How diversification is coordinately introduced in a population of cells that otherwise appears uniform is a central question in muscle development. Anderson et al. probe the early transcriptional events controlled by Shh signaling in limb morphogenesis and show that Shh drives the initial expression of Myf5 through the direct action of Gli activators. Different muscle precursor cell populations in the limb bud respond differentially to Shh signals, and myogenesis is activated in muscle precursor cells only when they reach the ventral limb mesenchyme. See also the related study by Hu et al. (in this issue).
Keywords: myogenesis, Shh, limb, Myf5
Abstract
How muscle diversity is generated in the vertebrate body is poorly understood. In the limb, dorsal and ventral muscle masses constitute the first myogenic diversification, as each gives rise to distinct muscles. Myogenesis initiates after muscle precursor cells (MPCs) have migrated from the somites to the limb bud and populated the prospective muscle masses. Here, we show that Sonic hedgehog (Shh) from the zone of polarizing activity (ZPA) drives myogenesis specifically within the ventral muscle mass. Shh directly induces ventral MPCs to initiate Myf5 transcription and myogenesis through essential Gli-binding sites located in the Myf5 limb enhancer. In the absence of Shh signaling, myogenesis is delayed, MPCs fail to migrate distally, and ventral paw muscles fail to form. Thus, Shh production in the limb ZPA is essential for the spatiotemporal control of myogenesis and coordinates muscle and skeletal development by acting directly to regulate the formation of specific ventral muscles.
A central question in muscle development is how and when diversification of muscle precursor cells (MPCs) takes place for the generation of distinct muscles in the body. Skeletal muscles of the limbs represent a useful paradigm to address this question. Limb muscles originate from the paraxial mesoderm and, more specifically, from a subpopulation of epithelial progenitor cells residing in the ventro–lateral dermomyotome of somites (Fig. 1A; Ordahl and Le Douarin 1992). At around embryonic day 10.0 (E10.0) in mouse embryos, MPCs at limb axial levels undergo an epithelial-to-mesenchymal transition and delaminate from the dermomyotome (see ① in Fig. 1A) under the action of c-Met, the receptor for hepatocyte growth factor/scatter factor (HGF/SF) (Bladt et al. 1995; Brand-Saberi et al. 1996; Dietrich et al. 1999). They then migrate to the limb mesenchyme along two routes: a lateral route that brings MPCs to the dorsal limb mesenchyme, and a ventral route heading toward the ventral limb mesenchyme (see ② in Fig. 1A). Migratory MPCs all express the homeobox-containing transcription factors Lbx1 and Pax3 and are thought not to be predestined to join ventral or dorsal muscles masses (Kardon et al. 2002). However, mutations in Lbx1 or in Cxcr4, the chemokine receptor for stromal cell-derived factor 1 (Sdf1), affect the formation of specific muscles (Schafer and Braun 1999; Brohmann et al. 2000; Gross et al. 2000; Vasyutina et al. 2005), suggesting that distinct MPC subpopulations are differentially regulated. Once in the ventral or dorsal limb muscle masses, MPCs expand through proliferation (see ③ in Fig. 1A) and only then initiate the myogenic program (see ④ in Fig. 1A). In mice, the onset of the myogenic program involves the sequential activation of Myf5 at E10.0 (Sassoon et al. 1989; Ott et al. 1991), followed by MyoD and Myogenin at E10.5–E11 (Sassoon et al. 1989). Mice lacking Myf5 and MyoD function do not form limb muscles (Kassar-Duchossoy et al. 2004). Thus, in contrast to trunk muscles, where early Mrf4 expression can compensate for loss of Myf5 and MyoD, Myf5 and MyoD are the key regulators of myogenic determination in the limb musculature (Kassar-Duchossoy et al. 2004). Myf5 and MyoD are expressed with a different spatiotemporal pattern in MPCs of the ventral and dorsal masses and in forelimbs and hindlimbs. Thus, activation of Myf5 in a subset of Pax3+ cells is the earliest sign of muscle diversification in the limb. Significant progress has been made in the elucidation of Myf5 and MyoD regulation (Mankoo et al. 1999; Carvajal et al. 2001; Chen et al. 2001; Hadchouel et al. 2003; Bajard et al. 2006; Buchberger et al. 2007; Giordani et al. 2007). However, the mechanism of Myf5 activation and muscle diversification and how these are coordinated in space and time with the patterned limb axes is unknown.
Figure 1.
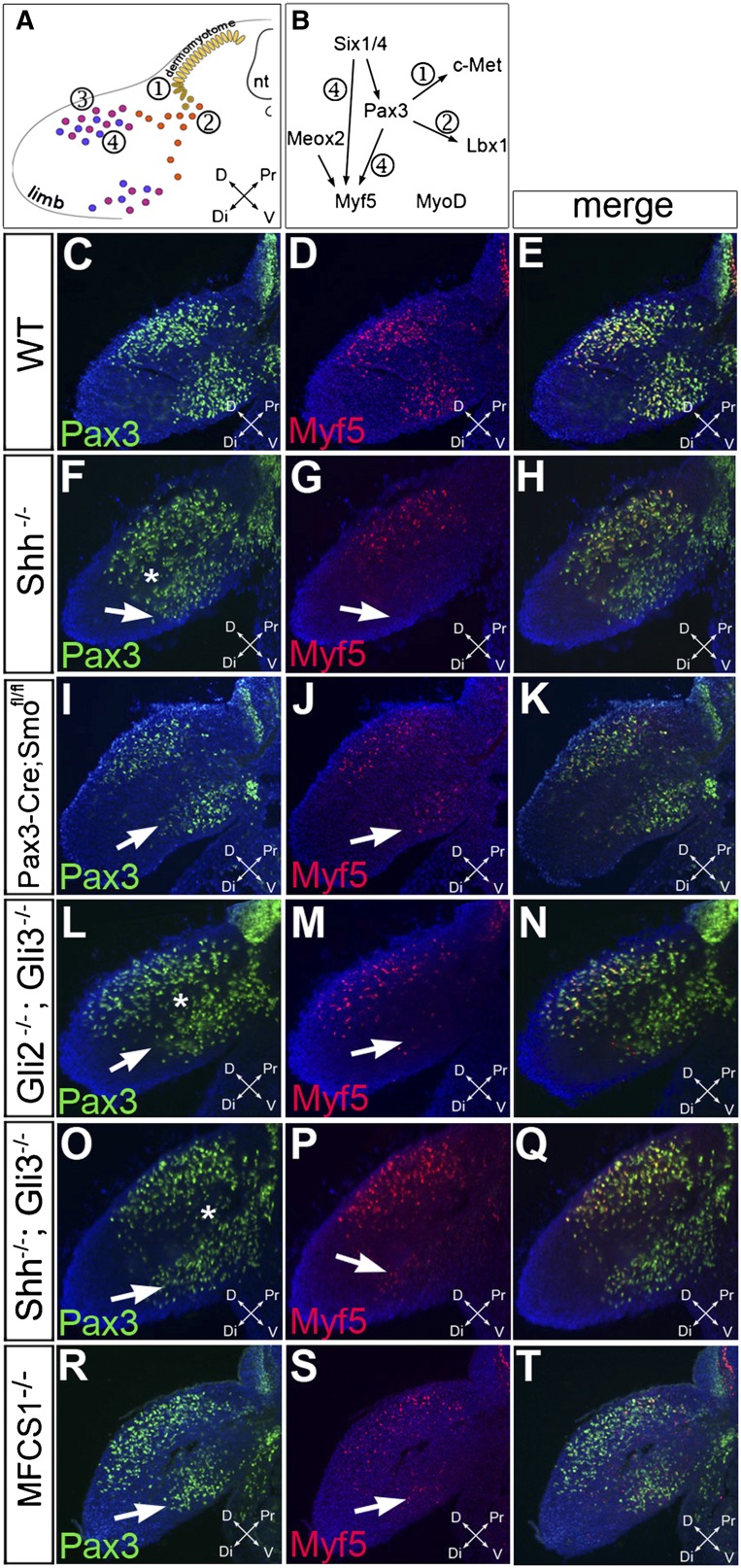
Shh signaling is required cell-autonomously for the initiation of myogenesis in ventral limb MPCs. (A) Schematic diagram representing the successive steps leading to limb myogenesis with delamination from the ventral dermomyotome (①), migration to the limb bud (②), proliferation and expansion of limb MPCs (③), and activation of the myogenic program with the activation of Myf5 and MyoD (④). (B) Gene network controlling limb myogenesis. (C–T) Immunofluorescence analysis of Pax3 (green) and Myf5 (red) expression in forelimb bud transverse sections from E10.5 wild-type (WT) (C–E), Shh−/− (F–H), Pax3:Cre;Smoflox/flox (I–K), Gli2−/−;Gli3−/− (L–N), Shh−/−;Gli3−/− (O–Q), and MFCS1−/− (R–T) embryos. White arrows indicate the location of ventral limb MPCs. White stars indicate incomplete segregation of Pax3+ MPCs in dorsal and ventral muscle masses. (nt) Neural tube; (D) dorsal; (V) ventral; (Pr) proximal; (Dt) distal. Magnification: 200×.
Among major regulators of limb patterning is Sonic Hedgehog (Shh), which is expressed in the zone of polarizing activity (ZPA) at the posterior margin of the limb bud and signals to adjacent cells to pattern the antero–posterior lateral plate-derived limb mesenchyme, including the cartilage and tendon elements (Riddle et al. 1993). Shh is also necessary for both maintenance of limb bud outgrowth—and thus the proximo–distal patterning of the limb (Niswander et al. 1994)—and aspects of dorso–ventral limb patterning (Tzchori et al. 2009). Numerous reports have suggested that Shh also regulates limb myogenesis in various ways, although it remains unknown whether these effects result from direct or indirect roles of Shh on MPCs (Amthor et al. 1998; Duprez et al. 1998; Kruger et al. 2001; Bren-Mattison and Olwin 2002; Li et al. 2004; Bren-Mattison et al. 2011).
Here, we use mouse genetics to disrupt Shh signaling in limb MPCs and show that Shh acts cell-autonomously on MPCs upon their entry into the limb bud to initiate Myf5 expression in the ventral muscle masses of E10.5 embryos. Shh control of Myf5 expression maps to four putative Gli-binding sites within the 5′ region of the Myf5 limb enhancer, which are essential for Gli-mediated enhancer activity in myoblasts and for reporter gene activity in ventral limb muscles in vivo. In addition, Shh signaling acts directly on limb MPCs to promote their distalward migration and the formation of muscles of the forepaws and hindpaws. Altogether, our data indicate a novel role for Shh/Gli signaling in the spatiotemporal control of Myf5 activation in a subset of limb MPCs and reveal the existence of a specific MPC subpopulation directed by Shh to particular muscle fates within the limb bud.
Results
The ventral limb myogenic program is delayed in the absence of Shh signaling
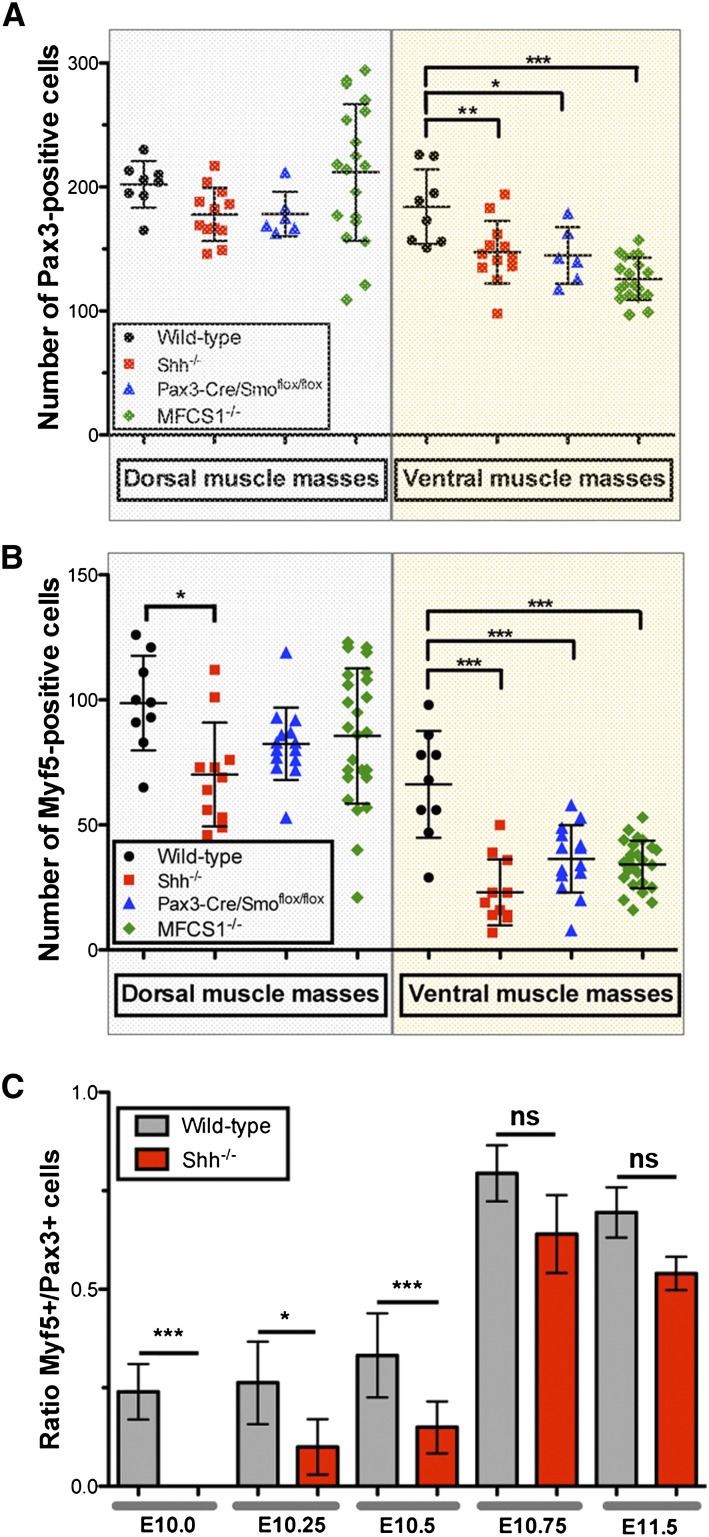
To investigate the role of Shh signaling in limb myogenesis, we examined limb MPCs at E10.5, half a day after their entry into the limb bud (Fig. 1). Immunofluorescence on transverse sections of wild-type embryos revealed Pax3+ MPCs entering the ventral and dorsal muscle masses of the forelimb mesenchyme (Fig. 1C,E). Myf5 protein was detected in MPCs within the dorsal and ventral muscle masses, marking the onset of the myogenic program (Fig. 1D,E; Ott et al. 1991). In E10.5 Shh−/− embryos, we observed a severe reduction in the number of Myf5+ cells in the ventral forelimb, whereas the numbers of Myf5+ and Pax3+ cells in the dorsal forelimb appeared to not be significantly changed (Fig. 1F–H). A similar defect was observed in the hindlimbs of Shh−/− embryos from E11.0 onward (Supplemental Fig. S1). To quantify this defect, we counted the number of Pax3+ and Myf5+ cells in the dorsal and ventral muscle masses of wild-type and Shh−/− forelimbs (Fig. 2A,B; Supplemental Table S1). Confirming our observation, there was a 65% reduction in the number of Myf5+ cells (P < 0.0001) in the ventral limb of Shh−/− embryos, accompanied by a more modest decrease (20%, P = 0.0085) in the number of Pax3+ cells (Fig. 2A–C; Supplemental Table S1). In the dorsal muscle mass of Shh−/− limbs, the number of Pax3+ and Myf5+ cells was generally not significantly different from controls, although a modest trend toward reduced Pax3 and Myf5 was apparent, likely to reflect the overall smaller size of Shh−/− embryos (Fig. 2A,B; Supplemental Table S1). Thus, the absence of Shh differentially affects dorsal and ventral muscle masses, causing defective myogenic progression in the ventral limb.
Figure 2.
A specific defect in Myf5 activation in the absence of Shh signaling. (A) Quantification of the number of Pax3+ cells per transverse section in E10.5 dorsal (gray shaded area) and ventral (yellow shaded area) forelimbs of mutant mouse embryos. (B) Quantification of the number of Myf5+ cells per transverse section in E10.5 dorsal (gray shaded area) and ventral (yellow shaded area) forelimbs of mutant mouse embryos. (C) Ratio of the number of Myf5+ to Pax3+ cells of wild-type (black) and Shh−/− (red) ventral forelimb muscle masses between E10.0 and E11.5. Statistical analysis is described in Supplemental Table S1.
The shortfall of Myf5+ MPCs in the absence of Shh does not appear to be due to a dorso–ventral patterning defect. Dorso–ventral patterning of the limb is established through mutually exclusive expression of Wnt7a in the dorsal ectoderm and En-1 in the ventral ectoderm, allowing for the dorsally restricted expression of Lmx1b, which specifies the dorsal limb mesenchyme (Supplemental Fig. S2A; Parr and McMahon 1995; Loomis et al. 1996; Cygan et al. 1997). As En1 expression requires signaling through the BMP-RI receptor and BMP expression is altered in Shh−/− mice (Ahn et al. 2001; Kruger et al. 2001; Ovchinnikov et al. 2006), it was possible that the failure in ventral limb myogenesis results from a dorso–ventral limb patterning defect in Shh−/− mice (Supplemental Fig. S2A). However, although we observed a loss of Bmp4 expression in the posterior limb mesenchyme associated with an expansion in the anterior limb mesenchyme (Supplemental Fig. S2B), there was no change in the respective ventral and dorsal expression domains of En1 (Supplemental Fig. S2C) and Wnt7a (Supplemental Fig. S2D) in Shh−/− compared with wild-type limbs. Consistent with this observation and with previous reports (Kraus et al. 2001), Lmx1b expression remained dorsally restricted in E10.5 Shh−/− limbs (Supplemental Fig. S2E), indicating that dorso–ventral patterning was essentially normal in early Shh−/− limbs.
Limb MPCs do not require Shh signaling to enter the limb bud, proliferate, or survive
The decrease in the number of Pax3+ MPCs in Shh−/− ventral forelimbs suggests that Shh signaling is required to maintain a subset of limb MPCs. The possibility that Shh signaling directly controls Pax3 expression in MPCs is unlikely because the known Pax3 hypaxial enhancer has no Gli-binding site, and transgenic mice expressing a reporter gene under the control of the hypaxial Pax3 enhancer have no defect when crossed into a Shh−/− background (Brown et al. 2005). Consistent with this, Pax3 mRNA expression in the dorsal and ventral muscle masses of Shh−/− embryos was not detectably different from that of wild-type embryos (Fig. 3A). This implies that Shh controls a process downstream from Pax3 expression in limb MPCs.
Figure 3.
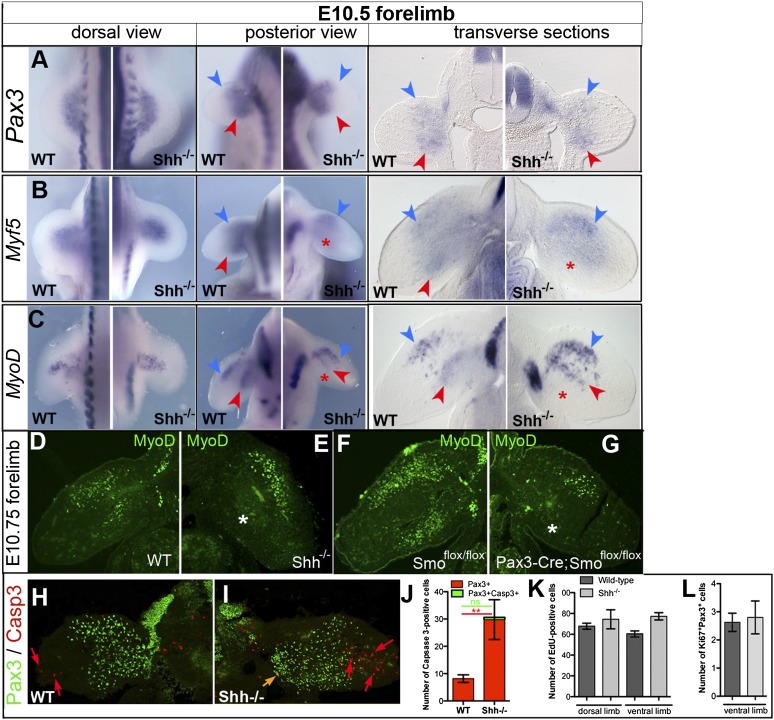
MPC failure to initiate the myogenic program in the absence of Shh is not due to defective proliferation or survival. Expression of Pax3 (A), Myf5 (B), and MyoD (C) was examined by in situ hybridization in E10.5 wild-type (WT) and Shh−/− forelimbs. Blue and red arrowheads indicate the dorsal and ventral muscle masses, respectively. Red asterisks indicate the loss of Myf5 and MyoD expression in the ventral muscle mass. Note the abnormal pattern of ventral MyoD expression in E10.5 Shh−/− forelimbs. Magnification: 200×. (D–G) MyoD distribution (green) in forelimbs from E10.5 wild-type (D), Shh−/− (E), Smoflox/flox (F), and Pax3:Cre;Smoflox/flox (G) mice. White asterisks indicate the loss of MyoD in ventral muscle masses. (H–J) Immunofluorescence analysis of Pax3 (green) and activated Caspase 3 (red) distribution in E10.5 wild-type (H) and Shh−/− (I) forelimbs. Red and orange arrows indicate apoptotic mesenchymal cells and MPCs, respectively. (J) Quantification of the average number of Caspase 3+ cells (red) and Pax3+Caspase 3+ cells (green) per section. One-way ANOVA analysis with Bonferroni post-test showed no significant difference in the number of Pax3+Caspase 3+ cells in wild-type and Shh−/− limbs. (K) Number of EdU+ cells per transverse section in the dorsal and ventral forelimb of E10.5 wild-type and Shh−/− embryos. Unpaired t-test analyses showed no significant differences. (L) Number of Pax3+Ki67+ cells per transverse section of E10.5 wild-type and Shh−/− ventral forelimbs.
Several separate populations of MPCs have been suggested to migrate into limb buds at successive developmental stages (van Swearingen and Lance-Jones 1993). We therefore examined the expression of genes implicated in MPC delamination from the dermomyotome and migration to the limb mesenchyme, including the tyrosine kinase receptor c-Met and its ligand, HGF/SF (Supplemental Fig. S3D,E); the chemokine receptor Cxcr4 and its ligand, Sdf1 (Supplemental Fig. S3G,H); and the transcription factor Lbx1 (Supplemental Fig. S3F; Jagla et al. 1995; Vasyutina et al. 2005). All genes presented similar levels of expression in wild-type and Shh−/− limbs at E10.5, indicating that delamination and migration were not grossly affected in the absence of Shh. Together, these data suggest that the reduced number of Pax3+ and Myf5+ MPCs in Shh−/− limbs does not result from a defect in delamination or migration into the limb.
Ventral Pax3+ MPCs may fail to proliferate. We therefore examined the expression pattern of transcription factors known to be important for MPC expansion prior to myogenic differentiation: Six1, Six4, and Meox2 (Mankoo et al. 1999, 2003; Laclef et al. 2003; Grifone et al. 2005). Despite an anterior expansion of Six1 and Six4 nonmyogenic expression domains (yellow brackets in Supplemental Fig. S3A,B), most likely a consequence of the antero–posterior patterning defect in Shh−/− limbs (Chiang et al. 2001), Six1 and Six4 expression was not affected in the dorsal and ventral muscle masses of Shh−/− embryos (blue and red arrowheads in Supplemental Fig. S3A,B). In contrast, both Meox2 expression domains in MPCs (black arrow in Supplemental Fig. S3C) and the distal mesenchyme (black arrowhead in Supplemental Fig. S3C) were noticeably reduced in Shh−/− limbs, suggesting that the smaller population of Pax3+ MPCs in Shh−/− limbs may result from an inefficient expansion of the MPC population in the ventral muscle masses. We next assayed the proliferation of limb MPCs by measuring the levels of EdU incorporation in wild-type and Shh−/− limbs. Overall, cell proliferation in Shh−/− forelimbs was not significantly different from that of wild-type forelimbs at E10.5 (Fig. 3K), and the same average number of Pax3+/Ki67+ cells was present in both wild-type and Shh−/− ventral forelimbs (Fig. 3L).
Finally, we also assessed the survival of limb MPCs in the absence of Shh (Fig. 3H–J). Although apoptosis increases in the distal limb mesenchyme in the absence of Shh (red arrows in Fig. 3H,I), we only detected occasional Pax3+ cells undergoing apoptosis (average one cell per section) (orange arrow in Fig. 3I,J). This suggests that neither the Meox2 down-regulation nor other effects in Shh-null embryos interfere with MPC proliferation or survival. As Meox2 binds Pax3 protein (Stamataki et al. 2001), it remains to be established whether the formation of this complex is important for Pax3 protein accumulation. Whatever the reason for Pax3 reduction, it is clear that lack of Shh causes a failure of normal Myf5 protein accumulation within Pax3+ cells of the ventral muscle mass (Fig. 2B).
ZPA-secreted Shh is required for ventral limb myogenesis
Considering that limb MPCs originate from the somite and Shh signaling has previously been shown to play essential roles in somite myogenesis (Borycki et al. 1999; McDermott et al. 2005), we wanted independent evidence that the defect in limb Myf5 activation was not a consequence of an earlier impairment at the time limb MPCs still reside in somites. To address this possibility, we examined the expression levels of Pax3 and Myf5 in the limbs of MFCS1−/− mice in which a distal enhancer that drives Shh expression in the limb bud ZPA has been deleted. MFCS1−/− mice completely phenocopied the limb patterning defect of Shh−/− embryos without showing any other Shh−/−-associated defects (Sagai et al. 2004, 2005). Dorsal muscle masses were not affected in MFCS1−/− mice (Figs. 1R–T, 2A,B; Supplemental Table S1). However, we observed a significant reduction in the number of Pax3+ (31.6%, P < 0.0001) and Myf5+ (48.3%, P < 0.0001) cells in the ventral forelimb of E10.5 MFCS1−/− embryos (Figs. 1R–T, 2A,B; Supplemental Table S1). This observation shows that the myogenic program in ventral limb MPCs depends on Shh signals produced in the limb itself and not in midline body tissues.
Shh signaling drives the initiation of limb myogenesis
To establish whether the lack of Myf5 in the absence of Shh reflected a failure of initiation or maintenance, Myf5 and Pax3 accumulation between E10.0 and E11.5 was examined and the ratio of Myf5+ to Pax3+ cells was determined in the ventral forelimb of wild-type and Shh−/− embryos (Fig. 2C). At E10.0, there was already a significant difference between wild-type and Shh−/− embryos. Although fewer Pax3+ MPCs were present in the Shh−/− limb compared with wild type, there was a total absence of Myf5+ cells in Shh-deficient embryos at E10.0, resulting in a nil Myf5+/Pax3+ ratio. This indicates that in the absence of Shh, Myf5 fails to be activated. Thereafter, Myf5 accumulation recovered somewhat so that by E11.5, only a 20% shortfall in Myf5+ MPCs was detected in Shh−/− limbs (Fig. 2C). Thus, an ongoing failure of Myf5 activation persists until at least E11.5 in the absence of Shh signaling.
Cell-autonomous requirement for Shh signaling in limb myogenesis
To address whether Shh signaling is required cell-autonomously in MPCs, we crossed mice carrying a conditional mutant allele of Smoothened (Smoflox), encoding a transmembrane protein essential for Shh signaling (Long et al. 2001), to a Pax3:Cre driver line in which the Cre recombinase gene was introduced into the Pax3 locus (Engleka et al. 2005) in order to eliminate Smo-mediated signaling specifically in all Pax3-expressing cells, including the limb MPCs. Pax3:Cre;Smoflox/flox limbs displayed a decrease in the number of Pax3+ cells in the ventral muscle mass comparable with that of Shh−/− limbs (22%, P = 0.0208 in the ventral limb) (Figs. 1I,K, 2A,B; Supplemental Table S1). In addition, there was a 45% (P = 0.0005) decrease in the number of Myf5+ cells in Pax3:Cre;Smoflox/flox ventral limbs (Figs. 1J,K, 2A,B; Supplemental Table S1), indicating that Myf5 activation in ventral limb MPCs requires functional Smo proteins in a cell-intrinsic manner. Therefore, Myf5 expression appears to require Shh signaling cell-autonomously in limb MPCs.
Sonic hedgehog exerts its effect on limb MPCs through a Gli activator function
Shh signaling exerts dual roles on cells by acting on Gli proteins to prevent the formation of repressor forms of Gli2 and Gli3 and to promote the activator forms of Gli1, Gli2, and Gli3 (Ribes and Briscoe 2009). To establish whether the defect observed in Shh−/− and Pax3:Cre;Smoflox/flox embryos was due to lack of Gli activator or persistence of Gli repressor forms, we compared the phenotype of Gli2−/−;Gli3−/− embryos, which lack all Gli activity (Bai et al. 2004; McDermott et al. 2005), with that of Gli3−/−;Shh−/− embryos, which lack the powerful repressor activity of Gli3. We found that Myf5 also failed to be activated in E10.5 Gli2−/−;Gli3+/− (data not shown) and Gli2−/−;Gli3−/− (Fig. 1L–N) ventral limbs, indicating that Shh signals are mediated through the Gli protein family. Furthermore, the loss of Myf5 in Gli3−/−;Shh−/− embryos (Fig. 1O–Q) suggests that Shh requirement in Myf5 expression involves a Gli activator function.
Shh controls directly Myf5 activation in ventral limb MPCs
As Myf5 is the earliest MRF to be expressed in limb MPCs and is delayed in the absence of Shh signaling, we asked whether Shh signaling directly controls Myf5 transcription in ventral limb MPCs. A 10-kb enhancer at −48 kb upstream of the Myf5 start site drives Myf5 expression in the limb. In particular, a 1.4-kb fragment in the 5′ part of the enhancer that contains two elements, H1 and H2, controls Myf5 transcription in limbs and in the myotome, respectively (see Fig. 4A; Buchberger et al. 2003, 2007; Bajard et al. 2006). As previously reported (Hadchouel et al. 2003), transgenic mice expressing the lacZ reporter gene under the control of the 10-kb Myf5 limb enhancer and Myf5 minimal promoter display high-level reporter gene activity in both dorsal and ventral limb muscle masses at E10.5 (Fig. 4B,C). In contrast, reporter gene expression was significantly reduced in the ventral limb muscle mass of transgenic embryos crossed into a Shh−/− background (P = 0.0003) (Fig. 4B,C) but was unchanged in the dorsal limb muscle mass. Thus, the ventral limb muscle phenotype encountered in Shh-deficient embryos maps to the Myf5 limb enhancer.
Figure 4.
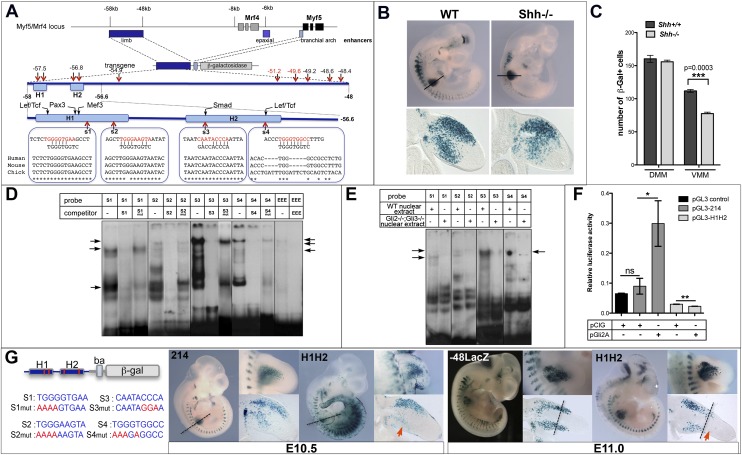
Gli-binding sites present in the Myf5 limb enhancer are essential for Myf5 activation in the ventral forelimb. (A) Schematic representation of the Myf5/Mrf4 genomic locus indicating the position of the limb (dark blue), epaxial (deep blue) and branchial arch (light blue) enhancers. An enlargement of the 10-kb limb enhancer indicating the position of the 1.4-kb 5′ region demonstrated previously to drive Myf5 expression in the limb is shown. Putative Gli-binding sites are shown in black (with mismatch) and red (no mismatch). An enlargement of the 1.4-kb region is also represented, indicating the position of the putative Gli-binding sites 1–4 in relation to previously identified binding sites. The sequence conservation of sites 1–4 in humans, mice, and chicks is shown. (B) β-Gal staining of E10.5 transgenic embryos expressing LacZ under the control of the 10-kb Myf5 limb enhancer in a wild-type (WT) background (left) and Shh−/− background (right). Transverse sections through the forelimb are shown below. Note the reduced number of β-gal+ cells in the ventral limb. (C) Quantification of the number of β-gal+ cells present in the dorsal muscle mass (DMM) and ventral muscle mass (VMM) of wild-type and Shh−/− transgenic embryos. Unpaired t-test analysis was performed. (D) EMSA analysis using oligonucleotides encompassing sites 1–4 and nuclear extract from E10.5 embryos in the presence or not of cold competitor oligonucleotides (wild type or with mutated Gli-binding sites). “EEE” shows the gel shift with an oligonucleotide encompassing the Gli-–binding site from the Myf5 epaxial enhancer. Arrows point to specific complexes that are not competed by cold oligonucleotides with mutated Gli sites. (E) EMSA analysis as in D in the presence of wild-type or Gli2−/−;Gli3−/− nuclear extract. Arrows indicate complexes that disappear in the presence of Gli2−/−;Gli3−/− nuclear extract. (F) Transactivation assay in C2C12 cells using the 1.4-kb Myf5 enhancer with (H1H2) or without (214) Gli-binding sites 1–4 mutated in a luciferase reporter construct. Relative luciferase activity was determined after cotransfection of a control vector (pCIG) or a construct expressing a constitutively active form of Gli2 (pGli2A). (G) β-Gal staining of E10.5 and E11.0 transgenic embryos expressing LacZ under the control of the 1.4-kb enhancer with mutated Gli sites1–4 (H1H2) compared with control transgenic mice containing either the 10-kb Myf5 enhancer (−48LacZ) or the wild-type 1.4-kb enhancer (214). The left panel depicts the H1H2 construct and the mutations introduced into sites 1–4. Dashed lines indicate the boundary between proximal and distal MPC domains. Red arrows show the total absence of β-gal staining in the distal MPC domain.
We identified a number of putative Gli-binding sites clustered at either end of the 10-kb Myf5 limb enhancer (Fig. 4A). These included four putative Gli-binding sites within a 1.4-kb region (−58 to 56.6) that includes H1 and H2, responsible for Myf5 activation in limb MPCs and myotome (Hadchouel et al. 2003). To test whether Shh signaling controls directly Myf5 activation in limb MPCs via the Gli proteins, we performed electrophoretic mobility shift assays (EMSAs) using radioactively labeled oligonucleotides encompassing sites 1–4, located within the 1.4-kb enhancer (Fig. 4A; Supplemental Table S2). All four oligonucleotides formed specific complexes in the presence of nuclear extract from E10.5 mouse embryos, including high-molecular-weight complexes with a gel retardation pattern similar to complexes forming in the presence of an oligonucleotide containing the Gli-binding site from the Myf5 epaxial enhancer previously found to bind Gli proteins (Fig. 4D; Gustafsson et al. 2002). Complex formation was competed by excess of unlabeled oligonucleotides but not by excess of mutated oligonucleotides in which the putative Gli-binding site was disrupted (Fig. 4D; Supplemental Table S2). Such high-molecular-weight complexes did not form when radiolabeled oligonucleotides were incubated in the presence of nuclear extract from Gli2−/−;Gli3−/− embryos (which also lack Gli1) (Fig. 4E; McDermott et al. 2005), suggesting that these high-molecular-weight complexes contain Gli proteins. To confirm that Gli proteins bind to and transactivate the Myf5 limb enhancer, we cloned the wild-type 1.4-kb Myf5 limb enhancer (named pGL3-214) and a mutated version (named pGL3-H1H2) in which all four putative Gli-binding sites were altered into the luciferase reporter construct pGL3. The wild-type enhancer (pGL3-214) yielded a weak increase in transcriptional activity compared with pGL3 alone in C2C12 myoblasts (Fig. 4F). In the presence of a construct expressing a constitutively active form of Gli2 (pGli2A), pGL3-214 transcriptional activity was greatly increased (Fig. 4F). In contrast, the mutated enhancer (pGL3-H1H2) displayed no transcriptional activity in either the presence or absence of pGli2A (Fig. 4F). Together, these data indicate that Gli protein-containing complexes can bind to and transactivate the 1.4-kb Myf5 limb enhancer.
We next generated transient transgenic mice carrying a transgene that contains the β-galactosidase reporter gene under the control of either the wild-type 1.4-kb Myf5 enhancer (214) or the mutant 1.4-kb enhancer (H1H2) (Fig. 4G). At E10.5, the mutated enhancer H1H2 failed to drive reporter gene expression in the ventral limb MPCs, and by E11.0, the most distal cells were especially affected compared with the control enhancer (Fig. 4G). These results confirm that Shh signaling directly controls Myf5 activation in ventral forelimb MPCs through the interaction of Gli activator proteins with one or more Gli-binding sites located in the 5′ domain of the Myf5 limb enhancer.
Impaired Shh signaling causes transient early loss of MyoD expression in the ventral forelimb and loss of specific autopod muscles at later stages
The absence of Myf5 in the ventral limb of E10.5 Shh−/− and Pax3:Cre;Smoflox/flox embryos suggests that some MPCs may fail to implement the myogenic program, including the activation of MyoD leading to terminal differentiation. MyoD activation at both the mRNA and protein levels in Shh−/− and Pax3:Cre;Smoflox/flox forelimbs was examined between E10.5 and E11.5. At E10.5, coinciding with the absence of Myf5 mRNA (Fig. 3B), MyoD transcripts were not detected in ventral MPCs but were aberrantly located to a central domain of the Shh−/− forelimb (Fig. 3C). This abnormal central expression pattern concurs with an apparent failure of Pax3+ MPCs to segregate completely in distinct dorsal and ventral muscle masses (white star in Fig. 1F,L,O). Consistent with the lack of MyoD transcripts, no MyoD protein was detected in the ventral forelimb of E10.75 Shh−/− and Pax3:Cre;Smoflox/flox embryos (Fig. 3E,G,J), although it was present in control embryos at this stage (Fig. 3D,F). In the dorsal muscle masses, MyoD mRNA and protein expression in wild type, Shh−/−, and Pax3:Cre;Smoflox/flox was indistinguishable (Fig. 3C–G). The ventral-specific defect in MyoD expression was transient, as the MyoD transcript and protein levels appeared normal at E11.0 and E11.5 (data not shown), respectively. Thus, loss of Myf5 in the absence of Shh signaling causes a delay in MyoD activation in the ventral forelimb, thereby generating a population of limb MPCs that fail to engage in the myogenic program at an early stage.
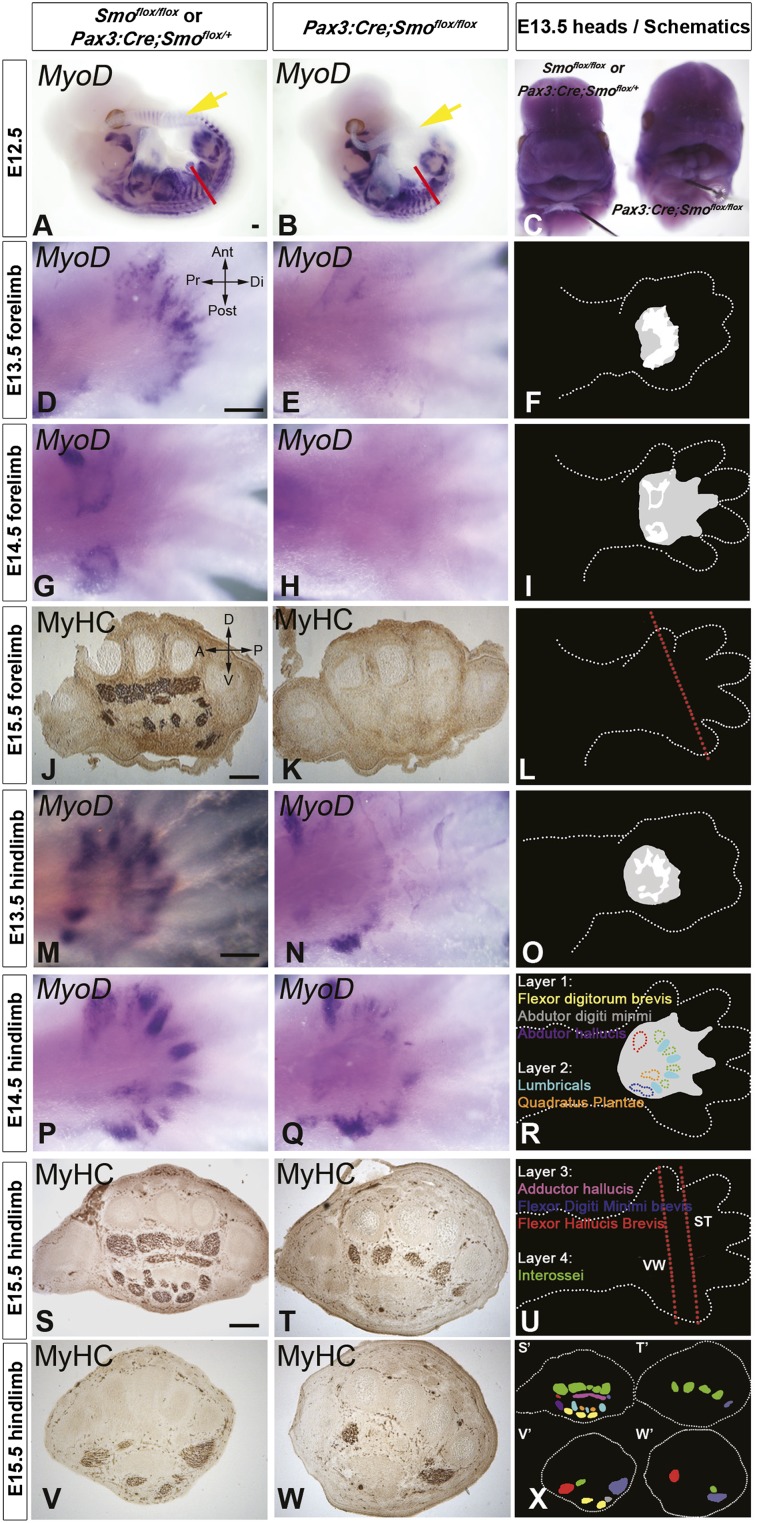
We next asked whether loss of Hedgehog signaling resulted in later limb myogenesis defects. Pax3:Cre;Smoflox/flox embryos were significantly smaller than wild type, with growth appearing to stop at E16.5 (data not shown), with some embryos surviving until E18.5. Consistent with the known requirement for Shh signaling in craniofacial development (Jeong et al. 2004), Pax3:Cre;Smoflox/flox embryos died before birth with severe craniofacial defects (Fig. 5C). Whole-mount MyoD in situ hybridization at E12.5 showed an absence of epaxial muscles in the tail and shortening of the Pax3:Cre;Smoflox/flox myotome thereafter, as previously reported in Shh−/− embryos (Fig. 5A,B; Borycki et al. 1999). Whereas most limb muscles formed in Pax3:Cre;Smoflox/flox embryos, forelimbs lacked MyoD expression in the autopod (Fig. 5D–I) and failed to produce differentiated muscle in the forepaws (Fig. 5J–L). Thus, Shh signaling is cell-autonomously required in limb MPCs for the formation of distal limb muscles in the paw, all of which derive from the ventral muscle mass.
Figure 5.
Sonic hedgehog signaling is required cell-autonomously in limb MPCs for the formation of specific limb muscles. Whole-mount MyoD in situ mRNA hybridization (A–E,G,H,M–Q) or pan-myosin immunodetection on cryosections (J,K,S–W) of sibling (left column) and Pax3:Cre;Smoflox/flox (middle column) mouse embryos. (A,B) Whole mount of MyoD mRNA on Pax3:Cre;Smoflox/flox embryos and sibling E12.5 embryos showing lack of expression in Pax3:Cre;Smoflox/flox tail somites (yellow arrow) and reduction in trunk somites (red line). (C) Heads of E13.5 embryos showing the cleft lip and palette phenotype in Pax3:Cre;Smoflox/flox. (D–W) Ventral (D–H,M–Q; schematized in F,I,O,R) or transverse (J,K,S–W; cut at positions indicated in L,U) views of the autopod showing regions of muscle development. (F,I,O,R) In schematic diagrams, gray areas represent regions of defective muscle, and white areas (shown in F,I) represent the location of MyoD+ cells in siblings. In R and X, colored areas represent the indicated muscles, with those absent in mutants shown as solid (in R). (X) Schematic of muscles lost in the hindlimb.
In hindlimbs, a milder, but nevertheless highly specific, phenotype was observed. At E13.5 and E14.5, there was a noticeable loss of some MyoD expression domains overlying digits 2–5 of the autopod (Fig. 5M–R). This correlated with the absence of specific distal muscles by E15.5 (Fig. 5S–W). The plantar (ventral) hindpaw musculature consists of four layers of muscles. At E15.5, compared with sibling embryos, layer 1 (abdutor hallucis, flexor digitorum brevis, and abdutor digiti minimi) and layer 2 (quadratus plantae and the lumbrical muscles) were completely missing from Pax3:Cre;Smoflox/flox embryos (Fig. 5S,T,V–X). Layer 3 (flexor hallucis brevis, adductor hallucis, and flexor digiti minimi brevis) and layer 4 (interossei) were still grossly present but reduced in size (see Fig. 5W for a comparison). The stylopod and zeugopod of both forelimbs and hindlimbs, although smaller, appeared to have a full complement of muscles present. We conclude that Shh signaling is cell-autonomously required for the initiation of the myogenic program in the ventral muscle masses and for the formation of specific limb muscles in the autopod.
Loss of Myf5 is not solely responsible for the autopod defect
We next investigated whether the early delay in Myf5 expression and myogenesis was linked to the later muscle defect. Although Myf5-deficient embryos have not been reported to have limb muscle defects (Kablar et al. 1997), we reasoned that a defect in autopod muscle formation in Myf5-deficient limbs would be consistent with a role for a specifically Myf5-dependent MPC population in the formation of distal limb muscles. Thus, we examined autopod muscle formation in E14.5 and E15.5 Myf5nLacZ mutant embryos (Tajbakhsh et al. 1996). At E14.5, we observed a reduction in MyoD expression and variable reduction of autopod muscles—in particular, interossei muscles (Supplemental Fig. S4). However, by E15.5, most of this phenotype was resolved; muscles were present, and paws did not show the severe defect observed in Smo-deficient embryos (data not shown). Thus, we concluded that early delay in Myf5 activation in the limb ventral muscle masses is not solely responsible for the loss of autopod muscles at later stages in the absence of Shh signaling.
Shh signaling promotes distalward migration of MPCs
Loss of autopod muscles could arise from failure of distal migration of limb MPCs. Consistent with this possibility, we noticed that the distal mesenchymal domain of Sdf1 expression was missing in Shh−/− forelimbs, whereas the proximal domain was unaffected at E11.5 (red arrows in Supplemental Fig. S5A). This was not due to Shh action on MPCs, as Sdf1 expression in Pax3:Cre;Smoflox/flox limbs was unaffected (data not shown). As Sdf1 signaling through Cxcr4 has been implicated in the distal migration of dorsal MPCs (Vasyutina et al. 2005), this finding suggests that the migration of distal, but not proximal, MPCs may be affected in Shh−/− embryos. Indeed, we observed that the MPC markers c-Met and Lbx1 mRNAs were reduced or absent from the distal limbs of E12.0 Shh−/− embryos (red arrows in Supplemental Fig. S5B,C), indicating that MPCs fail to populate the distal limb mesenchyme in the absence of Shh.
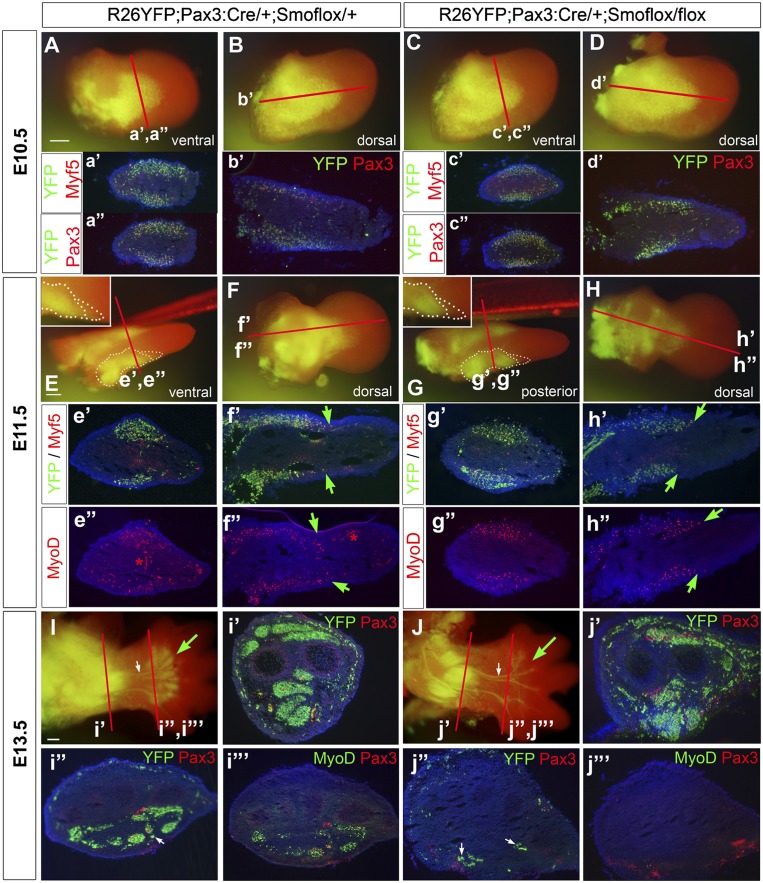
To investigate MPC migration, we examined the fate of MPCs lacking Smo by crossing Pax3:Cre;Smoflox/+ mice with the reporter Rosa26-EYFP mice. Whereas YFP+ Pax3-descendent cells migrated to the same extent in E10.5 wild-type and Smo-deficient limbs (Fig. 6A–D; Supplemental Fig. S6A–D), there was a noticeable absence of Smo-deficient YFP+ cells in the distal domain of E11.5 limbs compared with Pax3:Cre;Smoflox/+;Rosa26-EYFP controls (Fig. 6E,G). This absence of YFP+ Pax3-descendent cells coincided with a loss of Myf5- and MyoD-positive cells in the distal but not proximal domain of the ventral muscle mass (Fig. 6h′,h″). At E13.5, no YFP+ muscle cells could be detected in the autopod muscles, whereas stylopod and zeugopod muscles appeared normally patterned in Smo-deficient limbs (Figs. 6I,J; Supplemental Fig. S6I,J). Notably, YFP+ cells were observed associated with autopod median and ulnar nerves, which we attributed to Pax3+ neural crest-derived Schwann cell precursors (white arrows in Fig. 6J,j″; Kioussi et al. 1995). Both the neural YFP+ pattern and nerve branching were normal in Smo-deficient limbs, indicating that the muscle defect observed is autonomous of MPCs. Together, our findings indicate that Shh signaling is necessary for the distalward migration of MPCs and the formation of autopod muscles. Furthermore, our results show that early loss of Myf5 alone in the ventral muscle masses does not prevent migration of autopod MPCs and is therefore not solely responsible for the autopod defects observed when MPCs lack Shh signaling.
Figure 6.
Defects in forelimb autopod myogenesis in mice with Smo-deficient MPCs are first detected at E11.5 in the distal-most migratory MPC pool. (A–D) At E10.5, migration of Pax3 lineage cells, as revealed by Rosa26EYFP labeling, into the forelimb bud of Smo-deficient mice (C,D) is undistinguishable from that of control siblings (A,B). (E–H) At E11.5, a deficit in EYFP+ cells at the distal tip of the migrating ventral cohort of MPCs is observed in Smo-deficient forelimbs (G,H) compared with control forelimbs (E,F). The white dotted lines outline the ventral muscle mass and its distal tip. Note the ventral-specific migration delay of EYFP+ cells in Smo-deficient forelimbs (cf. green arrows in h′,h″ and f′,f′). (I,J) At E13.5, there is a complete absence of Pax3 lineage muscle cells (EYFP+) in the autopod of Smo-deficient mice (green arrow), whereas Pax3 lineage Schwann cell precursors are unaffected (white arrow). The loss of EYFP+ cells in distal muscles correlates with the absence of Pax3+ and MyoD+ cells in the autopod (cf. j″,j″′ and i″,i″′), but not in the zeugopod (i′,j′). Red lines indicate the level of transverse and longitudinal sections. Bar, 20 μm.
Discussion
Our study provides new insight into the role of Shh signaling in limb myogenesis. We show that Shh is required cell-autonomously in ventral limb MPCs for the spatiotemporal control of Myf5 activation and MPC migration and, subsequently, for the formation of muscles of the autopod. Therefore, Shh acts to promote myogenesis by a subpopulation of MPCs destined for particular muscle fates.
Shh signaling controls the onset of the myogenic program in ventral limb MPCs cell-autonomously
We demonstrate that Shh acts directly on a subset of limb MPCs in the ventral muscle mass. Previous studies investigating the role of Shh and Ihh in limb myogenesis have reported that limb MPCs are driven to proliferate (Duprez et al. 1998) or survive (Kruger et al. 2001; Bren-Mattison et al. 2011), change the balance between proliferation and differentiation (Amthor et al. 1998; Bren-Mattison and Olwin 2002), or undergo a slow specific myogenic program (Li et al. 2004) by Hedgehog signals. However, it remained unclear whether any of these effects of Shh act directly on MPCs or indirectly through the limb mesenchyme in vivo. Here, we used a combination of genetically altered mice, including a conditional allele that specifically eliminates the activity of Smoothened, the receptor essential for Hedgehog signal transduction in MPCs, to investigate the direct effects of Shh on limb MPCs. We show that in the mouse limb Shh is not required for the proliferation or survival of MPCs. Similar findings using a conditional allele that specifically eliminates Smo activity in limb MPCs are presented in the accompanying study (Hu et al. 2012). Together, these data indicate that the effects of Shh on proliferation and survival of MPCs in early limb buds are indirect and/or triggered by the gain-of-function approaches used previously. In contrast, our data demonstrate that limb-derived Shh directly promotes Myf5 transcription and MyoD accumulation in the earliest MPCs of the ventral muscle mass in a manner akin to the direct promotion by Hh of myogenic progression observed in early somitic muscle formation from zebrafish to mice (Blagden et al. 1997; Borycki et al. 1999; Wolff et al. 2003).
Shh acts preferentially on ventral muscle mass MPCs
Failure to initiate Myf5 activation was specifically observed in the ventral muscle mass of Shh-defective limbs, indicating that Shh acts on a subpopulation of limb MPCs to promote myogenesis. Two underlying mechanisms could account for the ventrally restricted defect observed in the absence of Shh signals. Shh could promote myogenesis by a subpopulation of MPCs that already has distinct properties, including distinct Shh responsiveness, prior to their ingression into the limb bud. Previous fate mapping studies have suggested that although some clones can contribute to the full complement of limb muscles, MPCs entering the chick limb bud at different stages may have restricted fates (van Swearingen and Lance-Jones 1993; Kardon et al. 2002; Rees et al. 2003). Such MPC heterogeneity would fit well with our finding that not all ventral muscle mass MPCs need Shh for Myf5 accumulation. Alternatively, local variation of signaling molecules (including Hedgehog) might act on ingressing limb MPCs and could induce a distinct subpopulation of cells in the ventral muscle mass from an otherwise homogeneous MPC population. Although neither Shh nor Ptch1 mRNAs appear unequally distributed between dorsal and ventral limb regions, a preferential effect on ventral limb MPCs of Shh signaling was previously noted in Shh−/− mice (Kruger et al. 2001) and in chick hindlimbs when Shh signaling was blocked by overexpressing a Gli1 repressor form (Bren-Mattison and Olwin 2002). Our study now shows that Shh mediates myogenesis by a subpopulation of ventral limb MPCs in a cell-autonomous manner that is likely to be conserved among amniotes. Furthermore, as Myf5 is induced in the dorsal muscle mass, we conclude that myogenesis proceeds without the aid of the Shh/Gli pathway in the dorsal limb and that Shh acts specifically on a subpopulation of limb MPCs.
Myf5 is a direct target of Shh signals in ventral limb MPCs
Consistent with our findings that Shh directly controls Myf5 transcription through Gli protein-mediated binding to the Myf5 limb enhancer, many limb MPCs are labeled in the inducible Gli-responsive reporter line Gli1-CreERT2 (Ahn and Joyner 2004). Pax3 and Six1/4, which also control the activity of the Myf5 limb enhancer (Bajard et al. 2006; Giordani et al. 2007), are already expressed in MPCs prior to their migration into the limb bud and are expressed similarly in dorsal and ventral MPCs. Additional signals and/or transcriptional activators are therefore required to coordinate Myf5 activation with the arrival of MPCs in the limb bud. We show that Shh is one such signal that acts directly on Myf5 transcription in ventral limb MPCs through the interaction of Gli proteins with one or more Gli-binding sites located within the 5′ region of the Myf5 limb enhancer. This provides a new framework to understand how the onset of myogenesis is regulated in limb MPCs (Fig. 7). In this model, the onset of Shh expression in the limb ZPA at E9.75 (Echelard et al. 1993) provides temporal control of Gli transcription factor activity to mediate the precise timing of Myf5 transcription, acting in combination with Pax3 and Six1/4. Whereas previous reports have uncovered mechanisms that inhibit the myogenic program in migrating MPCs (Bendall et al. 1999; Lee et al. 2004), no other study has identified a mechanism responsible for timing the onset of myogenesis in limb MPCs. Our finding that limb, not axially derived, Shh expression controls Myf5 provides a powerful mechanism to coordinate Myf5 transcription with the arrival of MPCs in the limb bud mesenchyme. Consistent with this, precocious development of forelimb muscles in marsupial embryos correlates with a much earlier onset of limb Shh expression (Keyte and Smith 2011).
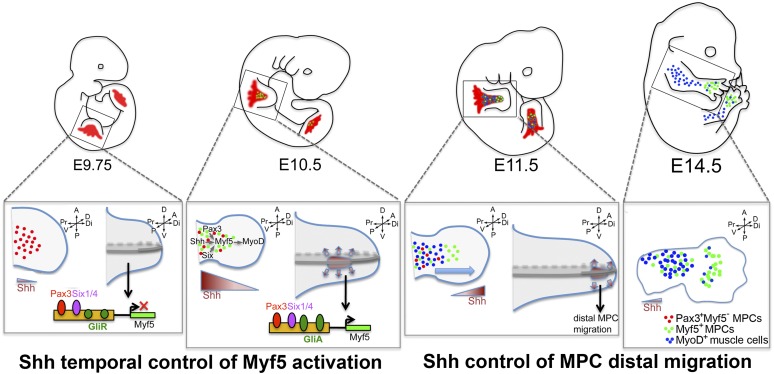
Figure 7.
Model of direct Shh signaling driving muscle diversification in the forelimb through spatiotemporal control of myogenesis and distal MPC migration. In E9.75 wild-type (WT) embryos, Shh is not expressed in the limb, and thus migrating MPCs (red) do not initiate the myogenic program despite the presence of known regulators of Myf5 expression (Pax3 and Six1/4). With Shh expression (brown) initiating in the limb ZPA around E10.0, Gli activator forms (GliA) are generated, which bind to and cooperatively activate the Myf5 limb enhancer together with Pax3 and Six1/4 in ventral limb MPCs (green). Myf5 activation allows for MyoD expression and entry into the myogenic program. At E11.5, both Myf5 and MyoD can now be activated via a Smo-independent mechanism (blue), allowing for a recovery of the myogenic program in Pax3:Cre;Smoflox/flox mice. At E11.5, Shh signaling acts also in a cell-autonomous manner to promote the distal migration of ventral MPCs, thus segregating a limb MPC population to a pool of MPCs destined to form autopod muscles at E14.5 (in green).
Myf5 and MyoD may define different myogenic stages in limb MPCs
Our data that Myf5 loss at E10.5 results in a transient loss of MyoD indicates that, as in the trunk, Myf5 acts upstream of MyoD to initiate the myogenic program. MyoD is likely to drive MPCs toward terminal differentiation, and, interestingly, its expression in the ventral forelimb was recently reported to be temporally controlled by Sim2 (Havis et al. 2012). Consistent with this, a Myogenin-lacZ transgene is not expressed in MyoD−/− limbs, whereas some expression occurs in Myf5−/− limbs (Kablar et al. 2003). Myogenin expression is also delayed in mice lacking the 258-base-pair (bp) MyoD core enhancer responsible for the early limb expression (Chen and Goldhamer 2004). Furthermore, transgenic mice expressing β-galactosidase under the control of the 258-bp MyoD core enhancer present a reduced transgene expression at E10.5 in Myf5−/− limbs and no expression in MyoD−/−;Myf5−/− limbs (Kablar et al. 1999). Interestingly, this regulatory relationship is mainly active during primary myogenesis, as by E11.5, both MyoD and Myf5 expression appear to be regulated by different cis-regulatory elements, most likely in preparation for secondary myogenesis (Kablar et al. 1997, 1999; Hadchouel et al. 2003). This may in part explain the “recovery” of Myf5 and MyoD expression observed in the present study, which coincides with the timing of Shh down-regulation in the ZPA (Echelard et al. 1993).
Shh-dependent limb MPCs are differentially recruited into muscles along the proximo–distal axis
We and Hu et al. (2012) now both report that mice in which limb MPCs are unable to respond to Shh lack all forelimb autopod muscles and specific muscles of the autopod in hindlimbs (Fig. 7). Nevertheless, most proximal muscles are present in these mice, again raising the issue of heterogeneity within the limb MPC population. Our analysis of the Myf5nLacZ mutant mice indicates that, while we cannot rule out a contribution of an early MPC subpopulation that fails to activate Myf5 to autopod muscles, it is unlikely that loss of such a MPC population is solely responsible for the distal muscle defect observed in Smo-deficient limbs. Although it is possible that the subpopulation of MPCs that forms autopod muscles may require longer/stronger Shh exposure, our data (and those of the accompanying study by Hu et al. [2012]) point to the absence of Pax3+YFP+ cells in the distal limb. This suggests that Shh signaling may be required for distalward migration of the subpopulation of MPCs destined to form autopod muscles. This is consistent with previous observations that MPCs fail to migrate and populate distal muscles in talpid3 chickens (Lee and Ede 1989), which have defective Shh responsiveness (Davey et al. 2006). Indeed, further studies by Hu et al. (2012) identified Net1 as a target of Shh signaling, mediating its migratory function in limb MPCs. Thus, a role of Shh from the ZPA is to segregate the limb MPC population in order to allocate a pool of MPCs to the distal limb for the formation of the autopod muscles.
Shh from the ZPA specifies an antero–posterior muscle pattern within the limb (Duprez et al. 1999). Although much of this action is likely to be through an indirect effect of Shh on limb mesenchyme, we observed MPC-autonomous preservation of hindlimb autopod muscles in the most anterior and posterior locations but loss of muscles at intermediate positions within the limb. Thus, Shh signals to MPCs subdivide the limb MPC pool not only in the dorso–ventral and proximo–distal dimensions, but also the antero–posterior. It seems possible that, during evolution of the amniote autopod, an existing system of direct control of MPC behavior was co-opted to play a more significant role in autopod myogenesis. Such findings may have important implications for our understanding of the molecular mechanisms underlying the great heterogeneity and diversity of muscles affected in muscle dystrophies (Emery 2002).
Materials and methods
Mouse lines
Shh+/− (Chiang et al. 1996), Gli2+/−;Gli3+/− (Mo et al. 1997), Shh+/−;Gli3+/−, MFCS1+/− (Sagai et al. 2005), Rosa26-YFP (Srinivas et al. 2001), Myf5nlacZ/+ (Tajbakhsh et al. 1996), B6;129-Pax3 tm1(cre)Joe/J (Engleka et al. 2005), and Smotm2Amc (Long et al. 2001) were maintained as described. −48Myf5nLacZ mice were maintained as heterozygous and bred to Shh+/− mice to generate −48Myf5nLacZ;Shh+/− mice. E0.5 was the day vaginal plugs were found. Embryos were harvested between E10.0 and E15.5 by caesarian section, and genotyping was performed on yolk sac DNA by PCR with primers described previously (Chiang et al. 1996; Mo et al. 1997; Long et al. 2001; Maynard et al. 2002; Engleka et al. 2005; Sagai et al. 2005). −48Myf5nLacZ mice were genotyped using the primers 48Wt-F (5′-GGCTGGGAGACTATTAATGACTGC-3′), 48Wt-R (5′-GGCCTATGTGCAACAGCAAGAG-3′), and pA-F2 (5′-CCCCTGAACCTGAAACATAAAATGAATGC-3′) to detect a 230-bp wild-type PCR product and a 260-bp transgene PCR product.
Whole-mount and section in situ hybridization
Embryos were fixed overnight at 4°C in 4% formaldehyde + 2 mM EGTA in PBS (pH 7.5) and processed for whole-mount in situ hybridization using DIG-labeled RNA probes as previously described (Anderson et al. 2009). Whole-mount images were captured on a stereomicroscope (Leica) using a Spot digital camera (Spot Insight Color). Transverse sections (80 μm) were cut from embryos embedded in 2% agarose in PBS using a Vibratome 1500 and mounted in Glycergel (Dako). Sections were photographed on a DMR microscope using DIC optics (Leica) and captured using a Leica camera, DC300FX. In situ hybridization on sections was performed on 15-μm cryosections. Probes used in this study are described in Supplemental Table S3.
Immunofluorescence
Immunofluorescence was essentially performed as described previously (Anderson et al. 2007). Antibodies used in this study are described in Supplemental Table S4. Images were captured on a BX51 microscope (Olympus) using a Hamamatsu digital camera or Zeiss Axiophot with Axiocam and Openlab image capture software (Improvisions). Images were treated using Photoshop (Adobe) software.
X-Gal staining
Embryos were fixed in 2% PFA for 30–60 min at room temperature, washed in lacZ rinse solution (5 mM EGTA, 0.01% sodium deoxycholate, 0.02% NP40 [Igepal], 2 mM MgCl2 in PBS), and stained for 8–16 h at 37°C in lacZ rinse solution containing 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside (X-Gal, Invitrogen).
EdU labeling
Pregnant female mice received an intraperitoneal injection of 1.33 mM EdU (Click-iT EdU kit, catalog no. C10339, Invitrogen) in 300 μL of PBS 90 min before harvesting the embryos. Embryos were cryo-embedded, and cryosections were taken as described above. Sections were rehydrated for 30 min in 1× PBS, washed twice for 5 min each in 3% BSA (bovine serum albumine) in PBS, permeabilized for 15 min in 0.5%Triton X-100 in PBS, and washed twice for 5 min each in 3% BSA in PBS before the incubation with the labeling reaction mix for 30 min according to the manufacturer's protocol. Following incubation, sections were washed twice for 5 min each in 3% BSA in PBS before immunodetection of Pax3.
EMSA
Nuclear extract was prepared from E10.5 C57BL/6 or E10.5 Gli2−/−;Gli3−/− mouse embryos, and EMSAs were performed using radiolabeled oligonucleotides (described in Supplemental Table S2) as previously described (Coy et al. 2011).
Transactivation assay
C2C12 myoblasts were cultured in DMEM medium (Gibco) supplemented with 10% FCS (HyClone Laboratories, Inc.). Cells were seeded at 6 × 105 cells per well in a six-well plate 16 h prior to transfection. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Plasmids used were pGL3-214 in which the −58-56.6 Myf5 enhancer was cloned into the firefly luciferase reporter construct (pGL3-Promoter, Promega), pGL3-H1H2 in which the four putative Gli-binding sites of pGL3-214 were mutated, and pGli2act in which constitutively active mouse Gli2 was cloned into pCIG vector. Transfection efficiency was assessed by cotransfecting pRL-SV40, which expresses the Renilla luciferase. Thirty-six hours after transfection, firefly luciferase and renilla luciferase activities were measured using the Dual-Luciferase Reporter (DLR) Assay System (Promega) according to the manufacturer's protocol on a luminometer (Berthold Sirius)
Generation of transgenic mice
The −58-56.6BAMyf5nLacZ transgene construct (Hadchouel et al. 2003) was mutated on all four putative Gli-binding sites by site-directed mutagenesis to generate the −58-56.6mutBAMyf5nLacZ transgene construct. Site-directed mutagenesis was carried out following the protocol described in Coy et al. (2011). Primers used are described in Supplemental Table S2. Transgene DNA purification was performed as described previously (Kelly et al. 1995). Pronuclear injections on single-cell embryos were carried out, and injected eggs were reimplanted into pseudo-pregnant foster females and staged as E0.5.
Acknowledgments
We thank Jimmy Hu and Cliff Tabin for sharing unpublished results. We are grateful to Susanne Dietrich, Thomas Braun, Christine Hartmann, Baljinder Mankoo, Martin Goulding, Carmen Birchmeier, Pascal Maire, Cliff Tabin, Delphine Duprez, Gabrielle Kardon, Chi-Chung Hui, Lisa Goodrich, and Andy McMahon for kindly providing us with mouse probes. We also thank Jonathan Epstein, Malcolm Logan, and Andrew McMahon for mouse strains. We thank Marysia Placzek for sharing her equipment. Thanks to Michelle Down, who contributed to early studies. A.G.B.'s laboratory was supported by grants from the European Commission Sixth Framework Programme (MYORES contract 511978) and from the AFM, which also supported work in M.E.B.'s laboratory. S.M.H. is an MRC Scientist with Programme Grant support. C.A. was supported by funding from the European Commission and by a Wellcome Trust Value in People award.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187807.112.
References
- Ahn S, Joyner AL 2004. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118: 505–516 [DOI] [PubMed] [Google Scholar]
- Ahn K, Mishina Y, Hanks MC, Behringer RR, Crenshaw EB 3rd 2001. BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal–ventral patterning of the limb. Development 128: 4449–4461 [DOI] [PubMed] [Google Scholar]
- Amthor H, Christ B, Weil M, Patel K 1998. The importance of timing differentiation during limb muscle development. Curr Biol 8: 642–652 [DOI] [PubMed] [Google Scholar]
- Anderson C, Winder SJ, Borycki AG 2007. Dystroglycan protein distribution coincides with basement membranes and muscle differentiation during mouse embryogenesis. Dev Dyn 236: 2627–2635 [DOI] [PubMed] [Google Scholar]
- Anderson C, Thorsteinsdottir S, Borycki AG 2009. Sonic hedgehog-dependent synthesis of laminin α1 controls basement membrane assembly in the myotome. Development 136: 3495–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Stephen D, Joyner AL 2004. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell 6: 103–115 [DOI] [PubMed] [Google Scholar]
- Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham ME 2006. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev 20: 2450–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall AJ, Ding J, Hu G, Shen MM, Abate-Shen C 1999. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development 126: 4965–4976 [DOI] [PubMed] [Google Scholar]
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C 1995. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376: 768–771 [DOI] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM 1997. Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes Dev 11: 2163–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP Jr 1999. Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development 126: 4053–4063 [DOI] [PubMed] [Google Scholar]
- Brand-Saberi B, Muller TS, Wilting J, Christ B, Birchmeier C 1996. Scatter factor/hepatocyte growth factor (SF/HGF) induces emigration of myogenic cells at interlimb level in vivo. Dev Biol 179: 303–308 [DOI] [PubMed] [Google Scholar]
- Bren-Mattison Y, Olwin BB 2002. Sonic hedgehog inhibits the terminal differentiation of limb myoblasts committed to the slow muscle lineage. Dev Biol 242: 130–148 [DOI] [PubMed] [Google Scholar]
- Bren-Mattison Y, Hausburg M, Olwin BB 2011. Growth of limb muscle is dependent on skeletal-derived Indian hedgehog. Dev Biol 356: 486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohmann H, Jagla K, Birchmeier C 2000. The role of Lbx1 in migration of muscle precursor cells. Development 127: 437–445 [DOI] [PubMed] [Google Scholar]
- Brown CB, Engleka KA, Wenning J, Min Lu M, Epstein JA 2005. Identification of a hypaxial somite enhancer element regulating Pax3 expression in migrating myoblasts and characterization of hypaxial muscle Cre transgenic mice. Genesis 41: 202–209 [DOI] [PubMed] [Google Scholar]
- Buchberger A, Nomokonova N, Arnold HH 2003. Myf5 expression in somites and limb buds of mouse embryos is controlled by two distinct distal enhancer activities. Development 130: 3297–3307 [DOI] [PubMed] [Google Scholar]
- Buchberger A, Freitag D, Arnold HH 2007. A homeo-paired domain-binding motif directs Myf5 expression in progenitor cells of limb muscle. Development 134: 1171–1180 [DOI] [PubMed] [Google Scholar]
- Carvajal JJ, Cox D, Summerbell D, Rigby PW 2001. A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development 128: 1857–1868 [DOI] [PubMed] [Google Scholar]
- Chen JC, Goldhamer DJ 2004. The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Dev Biol 265: 502–512 [DOI] [PubMed] [Google Scholar]
- Chen JC, Love CM, Goldhamer DJ 2001. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev Dyn 221: 274–288 [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA 1996. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383: 407–413 [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Harris MP, Simandl BK, Li Y, Beachy PA, Fallon JF 2001. Manifestation of the limb prepattern: Limb development in the absence of sonic hedgehog function. Dev Biol 236: 421–435 [DOI] [PubMed] [Google Scholar]
- Coy S, Caamano JH, Carvajal J, Cleary ML, Borycki AG 2011. A novel Gli3 enhancer controls the Gli3 spatiotemporal expression pattern through a TALE homeodomain protein binding site. Mol Cell Biol 31: 1432–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cygan JA, Johnson RL, McMahon AP 1997. Novel regulatory interactions revealed by studies of murine limb pattern in Wnt-7a and En-1 mutants. Development 124: 5021–5032 [DOI] [PubMed] [Google Scholar]
- Davey MG, Paton IR, Yin Y, Schmidt M, Bangs FK, Morrice DR, Smith TG, Buxton P, Stamataki D, Tanaka M, et al. 2006. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev 20: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C 1999. The role of SF/HGF and c-Met in the development of skeletal muscle. Development 126: 1621–1629 [DOI] [PubMed] [Google Scholar]
- Duprez D, Fournier-Thibault C, Le Douarin N 1998. Sonic Hedgehog induces proliferation of committed skeletal muscle cells in the chick limb. Development 125: 495–505 [DOI] [PubMed] [Google Scholar]
- Duprez D, Lapointe F, Edom-Vovard F, Kostakopoulou K, Robson L 1999. Sonic hedgehog (SHH) specifies muscle pattern at tissue and cellular chick level, in the chick limb bud. Mech Dev 82: 151–163 [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75: 1417–1430 [DOI] [PubMed] [Google Scholar]
- Emery AE 2002. The muscular dystrophies. Lancet 359: 687–695 [DOI] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA 2005. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol 280: 396–406 [DOI] [PubMed] [Google Scholar]
- Giordani J, Bajard L, Demignon J, Daubas P, Buckingham M, Maire P 2007. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc Natl Acad Sci 104: 11310–11315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P 2005. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 132: 2235–2249 [DOI] [PubMed] [Google Scholar]
- Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M 2000. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127: 413–424 [DOI] [PubMed] [Google Scholar]
- Gustafsson MK, Pan H, Pinney DF, Liu Y, Lewandowski A, Epstein DJ, Emerson Jr CP 2002. Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes Dev 16: 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadchouel J, Carvajal JJ, Daubas P, Bajard L, Chang T, Rocancourt D, Cox D, Summerbell D, Tajbakhsh S, Rigby PW, et al. 2003. Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development 130: 3415–3426 [DOI] [PubMed] [Google Scholar]
- Havis E, Coumailleau P, Bonnet A, Bismuth K, Bonnin MA, Johnson R, Fan CM, Relaix F, Shi DL, Duprez D 2012. Sim2 prevents entry into the myogenic program by repressing MyoD transcription during limb embryonic myogenesis. Development 139: 1910–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JKH, McGlinn E, Harfe BD, Kardon G, Tabin C 2012. Autonomous and nonautonomous roles of hedgehog signaling in regulating limb muscle formation. Genes Dev (this issue) doi: 10.1101/gad.187385.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagla K, Dolle P, Mattei MG, Jagla T, Schuhbaur B, Dretzen G, Bellard F, Bellard M 1995. Mouse Lbx1 and human LBX1 define a novel mammalian homeobox gene family related to the Drosophila lady bird genes. Mech Dev 53: 345–356 [DOI] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP 2004. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev 18: 937–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA 1997. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development 124: 4729–4738 [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Tapscott SJ, Goldhamer DJ, Rudnicki MA 1999. Myogenic determination occurs independently in somites and limb buds. Dev Biol 206: 219–231 [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Tajbakhsh S, Rudnicki MA 2003. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev Biol 258: 307–318 [DOI] [PubMed] [Google Scholar]
- Kardon G, Campbell JK, Tabin CJ 2002. Local extrinsic signals determine muscle and endothelial cell fate and patterning in the vertebrate limb. Dev Cell 3: 533–545 [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S 2004. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431: 466–471 [DOI] [PubMed] [Google Scholar]
- Kelly R, Alonso S, Tajbakhsh S, Cossu G, Buckingham M 1995. Myosin light chain 3F regulatory sequences confer regionalized cardiac and skeletal muscle expression in transgenic mice. J Cell Biol 129: 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyte AL, Smith KK 2011. Developmental origins of precocial forelimbs in marsupial neonates. Development 137: 4283–4294 [DOI] [PubMed] [Google Scholar]
- Kioussi C, Gross MK, Gruss P 1995. Pax3: A paired domain gene as a regulator in PNS myelination. Neuron 15: 553–562 [DOI] [PubMed] [Google Scholar]
- Kraus P, Fraidenraich D, Loomis CA 2001. Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mech Dev 100: 45–58 [DOI] [PubMed] [Google Scholar]
- Kruger M, Mennerich D, Fees S, Schafer R, Mundlos S, Braun T 2001. Sonic hedgehog is a survival factor for hypaxial muscles during mouse development. Development 128: 743–752 [DOI] [PubMed] [Google Scholar]
- Laclef C, Hamard G, Demignon J, Souil E, Houbron C, Maire P 2003. Altered myogenesis in Six1-deficient mice. Development 130: 2239–2252 [DOI] [PubMed] [Google Scholar]
- Lee KK, Ede DA 1989. The capacity of normal and talpid3 mutant fowl myogenic cells to migrate in quail limb buds. Anat Embryol 179: 395–402 [DOI] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C 2004. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304: 1675–1678 [DOI] [PubMed] [Google Scholar]
- Li X, Blagden CS, Bildsoe H, Bonnin MA, Duprez D, Hughes SM 2004. Hedgehog can drive terminal differentiation of amniote slow skeletal muscle. BMC Dev Biol 4: 9 doi: 10.1186/1471-213X-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP 2001. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128: 5099–5108 [DOI] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL 1996. The mouse Engrailed-1 gene and ventral limb patterning. Nature 382: 360–363 [DOI] [PubMed] [Google Scholar]
- Mankoo BS, Collins NS, Ashby P, Grigorieva E, Pevny LH, Candia A, Wright CV, Rigby PW, Pachnis V 1999. Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature 400: 69–73 [DOI] [PubMed] [Google Scholar]
- Mankoo BS, Skuntz S, Harrigan I, Grigorieva E, Candia A, Wright CV, Arnheiter H, Pachnis V 2003. The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development 130: 4655–4664 [DOI] [PubMed] [Google Scholar]
- Maynard TM, Jain MD, Balmer CW, LaMantia AS 2002. High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm Genome 13: 58–61 [DOI] [PubMed] [Google Scholar]
- McDermott A, Gustafsson M, Elsam T, Hui CC, Emerson CP Jr, Borycki AG 2005. Gli2 and Gli3 have redundant and context-dependent function in skeletal muscle formation. Development 132: 345–357 [DOI] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, et al. 1997. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124: 113–123 [DOI] [PubMed] [Google Scholar]
- Niswander L, Jeffrey S, Martin GR, Tickle C 1994. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature 371: 609–612 [DOI] [PubMed] [Google Scholar]
- Ordahl CP, Le Douarin NM 1992. Two myogenic lineages within the developing somite. Development 114: 339–353 [DOI] [PubMed] [Google Scholar]
- Ott MO, Bober E, Lyons G, Arnold H, Buckingham M 1991. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development 111: 1097–1107 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA, Selever J, Wang Y, Chen YT, Mishina Y, Martin JF, Behringer RR 2006. BMP receptor type IA in limb bud mesenchyme regulates distal outgrowth and patterning. Dev Biol 295: 103–115 [DOI] [PubMed] [Google Scholar]
- Parr BA, McMahon AP 1995. Dorsalizing signal Wnt-7a required for normal polarity of D–V and A–P axes of mouse limb. Nature 374: 350–353 [DOI] [PubMed] [Google Scholar]
- Rees E, Young RD, Evans DJ 2003. Spatial and temporal contribution of somitic myoblasts to avian hind limb muscles. Dev Biol 253: 264–278 [DOI] [PubMed] [Google Scholar]
- Ribes V, Briscoe J 2009. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: The role of negative feedback. Cold Spring Harb Perspect Biol 1: a002014 doi: 10.1101/cshperspect.a002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C 1993. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75: 1401–1416 [DOI] [PubMed] [Google Scholar]
- Sagai T, Masuya H, Tamura M, Shimizu K, Yada Y, Wakana S, Gondo Y, Noda T, Shiroishi T 2004. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh). Mamm Genome 15: 23–34 [DOI] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T 2005. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132: 797–803 [DOI] [PubMed] [Google Scholar]
- Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M 1989. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature 341: 303–307 [DOI] [PubMed] [Google Scholar]
- Schafer K, Braun T 1999. Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat Genet 23: 213–216 [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4 doi: 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamataki D, Kastrinaki M, Mankoo BS, Pachnis V, Karagogeos D 2001. Homeodomain proteins Mox1 and Mox2 associate with Pax1 and Pax3 transcription factors. FEBS Lett 499: 274–278 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Bober E, Babinet C, Pournin S, Arnold H, Buckingham M 1996. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev Dyn 206: 291–300 [DOI] [PubMed] [Google Scholar]
- Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA, Li L, Lewandoski M, Gorivodsky M, Love PE, Porter FD, et al. 2009. LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136: 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swearingen JM, Lance-Jones C 1993. Spatial and temporal patterns of muscle formation in the limb of the avian embryo. Prog Clin Biol Res 383B: 553–562 [PubMed] [Google Scholar]
- Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, Birchmeier C 2005. CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev 19: 2187–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C, Roy S, Ingham PW 2003. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol 13: 1169–1181 [DOI] [PubMed] [Google Scholar]