invading SpA-TGCs situated above the P-TGC layer (Supplemental Fig. 1B). However, by E9.5, the signal was predominantly restricted to small clusters of mature SpA-TGCs associated with maternal blood vessels (Supplemental Fig. 1C). Similarly, from E7.5 onward, immunohistochemical experiments reveal strong nuclear staining in the forming SpA-TGCs (Fig. 1A), with characteristic polyploid nuclei, which surround the incoming maternal arteries within the uterine stromal tissue (Adamson et al. 2002; Hemberger et al. 2003). At E9.5, robust expression continues in SpA-TGCs, whereas, in contrast, the secondary P-TGCs lack Blimp1 expression (Fig. 1B). Similarly, derivatives of chorionic trophoblast progenitors, including the S-TGCs and Syn-Ts that line the maternal labyrinth blood sinuses (Watson and Cross 2005; Simmons et al. 2008a), are devoid of expression (Fig. 1B).
Figure 1.
Characterization of Blimp1-expressing trophoblast cell subpopulations in the developing placenta. Immunohistochemical staining for nuclear Blimp1. (A) Midsections though the EPC at E7.5 and E8.5. Blimp1 expression (brown) is detectable in diploid trophoblasts (unfilled arrow) and TGCs (filled arrow) in the EPC. The position of the central maternal artery is outlined. High-magnification images of the boxed areas are shown below. (B–F) At E9.5, Blimp1 (red) is strongly expressed by the SpA-TGCs surrounding the maternal arteries (D) and diploid cells (unfilled arrow) and clusters of vacuolated Gly-T (filled arrow) (F) , but labyrinthine trophoblasts (E), P-TGCs (B, box in the bottom left corner) and Prdm1−/− mutant placenta lack Blimp1 expression. (G) PAS staining (pink), in combination with Blimp1 immunohistochemical staining (brown), confirms that double-positive vacuolated cells are Gly-Ts. (H) Numerous Ki67+ (brown) cells present in the labyrinth are Blimp1−, whereas Blimp1+ SpA-TGCs are quiescent (H,I). Ki67 staining confirms that ∼10%–20% of the diploid cells present within the spongiotrophoblast layer are double Blimp1+ (red)/Ki67+ (brown)-positive (unfilled arrow). Bars: A–C, 200 μm; G,H, 50 μm. (ma) Maternal artery; (SpT) spongiotrophoblast; (lab) labyrinth trophoblast.
Interestingly, from E7.5 onward, we consistently observed a subset of diploid Blimp1+ cells scattered within the forming spongiotrophoblast layer (Fig. 1A), as well as pockets of compact Blimp1+ cells having the highly characteristic morphology of Gly-T cells (Fig. 1F). Gly-T cells are readily identified by PAS staining (Bouillot et al. 2006; Tesser et al. 2010). Double-labeling experiments confirmed that Blimp1 expression marks all Gly-T cells in the forming spongiotrophoblast layer (Fig. 1G). Next, to assess the proliferative activities of the Blimp1+ diploid cells, we stained for Ki67. As expected, at E9.5 in double-labeling experiments, Blimp1+ mature SpA-TGCs are quiescent (Fig. 1I). Within the labyrinth, we observed a high proportion of Ki67+ cells but no Blimp1 expression (Fig. 1H). As shown in Figure 1I, ∼10%–20% of the Blimp1+ diploid cells scattered within the spongiotrophoblast layer are also Ki67+. These results strongly suggest that the Blimp1+ diploid population is indeed capable of self-renewal.
Blimp1 loss of function disrupts specification of the invasive SpA-TGC population
Blimp1-deficient embryos arrest at E10.5 due to placental insufficiency (Vincent et al. 2005). One day earlier, at E9.5, the characteristic morphological disturbances include extensive hemorrhaging of maternal blood within decidual tissues above the placenta (Fig. 2A). To further investigate the underlying causes of these abnormalities, we examined a panel of markers defining the major trophoblast subtypes. The transcription factor Gcm1 (Anson-Cartwright et al. 2000) and its target, SynB, are correctly expressed in E9.5 mutant placenta, but in contrast to wild type, the Gcm1+ Syn-T cell population fails to involute and form characteristic long narrow branches (Supplemental Fig. 2A). Expression of a paternally inherited Flk.LacZ allele marking endothelial tissue (Shalaby et al. 1995) confirmed that fetal capillaries fail to extend into the labyrinth tissue (Supplemental Fig. 2A). Rather, fetal blood cells identified by ɛ-globin expression are largely restricted to the base of the labyrinth (Supplemental Fig. 2A).
Figure 2.
Blimp1 inactivation eliminates trophoblast invasion into the maternal decidua, resulting in compaction of the labyrinth. (A) H&E sections of E9.5 wild-type and Blimp1-deficient placentae. (B–E) Expression of trophoblast lineage-restricted markers was analyzed at E9.5 by RNA in situ hybridization. Prl5a1 (B), Prl3b1 (C), Tpbpα (D), and Prl2a1 (E) identify both spongiotrophoblasts and the invading SpA-TGC population (red arrows). In Prdm1−/− mutants, expression is confined to the spongiotrophoblast layer, and the SpA-TGC population is absent. The position of the central maternal artery is outlined. (F) PAS staining reveals that Gly-T cells are correctly specified but are localized to the dense compacted spongiotrophoblast layer. Immunohistochemical labeling of CK (cytotrophoblasts) (G) and histochemical staining for AP (maternal cells) (H) highlight the absence of trophoblast invasion surrounding the maternal artery (traced by dotted line) in the Prdm1−/− placenta. Bar, 200 μm. (Dec) Maternal decidua; (ma) maternal artery; (SpT) spongiotrophoblast; (lab) labyrinth trophoblast.
Next, we examined expression of prolactin-related family members (Prls) selectively expressed by trophoblast subtypes in the spongiotrophoblast layer (Simmons et al. 2008b). Expression of Prl5a1 and Prl3b1 as well as Tpbpα by cells of the dense spongiotrophoblast cell layer (Simmons et al. 2008b) is unaffected (Fig. 2B–D). Similarly, expression of Prl2a1, a marker of Gly-T cells (Simmons et al. 2008b), is retained within the spongiotrophoblast layer (Fig. 2E). Additionally, PAS staining shows that the Gly-T cells differentiate appropriately (Fig. 2F). However, in the absence of Blimp1, this population becomes intermixed with cells of the compacted spongiotrophoblast layer. As assessed by expression of Prl3d1 (PL-1) or Prl2c2 (Plf) (Supplemental Fig. 2), Blimp1 functional loss has no noticeable effect on the formation of the secondary P-TGC population.
In striking contrast, the SpA-TGCs coexpressing Prl5a1, Prl2a1, Prl3b1, and Tpbpα, which normally invade the deciduum to surround the central maternal artery and ingressing spiral arteries, are entirely absent (Fig. 2A–E). The prominent cone-shaped spongiotrophoblast layer normally seen between the outer P-TGCs and the proximal labyrinth is abnormally flattened and unevenly distributed (Fig. 2A). To further investigate defective trophoblast invasion into the maternal tissue, we stained for cytokeratin (CK), a pan-trophoblast marker. The cone of invasive CK+ trophoblast cells that normally surrounds and migrates up the prominent invading central maternal artery is entirely absent (Fig. 2G). Additionally, we examined the pattern of alkaline phosphatase (AP) staining in maternal decidual tissue immediately surrounding the invading TGCs. In mutant placentae, maternal AP+ cells become closely and inappropriately juxtaposed to the central maternal artery (Fig. 2H). Collectively, these results demonstrate that Blimp1 expression is essential for specification of the SpA-TGCs.
Transcriptional profiling experiments reveal misregulated patterns of gene expression
To identify Blimp1-dependent transcripts and potentially new markers specific for the SpA-TGC lineage, we used the Illumina array platform to compare wild-type versus mutant transcripts in E9.5 placental tissue (Supplemental Fig. 3). We identified a modest number of differentially expressed genes (Illumina DiffScore >13, equivalent to P < 0.05) widely distributed throughout the genome (Supplemental Fig. 3A). Of these, 12 were up-regulated and 24 were down-regulated. None of these differentially expressed genes correspond to previously described Blimp1 targets (Doody et al. 2010). Limited evidence of common ontological function was noted among some of these genes (e.g., protease activity). However, functional annotation clustering analysis using DAVID 6.7 (Huang et al. 2009) did not identify significant enrichment of any ontological category due to the small number of significantly altered genes. Candidate misregulated genes were validated by quantitative PCR (Supplemental Fig. 3A).
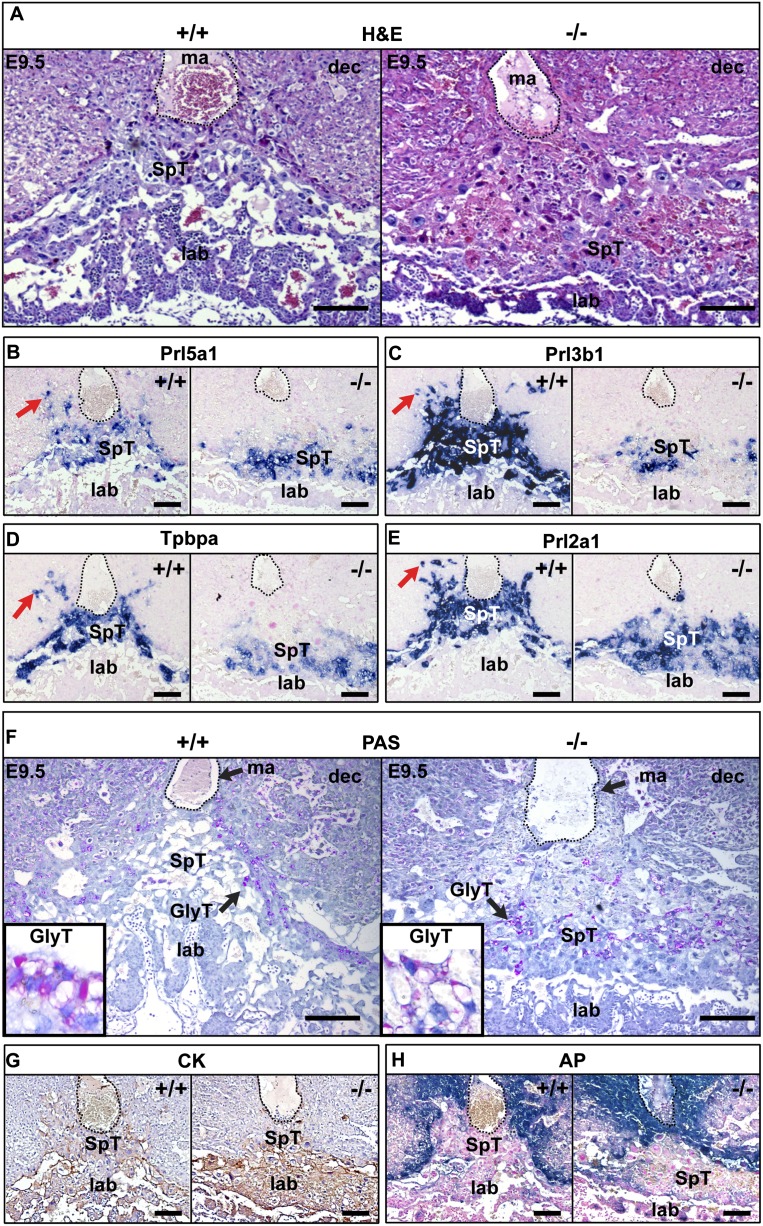
Changes in expression patterns were also assessed by in situ hybridization analysis (Fig. 3). Prl7b1 expression is initially induced in a subset of cells within the EPC at E7.5 (Simmons et al. 2008b) and by E9.5 marks cells at the periphery of the spongiotrophoblasts and invading SpA-TGCs (Fig. 3). Consistent with results above, Prl7b1 expression levels are >1000-fold reduced by quantitative PCR (Supplemental Fig. 3), and transcripts are undetectable in Blimp1-deficient placentae (Fig. 3). Interestingly, the regulator of G protein signaling family member Rgs5, a key regulator of vascular remodeling and pericyte maturation (Manzur and Ganss 2009), also shows significantly down-regulated expression. Rgs5 is also selectively expressed within the SpA-TGC trophoblasts at E9.5, and mutant placentae entirely lack Rgs5+ trophoblasts (Fig. 3). The matricellular protein Lipocalin 7 (Lnc7/Tinagl), known to contribute important proangiogenic functions in vascular microenvironments (Brown et al. 2010), is also robustly expressed by SpA-TGCs (Fig. 3) and largely absent in mutant placentae. Interestingly, proprotein convertase Pace4/Pcsk6 transcripts normally expressed in the invading SpA-TGCs are also undetectable in mutant sections (Fig. 3).
Figure 3.
Blimp1 function is essential for terminal differentiation of endovascular TGCs. RNA in situ hybridization reveals complete loss of Prl7b1, Rgs5, and Pace4/Pcsk6 and reduced expression of Lcn7/Tinagl, consistent with a failure to specify the SpA-TGC population. In contrast, Mmp1a expression is up-regulated but confined to the compacted spongiotrophoblast layer. Similarly, Cts8 expression, normally present in a subpopulation of migratory trophoblasts at the edges of the ecoplacental cone, is up-regulated and confined to the compacted mutant spongiotrophoblast layer. Bar, 200 μm.
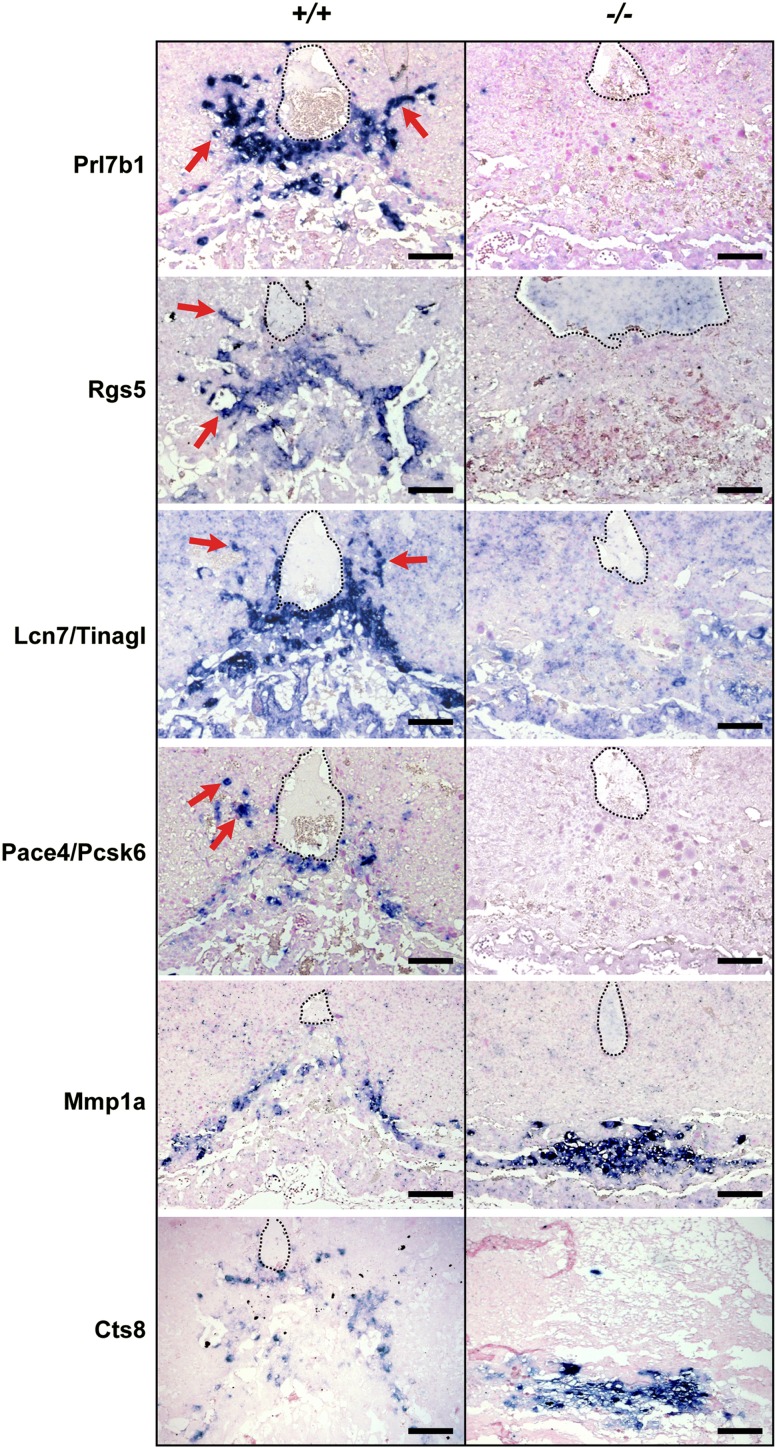
In contrast, loss of Blimp1 results in up-regulated expression of the metalloprotease Mmp1a (Fig. 3). Mmp1a transcripts normally mark a small population of spongiotrophoblast cells lying immediately proximal to the P-TGC layer (Fig. 3; Balbin et al. 2001). In contrast, here we observed increased numbers of Mmp1a+ cells in the flattened mutant spongiotrophoblast layer. The placenta-specific cathepsin Cts8, normally expressed in migratory trophoblasts (Hemberger et al. 2000; Screen et al. 2008), is also significantly up-regulated (Fig. 3; Supplemental Fig. 3). Cts8 overexpression in cultured TS cells results in increased cell and nuclear size, suggestive of giant cell differentiation (Screen et al. 2008). If Blimp1 normally represses Cts8 expression, its absence could potentially lead to precocious giant cell differentiation and loss of invasive potential. To examine this possibility, we compared nuclear sizes in the spongiotrophoblast layer of wild-type and mutant placentae (Fig. 4A). Nuance analysis of DAPI-stained sections at E9.5 revealed no significant differences in nuclear size profiles (Fig. 4B). Thus, Blimp1 functional loss has no global effect on endoreduplication and terminal giant cell differentiation.
Figure 4.
Loss of invasive behavior is not associated with precocious TGC differentiation. (A) H&E and DAPI staining of E9.5 placental sections demonstrates compaction and paucity of the spongiotrophoblast layer. High-magnification images of the boxed areas are shown below. Bar, 200 μm. (B) Morphometic analysis of DAPI-stained nuclei within the spongiotrophoblast layer demonstrates that the distribution of nuclear sizes in Prdm1+/+ and Prdm1−/− placenta are indistinguishable. (B) Data represent mean ± SEM of five fields. (A) Typical DAPI-stained fields from each genotype are indicated in the bottom panel.
Characterization of lineage-restricted Blimp1+ progenitors present within the spongiotrophoblast layer
Formation of the mature chorio-allantoic placenta depends on the continued ability of diploid trophoblast stem cells within both the spongiotrophoblast and labyrinth layers to undergo multiple rounds of proliferation. Intriguingly, at all stages examined, we consistently observed a subpopulation of diploid Blimp1+/Ki67+ cells present within the developing spongiotrophoblast layer (Fig. 1H,I).
Unfortunately, the Prdm1.Cre BAC transgene (Ohinata et al. 2005) that allowed fate mapping of Blimp1+ cells in the embryo proper (Robertson et al. 2007) is not expressed in the developing placenta. To test whether Blimp1+Ki67+ cells are lineage-restricted unipotent SpA-TGC progenitors or, alternatively, may give rise to multiple spongiotrophoblast subtypes, we generated a novel Prdm1.Cre.LacZ reporter allele (Prdm1Cre.LacZ) by inserting a Cre-IRES-nLacZ cassette into the first coding exon of the Prdm1 locus (Fig. 5A).
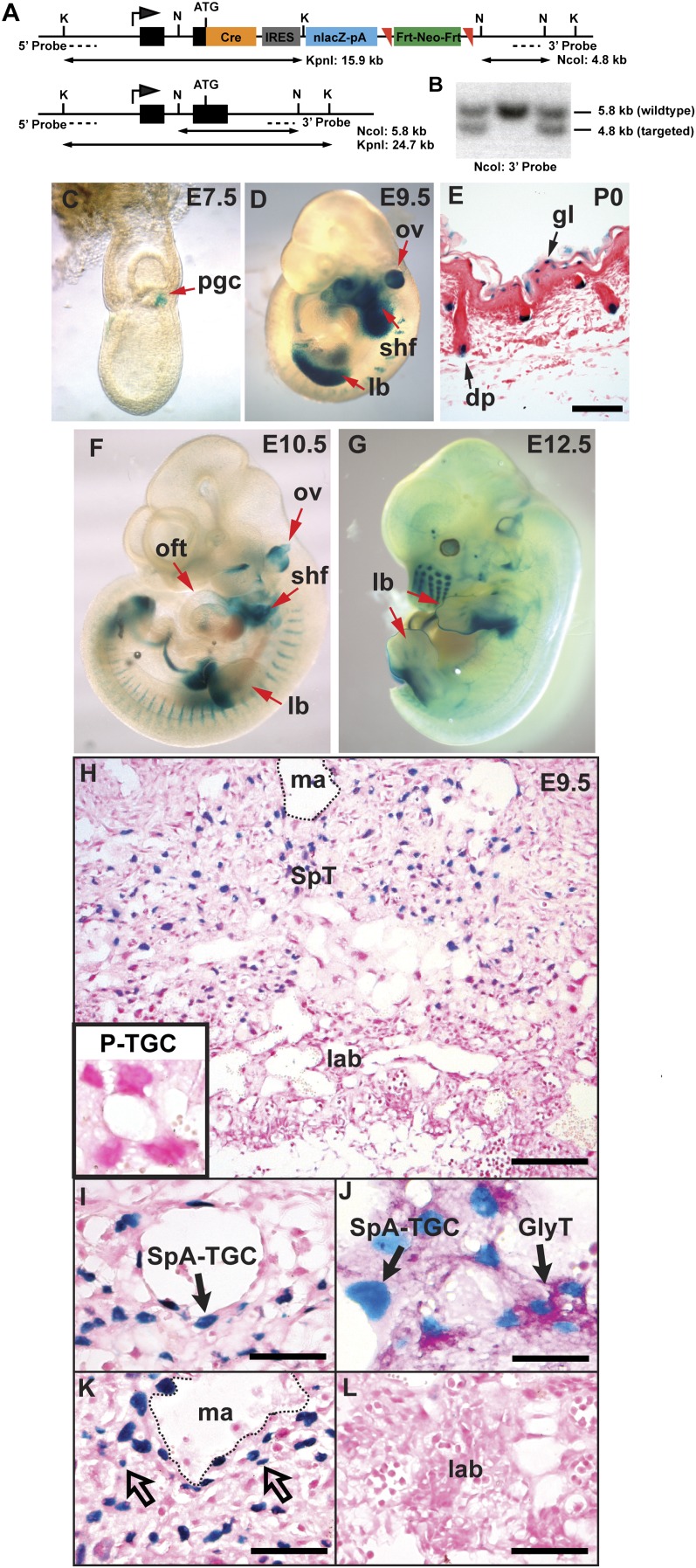
Figure 5.
Generation and validation of a dual-purpose Prdm1.Cre-LacZ reporter allele. (A) Targeting strategy for generating the Prdm1-Cre-IRES-nlacZ allele. Mutant (top) and wild-type (bottom) alleles with Southern blotting restriction enzyme sites, fragment sizes, and location of probes indicated. (K) KpnI; (N) NcoI; (ATG) Prdm1 translation initiation methionine codon. (B) Southern blot screening of embryonic stem (ES) cell clones. (C–G) X-gal staining of Prdm1 Cre-LacZ embryos to monitor endogenous Prdm1 transcriptional activity. (C) At E7.5, PGCs are readily visible. (D) E9.5 LacZ expression in the otic vesicle (ov), emerging forelimb bud (lb), and SHF faithfully reflects previously documented sites of mRNA expression. (E) LacZ staining of postnatal day 0 (P0) back skin identifies Prdm1-positive dermal papillae (dp) and granular layer keratinocytes (gl). Prdm1 Cre-LacZ reporter expression in E10.5 (F) and E12.5 (G) embryos demonstrating down-regulation of transcriptional activity in the forelimb and SHF. (H) LacZ staining of the E9.5 Prdm1 Cre-LacZ placenta shows faithful reporter gene expression. (H,L) The P-TGCs and labyrinth region are devoid of LacZ activity. (I) Nuclear LacZ expression marks the SpA-TGC associated with maternal blood vessels. (J) PAS staining identifies clusters of LacZ+ Gly-T cells. (K) Small diploid cells with blue nuclei (unfilled arrows) are interspersed with nonlabeled cells throughout the spongiotrophoblast layer. Bars: E,I,K,L, 100 μm; H, 200 μm; J, 50 μm. (Ma) Maternal artery; (SpT) spongiotrophoblast; (lab) labyrinth trophoblast.
To test expression of the nuclear LacZ reporter cassette, we mated Prdm1Cre.LacZ/+ males to wild-type females and stained the resulting embryos. As expected, at E7.5, whole-mount staining reveals LacZ activity confined to the Blimp1+ PGCs at the proximal end of the primitive streak (Fig. 5C). Later, at E9.5 (Fig. 5D), we also found that LacZ precisely mirrors dynamic patterns of endogenous Blimp1 expression within the second heart field (SHF), limb buds, and otic vesicle (Vincent et al. 2005; Robertson et al. 2007). In postnatal skin (Fig. 5E), LacZ activity identifies the dermal papillae of the hair follicles as well as the granular layer keratinocytes (Magnusdottir et al. 2007; Robertson et al. 2007). Importantly, we also observed at E10.5 that LacZ reporter activity accurately reflects down-regulated expression in the posterior forelimb and SHF (Fig. 5F; Robertson et al. 2007).
Similarly, we observed faithful expression of the nuclear LacZ reporter cassette in the developing placenta that tightly overlaps with endogenous Blimp1 immunoreactivity (Figs. 1B, 5H). At E9.5, LacZ precisely labels the polyploid nuclei of the SpA-TGCs surrounding the maternal blood vessels (Fig. 5I). Similarly, double staining for LacZ and PAS reactivity identifies the Gly-T cells (Fig. 5J). Scattered diploid nuclear LacZ cells are present within the spongiotrophoblast layer (Fig. 5K), but we observed no staining within the labyrinth (Fig. 5L).
Next, we examined stage-matched E10.5 and E12.5 Prdm1Cre.LacZ/+ embryos alongside those recovered from Prdm1Cre.LacZ/+animals crossed to R26R reporter mice (Soriano 1999). As expected, transient Prdm1.Cre expression accurately fate-maps descendents of the zone of polarizing activity (ZPA) that ultimately give rise to the bulk of the posterior tissue of the forelimbs and hindlimbs, as well as the descendents of the SHF giving rise to the right ventricle (Supplemental Fig. 4A). Notably, transient Prdm1.Cre expression in endothelial progenitors also efficiently marks the embryonic and allantoic vasculature (Supplemental Fig. 4A,B).
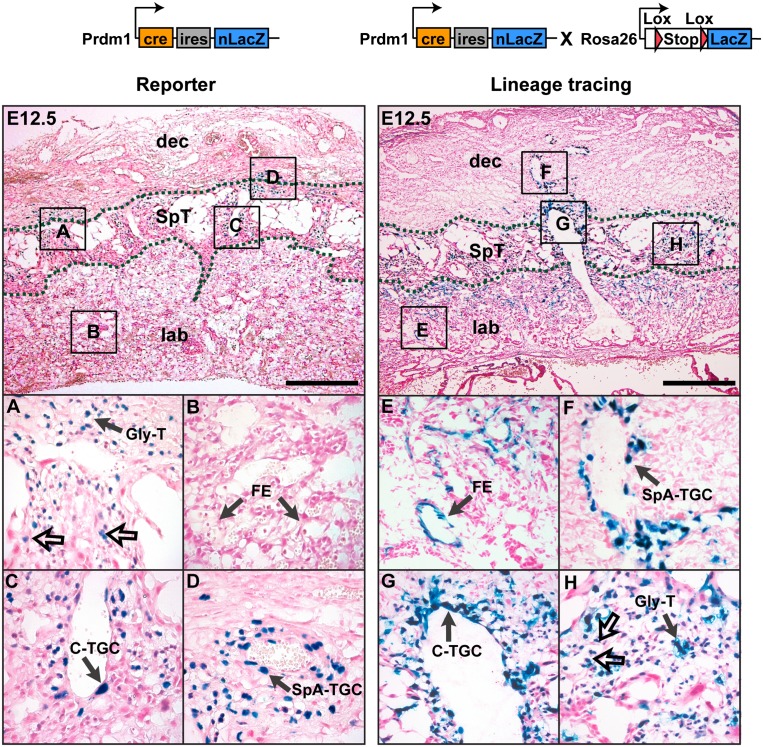
To fate-map Blimp1+ trophoblasts, we compared nuclear-specific LacZ-staining patterns of the Prdm1Cre.LacZ/+ reporter allele with cytoplasmic LacZ labeling seen in Prdm1Cre.LacZ/+:R26R placentae (Fig. 6; Supplemental Fig. 4C,D). At E12.5, we detected Prdm1Cre.LacZ/+ expression in all of the SpA-TGC as well as the Gly-T populations (Fig. 6A,D), also identified by Blimp1 immunostaining (Supplemental Fig. 4B). Interestingly, at this stage, C-TGCs also coexpress nuclear LacZ and endogenous Blimp1 protein (Fig. 6C; Supplemental Fig. 4B). Lineage tracing analysis shows Blimp1+ trophoblasts clearly only give rise to SpA-TGCs, fully differentiated C-TGCs, and both the spongiotrophoblast and interstitial Gly-T populations (Fig. 6F–H). Thus, diploid Blimp1+ cells comprise discrete lineage-restricted progenitors of multiple spongiotrophoblast subsets, including the invasive endovascular cells, Gly-Ts, and C-TGCs formed at different stages and tissue sites within the developing placenta. In contrast, spongiotrophoblasts and P-TGCs are formed from Blimp1− progenitors and represent a distinct trophoblast cell lineage.
Figure 6.
Lineage-restricted Blimp1+ multipotent progenitors give rise to the SpA-TGCs, Gly-Ts, and C-TGCs. At E12.5, the Prdm1 Cre-LacZ reporter allele identifies the SpA-TGCs lining the maternal blood vessels (D), the Gly-T cells (A), and the C-TGCs (C), and a population of small diploid cells (A, unfilled arrows) within the spongiotrophoblast layer. (B) There was no expression in the labyrinth. Cell fate-mapping studies (lineage tracing identified by cytoplasmic LacZ activity) (Supplemental Fig. 4D). Blimp1+ cells extensively contribute to the spiral artery trophoblast cells (F), canal (G), and interstitial/glycogen trophoblasts (H) in the E12.5 placenta. (H) Notably, diploid trophoblast cells (unfilled arrows) located in the spongiotrophoblast layer retain nuclear Prdm1Cre-LacZ expression. (E) The LacZ-positive cells in the labyrinth in Prdm1Cre-LacZ × R26R sections are fetal endothelial cells that transiently express Blimp1 at earlier stages (Supplemental Fig. 4A,B). Bar, 500 μm. (Dec) Maternal decidua; (ma) maternal artery; (SpT) spongiotrophoblast; (lab) labyrinth trophoblast; (FE) fetal endothelium.
Discussion
In eutherian mammals, the placenta is essential to support fetal development in utero. Derivatives of the trophoblast cell lineage grow in concert with the fetal capillary network and the maternal vasculature to construct this highly specialized organ. In both humans and rodents, invasive TGCs migrate through the uterine epithelium to infiltrate the maternal blood vessels. Degradation of the maternal endothelium and smooth muscle layers and replacement by endovascular TGCs result in vessel dilation and promote the formation of maternal blood sinuses (Hemberger et al. 2003; Screen et al. 2008). Blimp1/Prdm1 mutant embryos arrest at around E10.5 due to placental insufficiency. Here we investigate the underlying causes of this post-implantation lethality.
The present experiments demonstrate that Blimp1 is required for specification of SpA-TGCs, a crucial TGC subpopulation that normally migrates away from the edges of the EPC to surround the incoming maternal arteries (Adamson et al. 2002). Our findings clearly reveal for the first time essential contributions made by this functionally discrete population of terminally differentiated TGCs. Focal loss of the SpA-TGCs disrupts formation of the maternal blood sinuses and severely compromises the ability of the spongiotrophoblast and labyrinth layers to expand appropriately.
Migrating SpA-TGCs coexpress several closely related Prl family members, including Prl2a1, Prl3b1, Prl5a1, and Prl7b1 (Figs. 2, 3; Simmons et al. 2008b), but Prl7b1 is exclusively expressed in mature SpA-TGCs associated with the maternal spiral arteries (Simmons et al. 2008b). Consistent with the focal loss of this cell population, we observed that Prl7b1 expression is undetectable in Blimp1-deficient placentae. Our transcriptional profiling experiments also identified several new markers specific for the SpA-TGC lineage, including the pericyte marker Rgs5 and the proangiogenic matricellular protein Lcn7 that potentially contribute to blood vessel remodeling. Interestingly, SpA-TGCs robustly express the proprotein convertase Pace4, required at early implantation stages for local signaling by the TGFβ growth factor Nodal during establishment of the anterior–posterior axis (Ben-Haim et al. 2006). Nodal signaling from the embryo also maintains the TS cell compartment within the early ExE (Guzman-Ayala et al. 2004). At later stages, Nodal expression within the spongiotrophoblast layer potentially prevents default differentiation of secondary P-TGCs (Ma et al. 2001). Pace4 expressed by SpA-TGCs may activate Nodal signaling within the underlying spongiotrophoblasts to regulate the balance between cell proliferation and differentiation.
Placental growth depends on ongoing recruitment of SpA-TGCs for remodeling of the maternal vasculature. Similarly, C-TGCs are continuously generated to surround the expanding surface area of the large blood canals within the spongiotrophoblast layer. Increasing numbers of glycogen-rich Gly-Ts form within the spongiotrophoblast layer from early stages onward and emerge to invade into interstitial maternal tissues beginning around E12.5. Interestingly, we found that Gly-Ts also express Blimp1 but in its absence are correctly specified. As judged by PAS staining, comparable numbers of Gly-T cells are present at E9.5 in both wild-type and mutant placentae. However, Blimp1-deficient Gly-T cells cannot migrate appropriately. Similarly, invasive trophoblast cells marked by Mmp1a and Cts8 expression (Balbin et al. 2001; Screen et al. 2008) differentiate normally but also fail to infiltrate maternal tissues. One possibility is that Blimp1 directly regulates gene expression profiles controlling their migratory behavior. Alternatively, defective migration may be a secondary consequence due to focal loss of the SpA-TGC population. It is tempting to speculate that trophic factors produced by the SpA-TGC are required for recruitment of these subjacent trophoblast cell subtypes to promote invasion into the maternal tissue.
Intrinsic and extrinsic cues that maintain Blimp1 expression within the SpA-TGCs as well as in the small proliferating cells and terminally differentiated C-TGC and Gly-T subtypes have yet to be elucidated. Recent experiments demonstrate that TLR/NFκB signals activate Blimp1 expression during plasma cell differentiation via consensus binding sites located upstream of the proximal exon 1A promoter (Morgan et al. 2009). However, cis-acting regulatory elements that direct dynamic patterns of tissue-specific expression remain largely unknown. Expression in committed PGCs at the base of the allantois is controlled by dose-dependent Bmp4–Smad signals (Bikoff et al. 2009). Bmp family members, notably Bmp2, are robustly expressed in the decidual tissue surrounding the EPC starting at E7.5 (Paria et al. 2001). We speculate that induction of Blimp1 expression in the SpA-TGCs may be controlled in part by Bmp signaling from maternal decidual cells surrounding the forming EPC.
Here we identified a novel population of lineage-restricted Blimp1+ Ki67+ diploid cells present within the spongiotrophoblast layer. Fate-mapping studies establish that these represent bona fide multipotent stem cells that give rise to mature SpA-TGCs as well as C-TGCs and Gly-Ts. In striking contrast, spongiotrophoblasts and secondary P-TGCs are derived from Blimp1− progenitors and represent a distinct trophoblast lineage. Previous fate-mapping studies reveal contributions made by the Tpbpα+ spongiotrophoblast cells to the SpA-TGCs, Gly-Ts, and a subset of P-TGCs and C-TGCs (Simmons et al. 2007). Cre-medated ablation of the Tpbpα+ population broadly disrupts the entire spongiotrophoblast layer and SpA-TGCs and arrests development at around E11.5 (Hu and Cross 2011). It will be important to learn more about regulatory networks governing underlying stage-dependent allocation of diverse trophoblast subtypes.
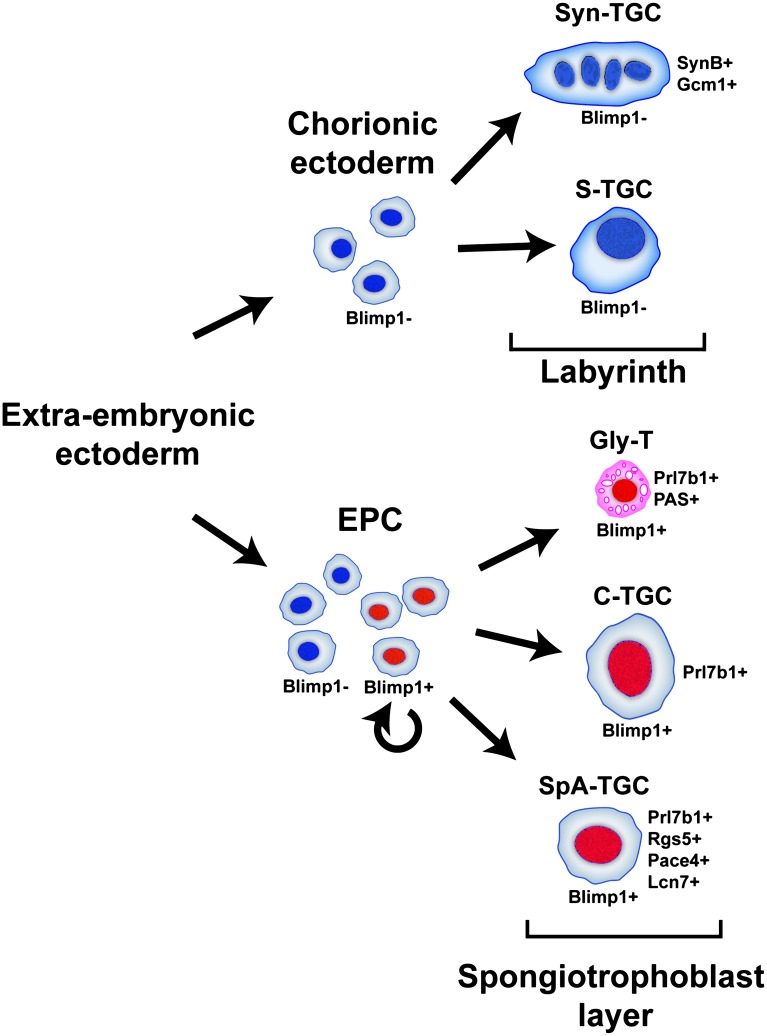
As outlined in Figure 7, Blimp1+ progenitors, present from early EPC stages onward, give rise to both the SpA-TGCs and C-TGCs as well as the diploid glycogen cell population. Cell fate choices may be determined in part by local signaling cues. Thus, Blimp1+ progenitors adjacent to blood sinuses may selectively give rise to C-TGCs, whereas those situated more proximally adopt Gly-T fates. In contrast, Blimp1− progenitors residing within both the spongiotrophoblast and labyrinth layers give rise to the remaining trophoblast lineages of the mature placenta, including the spongiotrophoblasts, P-TGCs, and S-TGCs and syncitial trophoblasts. Collectively, these diverse trophectodermal derivatives orchestrate the dramatic growth of the placenta during pregnancy (Fig. 7).
Figure 7.
Blimp1 functional roles in the mouse placenta. Diploid Blimp1+ spongiotrophoblasts capable of self-renewal give rise to mature Blimp1+; Prl7b1+; Rgs5+; Pace4+ and Lcn7+ SpA-TGCs that invade the maternal decidua to surround and remodel maternal blood vessels, as well as Blimp1+; Prl7b1+; PAS+ Gly-Ts and Blimp1+; Prl7b1+ C-TGCs. In striking contrast, trophoblast subtypes (Syn-Ts and S-TGCs) that form the labyrinth compartment, derived from the Blimp1− progenitors, represent a distinct lineage.
Divergent placental structures have evolved in mammals to support viviparity. Human and mouse placentae share many functional similarities. Both are characterized by the highly invasive nature of the trophoblast lineage. In humans, the so-termed extravillous or cytotrophoblasts (counterparts of the murine SpA-TGC, C-TGC, and Gly-T) invade the uterine stroma to form both interstitial and endovascular derivatives (Moffett and Loke 2006). Remodeling and dilation of maternal arteries is also necessary for efficient maternal–fetal exchange. Defects in cytotrophoblast invasion and differentiation are causally associated with intrauterine growth retardation and pre-eclampsia (Moffett and Loke 2006). Microarray analysis of human placental samples demonstrate complex stage- and tissue-specific patterns of gene expression encompassing a myriad of functional categories (Winn et al. 2007). It will be interesting to learn more about Blimp1 requirements in human endovascular lineages and whether Blimp1 similarly defines TS cells driving expansion of cytotrophoblasts throughout pregnancy.
Materials and methods
Animals
Prdm1BEH (Vincent et al. 2005), Blimp1gfp (Kallies et al. 2004), Flk.LacZ (Shalaby et al. 1995), and R26R reporter (Soriano 1999) mouse strains have been described. Prdm1BEH/+ animals were intercrossed to generate null placental tissue. The Prdm1Cre-LacZ allele was generated by introducing a cassette containing a codon-optimized Cre (Shimshek et al. 2002) upstream of IRES-nlacZ followed by a FRT-flanked neo cassette (iCre-IRES-nLacZ-FRT-neo-FRT) into the ATG translational site in exon 3 via homologous recombination in embryonic stem (ES) cells. Correctly targeted ES cell clones were transiently transfected with a FLP expression construct. Drug-excised subclones were injected into blastocysts to generate germline chimeras. All experiments using mice were performed in accordance with Home Office regulations.
Histology, in situ hybridization, and immunohistochemistry
For H&E staining, immunohistochemistry, and in situ hybridization, tissue was fixed overnight in 4% paraformaldehyde (PFA), dehydrated in ethanol, embedded in paraffin wax, and sectioned (6 μm). RNA in situ hybridization was performed according to standard protocols (Nagy et al. 2003) using paraffin sections collected from the mid-plane of the placenta. Probes for Tpbpα, Gcm1, Prl3d1, Prl5a1, Prl2a1, Prl3b1, Prl7b1 (Simmons et al. 2007, 2008b), and Pace4 (Constam and Robertson 2000) have been described. SynB, Rgs5, and Lcn7/Tinagl probes were generated by PCR.
For immunohistochemistry, dewaxed sections subjected to antigen retrieval by boiling for 30 min in either Target Retrieval Solution (S1699, DAKO) or Tris/EDTA (pH 9.0) were permeabilized for 10 min in 0.2% Triton X-100 in TBS and subsequently treated with peroxidase blocking buffer (K4011, DAKO) for 15 min, followed by 30 min in 10% goat serum. Sections incubated in primary antibodies overnight at 4°C were subsequently developed with the appropriate DAKO peroxidase-labeled polymer kit or VectaStain ABC System (Vector Laboratories) using DAB or Vector Red substrate (Vector Laboratories). Primary antibodies were rat monoclonal anti-Blimp1 (1:500 dilution; sc-130917, Santa Cruz Biotechnology), rabbit polyclonal anti-Ki67 (1:200 dilution; Ab1558, Abcam), and a mouse monoclonal anti-Pan CK (1:100 dilution; C-2562, Sigma-Aldrich).
For double-labeling experiments, Ki67-stained sections were then subjected to a second round of antigen retrieval, blocked, and incubated with rat anti-Blimp1 (1:500 dilution; sc-130917, Santa Cruz Biotechnology) overnight at 4°C. The signal was amplified with biotinylated rabbit anti-rat IgG (1:200 dilution; E0468, DAKO) followed by AP-conjugated biotin/streptavidin (ABC-AP, Vector Laboratories) and Vector Red substrate (Vector Laboratories). Sections were lightly counterstained with haemotoxylin. PAS staining was performed using a commercially available kit (Sigma-Aldrich). Glycogen-specific reactivity on paraffin sections was enhanced by acetylation prior to PAS staining as previously described (Tesser et al. 2010). Morphometric analysis of nuclear area was performed using DAPI-stained paraffin sections (one section each from five placenta per genotype) using a Nuance MSI camera (LOT, Oriel). Sections were post-stained with H&E for imaging.
X-gal staining of intact embryos was performed as described (Nagy et al. 2003). Cryosections (10 μm) of placental tissue were stained as described (Navankasattusas et al. 2008) and counterstained with Nuclear Fast Red or PAS. AP staining was carried out as described (Bissonauth et al. 2006).
Acknowledgments
We thank Ahmed Salman for help with genotyping, Jay Cross for in situ probes, Janet Rossant for the Flk.LacZ mice, Cordelia Langford and Peter Ellis for performing the arrays, and Sebastian Arnold for help with the initial microarray experiments. This work was funded by grants from the Wellcome Trust. E.J.R. is a Wellcome Trust Principal Research Fellow.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.199828.112.
References
- Adamson SL, Lu Y, Whiteley KJ, Holmyard D, Hemberger M, Pfarrer C, Cross JC 2002. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev Biol 250: 358–373 [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA 2006. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA, Cross JC 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet 25: 311–314 [DOI] [PubMed] [Google Scholar]
- Arnold SA, Sugnaseelan J, Groszer M, Srinivas S, Robertson EJ 2009. Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis 47: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbin M, Fueyo A, Knauper V, Lopez JM, Alvarez J, Sanchez LM, Quesada V, Bordallo J, Murphy G, Lopez-Otin C 2001. Identification and enymatic characterization of two diverging murine counterparts of human intersitial collagenase (MMP-1) expressed at sites of embryonic implantation. J Biol Chem 276: 10253–10262 [DOI] [PubMed] [Google Scholar]
- Ben-Haim N, Lu C, Guzman-Ayala M, Pescatore L, Mesnard D, Bischofberger M, Naef F, Robertson EJ, Constam DB 2006. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell 11: 313–323 [DOI] [PubMed] [Google Scholar]
- Bikoff EK, Morgan MA, Robertson EJ 2009. An expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev 19: 1–7 [DOI] [PubMed] [Google Scholar]
- Bissonauth V, Roy S, Gravel M, Guillemette S, Charron J 2006. Requirement for Map2k1 (Mek1) in extra-embryonic ectoderm during placentation. Development 133: 3429–3440 [DOI] [PubMed] [Google Scholar]
- Bouillot S, Rampon C, Tillet E, Huber P 2006. Tracing the gylcogen cells with protocadherin 12 during mouse placenta development. Placenta 27: 882–888 [DOI] [PubMed] [Google Scholar]
- Brown LJ, Alawoki M, Crawford ME, Reida T, Sears A, Torma T, Albig AR 2010. Lipocalin-7 is a matricellular regulator of angiogenesis. PLoS ONE 5: e13905 doi: 10.1371/journal.pone.0013905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ 2000. SPC4/PACE4 regulates a TGF β signalling network during axis formation. Genes Dev 14: 1146–1155 [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Johnston RJ, Schoenberger SP 2010. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol 11: 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doody GM, Care MA, Burgoyne NJ, Bradford JR, Bota M, Bonifer C, Westhead DR, Tooze RM 2010. An extended set of PRDM1/BLIMP1 target genes links binding motif type to dynamic repression. Nucleic Acids Res 38: 5336–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P 2003. Cyclin E ablation in the mouse. Cell 114: 431–443 [DOI] [PubMed] [Google Scholar]
- Gonzalez MA, Tachibana KE, Adams DJ, van der Weyden L, Hemberger M, Coleman N, Bradley A, Laskey RA 2006. Geminin is essential to prevent endoreduplication and to form pluripotent cells during mammalian development. Genes Dev 20: 1880–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL 1994. Essential role of Mash-2 in extraembryonic development. Nature 371: 333–336 [DOI] [PubMed] [Google Scholar]
- Guzman-Ayala M, Ben-Haim N, Beck S, Constam DB 2004. Nodal protein processing and fibroblast growth factor 4 synergize to maintain a trophoblast stem cell microenvironment. Proc Natl Acad Sci 101: 15656–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J, Mould A, Andrews RM, Bikoff E, Robertson EJ 2011. The transcriptional repressor Blimp1/Prdm1 regulates postnatal reprogramming of intestinal enterocytes. Proc Natl Acad Sci 108: 10585–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Himmelbauer H, Ruschmann J, Zeitz C, Fundele R 2000. cDNA subtraction cloning reveals novel genes whose temporal and spatial expression indicates association with trophoblast invasion. Dev Biol 222: 158–169 [DOI] [PubMed] [Google Scholar]
- Hemberger M, Nozaki T, Masutani M, Cross JC 2003. Differential expression of angiogenic and vasodilatory factors by invasive trophoblast giant cells depending on depth of invasion. Dev Dyn 227: 185–191 [DOI] [PubMed] [Google Scholar]
- Horsley V, O'Carroll DO, Tooze R, Ohinata Y, Saitou M, Obukhanych T, Nussenzweig M, Tarakhovsky A, Fuchs E 2006. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell 126: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Cross JC 2010. Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol 54: 341–354 [DOI] [PubMed] [Google Scholar]
- Hu D, Cross JC 2011. Ablation of Tpbpa-positive trophoblast precursors leads to defects in maternal spiral artery remodeling in the mouse placenta. Dev Biol 358: 231–239 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Kallies A, Nutt SL 2007. Terminal differentiation of lymphocytes depends on Blimp-1. Curr Opin Immunol 19: 156–162 [DOI] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Tarlinton DM, Dietricch W, Corocoran LM, Hodgkin PD, Nutt SL 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med 200: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Carotta S, Huntington ND, Bernard NJ, Tarlinton DM, Smyth MJ, Nutt SL 2011. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood 117: 1869–1879 [DOI] [PubMed] [Google Scholar]
- Ma GT, Soloveva V, Tzeng SJ, Lowe LA, Pfendler KC, Iannoccone PM, Kuehn MR, Linzer DIH 2001. Nodal regulates trophoblast differentiation and placental development. Dev Biol 236: 124–135 [DOI] [PubMed] [Google Scholar]
- Magnusdottir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K 2007. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci 104: 14988–14993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzur M, Ganss R 2009. Regulator of G protein signalling 5: A new player in vascular remodelling. Trends Cardiovasc Med 19: 26–30 [DOI] [PubMed] [Google Scholar]
- Martins G, Calame K 2008. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol 26: 133–169 [DOI] [PubMed] [Google Scholar]
- Moffett A, Loke C 2006. Immunology of placentation in eutherian mammals. Nat Rev Immunol 6: 584–594 [DOI] [PubMed] [Google Scholar]
- Morgan MA, Magnusdottir E, Kuo TC, Tunyaplin C, Harper J, Arnold SJ, Calame K, Robertson EJ, Bikoff EK 2009. Blimp-1/Prdm1 alternative promoter usage during mouse development and plasma cell differentiation. Mol Cell Biol 29: 5813–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navankasattusas S, Whitehead KJ, Suli A, Sorensen LK, Lim AH, Zhao J, Park KW, Wythe JD, Thomas KR, Chien CB, et al. 2008. The netrin receptor UNC5B promotes angiogenesis in specific vascular beds. Development 135: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R 2003. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Lab Press, Cold Spring Harbor, NY [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123: 917–929 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Fairfax KA, Kallies A 2007. BLIMP1 guides the fate of effector B and T cells. Nat Rev Immunol 7: 923–927 [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213 [DOI] [PubMed] [Google Scholar]
- Paria BC, Ma W, Tan J, Raja S, Das SK, Dey SK, Hogan BLM 2001. Cellular and molecular responses of the uterus to embryo implantation can be elicited by locally applied growth factors. Proc Natl Acad Sci 98: 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi T, Beck AR, Rougier N, McNeil T, Lucian L, Werb Z, Amati B 2003. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J 22: 4794–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC 1998. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet 18: 271–275 [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Charatsi I, Joyner CJ, Koonce CH, Morgan M, Islam A, Paterson C, Lejsek E, Arnold SJ, Kallies A, et al. 2007. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development 134: 4335–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. 2000. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404: 95–99 [DOI] [PubMed] [Google Scholar]
- Scott IC, Anson-Cartwright L, Riley P, Reda D, Cross JC 2000. The Hand1 basic helix–loop–helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol Cell Biol 20: 530–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screen M, Dean W, Cross JC, Hemberger M 2008. Cathepsin proteases have distinct roles in trophoblast function and vascular remodelling. Development 135: 3311–3320 [DOI] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC 1995. Failure of blood-island formation and vasculagenesis in Flk-1-deficient mice. Nature 376: 62–66 [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hübner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R 2002. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 34: 208–214 [DOI] [PubMed] [Google Scholar]
- Simmons DG, Cross JC 2005. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol 284: 12–24 [DOI] [PubMed] [Google Scholar]
- Simmons DG, Fortier AL, Cross JC 2007. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol 304: 567–578 [DOI] [PubMed] [Google Scholar]
- Simmons DG, Natale DRC, Begay V, Hughes M, Leutz A, Cross JC 2008a. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 135: 2083–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DG, Rawn S, Davies A, Hughes M, Cross JC 2008b. Spatial and temporal expression of the 23 murine prolactin/placental lactogen-related genes is not associated with their position in the locus. BMC Genomics 9: 352–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Maurin M, Cho HI, Becknell B, Freud AG, Yu J, Wei S, Djeu J, Celis E, Caligiuri MA, et al. 2010. PRDM1/Blimp1 controls effector cytokine production in human NK cells. J Immunol 185: 6058–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71 [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132: 2093–2102 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gertsenstein M, Rossant J, Nagy A 1997. Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev Biol 190: 55–65 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J 1998. Promotion of trophoblast stem cell proliferation by FGF2. Science 282: 2072–2075 [DOI] [PubMed] [Google Scholar]
- Tesser RB, Scherholz PLA, do Nascimento L, Katz SG 2010. Trophoblast glycogen cells differentiate early in the mouse ectoplacental cone: Putative role during placentation. Histochem Cell Biol 134: 83–92 [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ 2005. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 132: 1315–1325 [DOI] [PubMed] [Google Scholar]
- Watson ED, Cross JC 2005. Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 20: 180–193 [DOI] [PubMed] [Google Scholar]
- Welch RM 2009. Blimp hovers over T cell immunity. Immunity 31: 178–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, Feng KT, Bernlohr DA, McDonagh S, Pereira L, et al. 2007. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 148: 1059–1079 [DOI] [PubMed] [Google Scholar]