Hybridization of Phragmites has occurred in the Gulf Coast and likely is occurring elsewhere in North America. However, detection failure may be due to limited genetic tools. Additionally, nomenclature confusion necessitates a revision of the current classification system.
Abstract
Background and aims
We review evidence for hybridization of Phragmites australis in North America and the implications for the persistence of native P. australis ssp. americanus populations in North America. We also highlight the need for an updated classification system, which takes P. australis intraspecific variation and hybridization into account.
Methodology
We reviewed available published, in press and in preparation literature to assess the likelihood of hybridization and interbreeding in genotypes of P. australis present in North America.
Principal results
Experimental results demonstrate that hybridization among introduced and native haplotypes is possible within the genus Phragmites, yet evidence that hybridization has occurred naturally is only starting to emerge. The lag in identifying hybridization in Phragmites in North America may be related to under-sampling in some parts of North America and to a lack of molecular tools that provide the capability to recognize hybrids.
Conclusions
Our understanding of the gene flow within and between species in the genus Phragmites is moving at a fast pace, especially on the east and Gulf coasts of North America. More attention should also be focused on the Great Lakes region, the southwestern and the west coast of the USA, where sympatry has created opportunities for hybridization. Where hybridizations have been detected, there are currently no published data on how hybridization affects plant vigour, morphology, invasiveness or conservation of the genetic integrity of the North American native subspecies. We conclude that the detection of more hybridization is highly likely and that there is a need to develop new markers for the different Phragmites species and lineages to fill current knowledge gaps. Finally, we suggest that the classification system for P. australis should be updated and published to help clarify the nomenclature.
Introduction
As an ecologically and economically globally important species, Phragmites australis has been of significant interest to researchers for decades (e.g. Harris and Marshall 1960; Haslam 1969; Hauber et al. 1991; van der Putten 1997; Brix 1999; Chambers et al. 1999; Meyerson et al. 1999; Orson 1999). Because of its global distribution, its ability to thrive in a wide range of environmental conditions (Meyerson et al. 2000a, b), sexual and clonal reproductive strategies (Brisson et al. 2010; Saltonstall et al. 2010) and high genetic diversity within the species (McCormick et al. 2010a, b; Saltonstall 2011; Lambertini et al. 2012a), Phragmites is increasingly used as a model species in a variety of ecological and genetic research. The identification of three distinct lineages of P. australis in North America (i.e. North American native, introduced and Gulf Coast) and the development of species-specific chloroplast and nuclear markers catalysed research on the ecology, evolution and success of different P. australis haplotypes (Saltonstall 2002, 2003). The current genetic knowledge of Phragmites worldwide is largely based on this original set of markers.
One area of particular interest for ecology and evolution is whether genotypes of this cosmopolitan grass are able to disperse across continents and interbreed within P. australis, as well as hybridize across species within the genus Phragmites. It has been speculated that hybridization in Phragmites could potentially result in offspring with even greater vigour than the highly invasive genotypes that are currently expanding across North America, and that pollen swamping or outbreeding depression could hasten the decline of North American native populations (Meyerson et al. 2010a). Phragmites australis is self-compatible (e.g. Ishii and Kadono 2002), but Kettenring et al. (2011) clearly demonstrated that in the Chesapeake Bay P. australis needs to outcross in order to produce significant amounts of viable seed. This need for outcross pollen would seem to greatly increase the likelihood of hybridization, especially in newly invaded areas where within-species pollen may not be available but where pollen from related species (or subspecies) might be abundant. Despite evidence that native and introduced populations can interbreed under controlled conditions (Meyerson et al. 2010a), no convincing data have been published that demonstrate wild hybrids resulting from crosses of the North American native and introduced Phragmites (Saltonstall 2011). Recently, however, conclusive evidence for hybridization between the introduced and the more distantly related Gulf Coast lineage has been confirmed using different molecular markers (Lambertini et al. 2012a) and that suggests that detection of interbreeding between the native and introduced lineages and native and Gulf Coast lineages is only a matter of time.
In this paper, we review evidence for hybridization of P. australis in North America and the implications for the persistence of native Phragmites populations. We also highlight the need for an updated classification system that takes P. australis intraspecific variation and hybrids into account, and the need for new molecular markers to facilitate hybrid identification.
Overview of the different lineages present in North America
A growing body of published literature from the last decade describes the ecology and genetics of both the native and introduced (haplotype M) lineages of P. australis in North America, particularly on the Atlantic coast. Fewer papers have focused on the Gulf Coast type I and the invasion of type M to the Gulf Coast (Howard et al. 2008; Hauber et al. 2011; Lambertini et al. 2012a), and only two publications have described the additional haplotypes that have recently been found in the Gulf Coast (Hauber et al. 2011; Lambertini et al. 2012a). The literature describing Phragmites in the western USA is growing, particularly in the southwest where haplotype M is sympatric with the native lineage and with haplotype I (e.g. Kulmatiski et al. 2010; Meyerson et al. 2010b; Saltonstall 2002). However, there has been very little published on Phragmites on the Pacific Coast of North America, which is colonized by both the North American native and Eurasian introduced haplotypes (Saltonstall 2002). Below, we briefly describe each of the identified lineages present in North America (summarized in Table 1) and then discuss the evidence for hybridization in some of these lineages and the likelihood that it is occurring in others.
Table 1.
Identified types of P. australis in North America. This table summarizes the origins and ranges of different haplotypes identified in the North American native, introduced and Gulf Coast lineages. Note, however, that some North American haplotypes are common and widespread, such as E, while others are relative rare and geographically localized, such as AB. The three ‘Greeny’ Phragmites types have also been found in Europe, but they may have originated elsewhere and also been introduced to Europe relatively recently. Question marks indicate ‘origin’ is probably still under investigation. 1Saltonstall (2002), 2Meadows and Saltonstall (2007), 3Hauber et al. (2011), 4Lambertini et al. (2012a, b), 5L. A. Meyerson and J. T. Cronin, in review. Morphology of the different lineages is detailed in Swearingen and Saltonstall (2010).
| Common designation | Haplotype | Origin | North America range |
|---|---|---|---|
| North American native | (A-H, S, Z, AA, AB, AC, E1/E2, E3, E4)1,2 | North America | Widely distributed |
| North American introduced | M1, L5 | Eurasia | Widely distributed |
| New European-related introductions to the Gulf Coast | M1 (Delta)3,4 Greeny 1 (M)4 Greeny 2 (AD)3,4 Greeny 3 (AI)4 |
Mediterranean region (South Europe, North Africa, Middle East) | Mississippi ‘birdfoot’ Delta, sporadically in Terrebonne Bay, LA and Grand Isle State Park, LA. Two samples in Florida |
| Europe? | Atlantic Coast, Great Lakes, Mississippi ‘birdfoot’ Delta | ||
| North America? Or Europe? | Mississippi ‘birdfoot’ Delta | ||
| South Africa? | Mississippi ‘birdfoot’ Delta | ||
| Or Europe? | |||
| North American Gulf Coast | Land (I)4 | South America (Colombia, Ecuador, Peru) or | Gulf Coast Texas to Florida, South West (California) |
| Africa (Uganda Burkina Faso, Senegal) |
Geographic distribution of Phragmites genotypes in North America
North American lineage
North American native P. australis haplotypes are distributed throughout Northern Quebec to North Carolina and west of the Great Lakes, the Pacific northwest of the USA and southern British Columbia, and the southwestern USA (Table 1). Native haplotypes of P. australis do not occur south of North Carolina on the east coast or Gulf Coast of the USA. The native haplotypes appear very closely related to each other (Saltonstall 2002; Lambertini et al. 2006, 2012a; Vachon and Freeland 2011; Saltonstall and Lambertini 2012) and are considered one single lineage in this review, though their origin is still unknown. Their closest relative appears to be haplotype Q, distributed in Asia and Australia (Saltonstall 2002; Chu et al. 2011; Saltonstall and Lambertini 2012). Lambertini et al. (2006) detected a weak nuclear relationship with Phragmites japonicus in the Far East. However, this relationship was not evident in Lambertini et al. (2012a) where North American native P. australis ssp. americanus appeared to have evolved from within P. australis. Another relationship detected recently is with Phragmites mauritianus in Zambia (Lambertini et al. 2012a), which shares a mutation in the trnT-trnL region with the native North American lineage. Phragmites diversity in Asia and Africa has so far been under-represented in phylogeographic studies at the global scale (Saltonstall 2002; Lambertini et al. 2006, 2012a). Collection and analysis of more samples from these continents promise to disclose the origin of the genus (Lambertini et al. 2006) and the history of the North American lineage.
Eleven P. australis haplotypes considered native to North America were first identified by Saltonstall in 2002 and since that time five additional native haplotypes have been added. Meadows and Saltonstall (2007) added haplotypes AB and AC, and Vachon and Freeland (2011) added haplotypes E2, E3 and E4. However, of these, only E4 is identified as a new haplotype based on Saltonstall's classification system, which does not consider cp-microsatellite variants (Saltonstall 2002).
Specifically, Vachon and Freeland (2011) submitted two identical trnT-trnL sequences that they identified as E1 and E2, but these sequences are a cp-microsatellite variant of haplotype AB (Meadows and Saltonstall 2007) following Saltonstall (2002). Similarly, haplotype E3 (Vachon and Freeland 2011; Freeland and Vachon 2012) corresponds to a cp-microsatellite variant of haplotype E, again following Saltonstall (2002). Haplotype E4 (Vachon and Freeland 2011; Freeland and Vachon 2012) is a new haplotype that would be given a new letter in the classification that Saltonstall initiated (Saltonstall 2002). Adding more complexity, there is yet another haplotype E4 that was deposited in GenBank by Chu et al. (2011) that was found in South Korea. In GenBank it is identified as P. australis, but it is thought to be P. japonicus, a haplotype closely related to haplotype AM (Lambertini et al. 2012a, b). The implications of these examples for Phragmites classification are discussed in the concluding section.
Euroasiatic lineage
Until relatively recently, it was believed that there was only a single type of introduced P. australis from Eurasia introduced to North America, haplotype M. This haplotype has been detected throughout North America, overlaps the range of native P. australis (described above) and extends into the Gulf Coast of the USA, where it is known as a ‘short form’ of P. australis (Hauber et al. 2011) or the EU-type (Lambertini et al. 2012a). However, more recently, a cp-microsatellite variant of haplotype M, described as haplotype M1 or the Delta-type (Hauber et al. 2011; Lambertini et al. 2012a, b), has been detected in the Mississippi Delta and Gulf Coast (described below in the section Gulf Coast lineages), raising the possibility that some populations have been misidentified as type M. M1 differs from haplotype M in the number of repeats in one microsatellite in the trnT-trnL region (Hauber et al. 2011; NCBI accession no. JF271678). It is, therefore, very closely related to haplotype M and is thought to originate from the Mediterranean region, extending throughout North Africa, the Middle East and Southern Europe (Lambertini et al. 2012a, b). Another introduction to North America of haplotype L (most likely from Europe) was found in Quebec, Canada, providing conclusive evidence of multiple introductions of P. australis to North America (L. A. Meyerson and J. T. Cronin, in review).
Gulf Coast lineages
Similar to the evolving understanding of the Euroasiatic lineage, Phragmites researchers had evidence for only one other lineage colonizing the Gulf Coast of the USA:haplotype I. Haplotype I was also detected in the southwestern USA (Meyerson et al. 2010b). However, multiple other haplotypes (Table 1) were recently found in the Mississippi Delta and surrounding marshes, and one sample of M1 was found also in Florida, which makes the story of Phragmites in North America more complicated and suggests additional opportunities to detect interbreeding.
Haplotype I
As with the Eurasian haplotypes (M, M1), haplotype I also exhibits cp-microsatellite variation. Gulf Coast Phragmites is one such cp-variant (also called the ‘land type’; Lambertini et al. 2012a; NCBI accession no. HQ664450) and was detected along the Gulf Coast of the USA from Texas to Florida and in the Mississippi River Delta. This haplotype is shared with a population of P. australis in South America (Ecuador, Peru) and with the species P. mauritianus in Uganda and Burkina Faso (Lambertini et al. 2012a). Nuclear alleles indicate a hybrid origin for both the Gulf Coast and the South American populations from a cross between the two species P. mauritianus and P. australis. As the current distribution ranges of these species overlap only in tropical Africa, an African origin has been suggested (Lambertini et al. 2012a). However, given the similarities between the Gulf Coast and South American populations and their long establishment in the Americas, a different earlier distribution range of P. mauritianus could also entail an autochthonous American origin. With the data available, it is not possible to distinguish between an old accidental introduction and the radiation of Phragmites species (Lambertini et al. 2012a).
European-related haplotypes
Three other recently detected haplotypes of P. australis are named for the special blue-green colour of their leaves: Greeny 1 (haplotype M), Greeny 2 (haplotype AD) and Greeny 3 (haplotype AI). Haplotype AI differs from haplotype K (Saltonstall 2002) in one single substitution in the rbcL-psaI region (Lambertini et al. 2012a; NCBI accession no. HQ664451; see Table 1). Although the three Greeny genotypes have three distinct haplotypes, they share the same European nuclear alleles (alleles 195 and 197 at locus PaGT 22, which are distinctive in this group and are shared, along with many more alleles, among the European and North American introduced genotypes). Given the high nuclear similarities among the three Greeny types, their most likely origin is somewhere in Europe. All three haplotypes (M, AD and AI) have, in fact, also been found in Europe (Lambertini et al. 2012b). However, the Greeny 2 haplotype (AD) is closely related to the native North American haplotypes, whereas the best candidate for the origin of Greeny 3 is the South African population of P. australis with haplotype K (Lambertini et al. 2012a). This suggests that the three Greeny types may also have been previously introduced to Europe as well and this possibility further clouds an identification of the historical introduction pathways.
Hybridization of Phragmites in North America
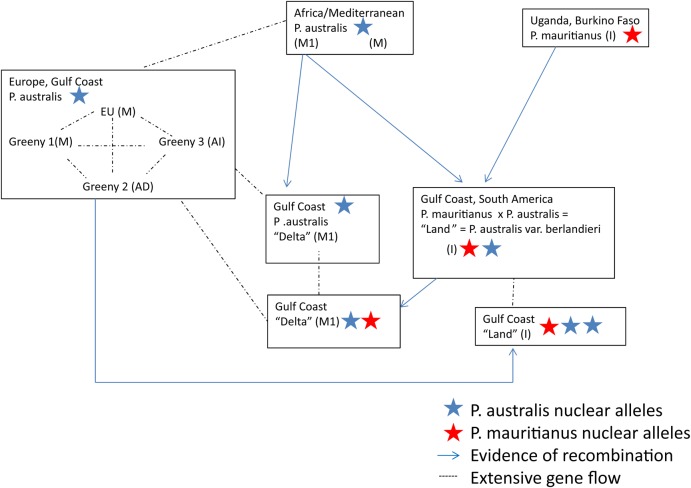
Does Phragmites hybridize in the wild in North America? The answer is probably yes, but so far the conclusive evidence is limited to the Gulf Region of the USA (Fig. 1, Table 1). An interspecific hybrid between the tropical African species P. mauritianus and P. australis became established long ago in South America and on the Gulf Coast of the USA. The hybrid is the ‘land-type’, previously described as P. australis var. berlandieri. Being an interspecific hybrid, the specific epithet australis does not appear appropriate any longer and should be dropped and renamed when the variation within haplotype I, including its hybrids, is further resolved and better understood.
Fig. 1.
Hybridization of Phragmites in the Gulf Coast of the USA. The stars represent P. mauritianus (red) and P. australis (blue) nuclear alleles and indicate how they are recombined in the hybrids. Arrows indicate parent–offspring relationships and gene flow direction detected. Dashed lines refer to high nuclear similarities among lineages which probably imply extensive gene flow. In each box, Phragmites species, haplotype and geographic location of populations involved in gene flow are indicated.
The recent introductions of the European-related haplotypes of P. australis (M, M1, AD and AI) to the Mississippi River Delta have brought the hybrid in sympatry with its paternal species P. australis. Hybridization in the Gulf Coast appears to be due to back-crossing of the P. mauritianus × P. australis hybrid (haplotype I) with P. australis haplotypes (M, M1, AD and AI) (Fig. 1). Given the high similarities in nuclear markers among haplotypes M, AD and AI, and their sympatry in Europe, it has not been possible to assign haplotype to the European alleles that introgressed into land-type Phragmites. For this reason, in Fig. 1, the dotted line refers to high nuclear similarities among haplotypes, which probably imply extensive gene flow. In this case, evidence against gene flow should be provided to exclude interbreeding.
Another interesting case suggesting gene exchange is given by the Greeny 2 genotypes of haplotype AD. Haplotype AD shares a mutation in the trnT-trnL region that appears exclusive to the native North American haplotypes, and shares the nuclear alleles with the Euroasiatic genotypes of haplotype M (Lambertini et al. 2012a). Further investigations of this group could reveal another history of hybridization.
Why has hybridization not been detected previously?
Since 2002, multiple papers have reported the failure to detect intra- or interspecific breeding in the genus Phragmites (e.g. Saltonstall 2002, 2011; Meyerson et al. 2010a, b) in the wild, despite evidence that it can occur (Meyerson et al. 2010a). Paul et al. (2010) detected possible hybrids in Canadian populations where native and introduced lineages are sympatric, but recombining alleles, providing evidence of interbreeding between the two lineages, have not been found. Recent studies by Chu et al. (2011) and Lambertini et al. (2012a) have identified an explanation for this failure. Chu et al. detected hybrids between P. japonicus and P. australis in the sequences of the PhaHKT1 gene (high-affinity K+ transporter gene). Lambertini et al. detected two hybridization events between P. mauritianus and P. australis, one where P. mauritianus is the seed parent (in the Gulf Coast and South America) and one where P. australis is the seed parent (in Senegal), in nuclear DNA fragments amplified by the grass-waxy gene primers. Introgression in P. australis in the Gulf Coast was recognized by distinguishing ancestral alleles, shared with the native populations, from newly evolved alleles, shared among haplotypes in the Gulf Coast areas but absent in the native populations and therefore probably acquired by gene flow (Lambertini et al. 2012a). Lambertini et al.'s approach, involving a large geographic and taxonomic sampling and the integration of several DNA sources, showed that microsatellite data alone may fail to detect hybridization.
The reason for this failure may be our reliance on the original set of microsatellite primers specifically developed by Saltonstall (2003) to study variation in the nuclear DNA of P. australis in North America. These markers were designed based on variation in the Euroasiatic introduced haplotype M (Saltonstall 2011), and therefore may not be optimally transferrable across species (Barbara et al. 2007) and across Phragmites haplotypes. Meyerson et al. (2010a) produced hybrids with native chloroplast but detected alleles from the Euroasiatic lineage using the microsatellite primers, yet the same microsatellites did not detect native alleles when the hybrid had a chloroplast from the Euroasiatic lineage. Microsatellites specifically designed for the maternal and paternal lineages should optimally be combined to detect hybrids (Symonds et al. 2010). However, this will only increase the support for hybridization hypotheses and will not provide compelling evidence, at least until a sufficiently wide part of the genome can be screened for hybridization. Other approaches, like the aforementioned PhaHKT1 gene or the grass-waxy primers, may work but more markers need to be developed to detect Phragmites hybrids. Until then, amplified fragment length polymorphisms (AFLPs) appear to be a simple and low-cost solution (Lambertini et al. 2006, 2012a; Kettenring and Mock 2012) to evaluate hybridization on a case-by-case basis in combination with microsatellites or other nuclear markers. Technical advances to the protocol introduced by Vos et al. (1995) have presented new opportunities for data analysis (Bensch and Åkesson 2005; Meudt and Clarke 2007), among which are adaptations for the study of hybrids (Vela et al. 2011).
Another reason that microsatellites have failed to detect hybrids may be that polysomic variation (samples with more than two alleles at a microsatellite locus) has so far been largely disregarded. Microsatellite software programs are mostly designed for diploid organisms, so three or more co-dominant alleles cannot be analysed in two-entry matrices. Binary matrices are an alternative for the analysis of polysomic markers and a few programs for tetraploids have been developed (AUTOTET, Thrall and Young 2000; TETRA, Liao et al. 2008; TETRASAT, Markwith et al. 2006; ATETRA, van Puyvelde et al. 2010) and for polyploids with different ploidy levels (PopDist, Guldbrandsten et al. 2000). Given the different ways of handling heterozygotes, calculations of Fst statistics are determined according to ploidy level and should be taken into account when interpreting the results (van Tienderen and Meirmans 2012). While difficult to analyse, polysomic variation may in fact provide evidence of hybridization. Polysomies reflect genomes of recent polyploid origin (which might include F1 hybrids and allopolyploids) that have not yet undergone diploidization (Otto and Whitton 2000) and/or that have somatic instability in chromosome number (Li et al. 2010). An excellent review on polyploidy, hybridization and invasion was recently published by te Beest et al. (2012).
Interbreeding between European and North American P. australis
Meyerson et al. (2010a) showed that no phenological or genetic barriers existed between the North American native and European (M) lineages when the populations were hand-crossed. The recent work by Lambertini et al. (2012a) and the earlier evidence provided by Meyerson et al. (2010a) make the likelihood of conclusive evidence of wild hybrids of the North American and European lineages a near certainty. Saltonstall (2011) showed that despite multiple threats, the genetic diversity in extant populations of native P. australis in eastern North America is being maintained. However, it would be worthwhile to re-analyse these populations for evidence of gene flow using different molecular approaches.
Conclusions and forward look
Our understanding of the gene flow within and between species in the genus Phragmites is moving at a fast pace. The new approaches that have confirmed Phragmites hybridization in the Gulf Coast represent significant progress and promise to provide insights for Phragmites gene flow throughout North America. While the east coast of North America is likely to be a focal point for research because of the extensive sympatry of North American native and Eurasian introduced P. australis, the Great Lakes region, the southwest and west coast deserve more attention. Furthermore, we do not yet have data on how hybridization will affect vigour, morphology and invasiveness of the introduced types or alter conservation strategies for the native Phragmites lineage, but these clearly warrant additional investigations, as highlighted by Schierenbeck and Ellstrand in their 2009 review of hybridization and invasion. In addition, there is a need to develop new markers for the different Phragmites species and lineages.
The lack of a published standardized classification system has resulted in a confused nomenclature. Several sequences are deposited in GenBank that are identified using letters that should indicate haplotype but do not follow the classification system implemented by Saltonstall (2002) and therefore are misleading and can be misinterpreted. In addition, often only one of the two sequences needed to identify Phragmites haplotypes is deposited (e.g. either trn-T or rbc-L) and no indication of the haplotype of the other sequence is provided in GenBank or in publications. Therefore, haplotypes already deposited in GenBank should be revised as needed and meta-data, such as information on the sample collection site, would be helpful.
Furthermore, Phragmites researchers must reach consensus on whether the microsatellite variations in the trnT-trnL and rbcL-psaI regions that are frequently detected constitute new haplotypes (requiring new labels) or whether the cp-microsatellite variants simply represent intra-haplotype variation. In the latter case, these variants should also be coded consistently. Finally, developing an accessible common published classification system would greatly increase the understanding of Phragmites distribution and phylogeography worldwide. While K. Saltonstall and C. Lambertini (pers. commun.) have begun to examine this issue, contributions from the wider research community would make this effort more robust.
A revision of the taxonomic and systematic classification of Phragmites is also needed, but also needed are morphological characters and nuclear markers to describe and identify Phragmites hybrids. It is especially relevant to further investigate DNA variation within haplotypes, particularly within haplotype I, which was recently shown to hybridize liberally (Lambertini et al. 2012a, b). These missing pieces of the puzzle are critical to ascertain the most appropriate classification system for species that readily interbreed and cannot be classified into separate species based on biological species concept (i.e. reproductive barriers, Mayr 1942).
The genus Phragmites is an excellent model system for studying ecology, evolution and species invasions, and is particularly interesting from the perspective of inter- and intraspecific hybridization and reverse evolution. Dogged pursuit by researchers to solve the issues raised in this paper will yield insights and opportunities for future studies.
Sources of funding
Funding for this paper was provided in part to L.A.M. by the US National Science Foundation DEB Award 1049914; the Fulbright Commission of the United States and the Czech Republic; and the University of Rhode Island College of Environment and Life Sciences Agricultural Experiment Station Project RI00H-332, 311000-6044. Funding to C.L. was provided by the Danish Council for Independent Research, Natural Sciences project 10-083195. Funding to M.K.M. and D.F.W. was provided in part by National Oceanic and Atmospheric Administration – Center for Sponsored Coastal Ocean Research grant #NA09NOS4780214.
Contributions by the authors
L.A.M. and C.L. contributed the greatest effort to early drafts of the manuscript, but all authors contributed to the various revisions. All authors have seen and agreed to the submitted manuscript.
Conflict of interest statement
None declared.
Acknowledgements
L.A.M. wishes to thank Professor Petr Pyšek at the Institute of Botany in the Academy of Sciences of the Czech Republic for support. C.L. thanks Hans Brix at the Department of Bioscience, Plant Biology, Aarhus University, for postdoctoral support.
References
- Barbara T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C. Cross-species transfer of nuclear microsatellite markers: potential and limitations. Molecular Ecology. 2007;16:3759–3767. doi: 10.1111/j.1365-294X.2007.03439.x. [DOI] [PubMed] [Google Scholar]
- Bensch S, Åkesson M. Ten year of AFLP in ecology and evolution: why so few animals? Molecular Ecology. 2005;14:2899–2914. doi: 10.1111/j.1365-294X.2005.02655.x. [DOI] [PubMed] [Google Scholar]
- Brisson J, de Blois S, Lavoie C. Roadsides as invasion pathway for common reed (Phragmites australis) Invasive Plant Science and Management. 2010;3:506–514. [Google Scholar]
- Brix H. Genetic diversity, ecophysiology and growth dynamics of reed (Phragmites australis) Aquatic Botany. 1999;64:179–184. [Google Scholar]
- Chambers RM, Meyerson LA, Saltonstall K. Expansion of reed into tidal wetlands of North America. Aquatic Botany. 1999;64:261–273. [Google Scholar]
- Chu H, Cho WK, Jo Y, Kim W, II, Rim Y, Kim R-Y. Identification of natural hybrids in Korean Phragmites using haplotype and genotype analyses. Plant Systematics and Evolution. 2011;293:247–253. [Google Scholar]
- Freeland J, Vachon N. Repetitive sequences in phylogeographic inference: a reply to Saltonstall and Lambertini. 2012. Molecular Ecology Resources. 2012;12:586–589. doi: 10.1111/j.1755-0998.2012.03146.x. [DOI] [PubMed] [Google Scholar]
- Guldbrandsten B, Tomiuk J, Loeschcke V. Popdist, Version 1.1.1: a program to calculate population genetic distance and identity measures. Journal of Heredity. 2000;91:178–179. doi: 10.1093/jhered/91.2.178. [DOI] [PubMed] [Google Scholar]
- Harris SW, Marshall WH. Experimental germination and seed establishment of seedlings of Phragmites communis. Ecology. 1960;41:395. [Google Scholar]
- Haslam SM. Stem types of Phragmites communis Trin. Annals of Botany. 1969;33:127–131. [Google Scholar]
- Hauber DP, White DA, Powers SP, DeFrancesch FR. Isozyme variation and correspondence with unusual infrared reflectance patterns in Phragmites australis (Poaceae) Plant Systematics and Evolution. 1991;178:1–8. [Google Scholar]
- Hauber DP, Saltonstall K, White DA, Hood CS. Genetic variation in the common reed, Phragmites australis, in the Mississippi River Delta marshes: evidence for multiple introductions. Estuaries and Coasts. 2011;34:851–862. [Google Scholar]
- Howard R, Travis JSE, Stiles BA. Rapid growth of a Eurasian haplotype of Phragmites australis in a restored brackish marsh in Louisiana, USA. Biological Invasions. 2008;10:369–379. [Google Scholar]
- Ishii J, Kadono Y. Factors influencing seed production of Phragmites australis. Aquatic Botany. 2002;72:129–141. [Google Scholar]
- Kettenring KM, Mock KE. Genetic diversity, reproductive mode, and dispersal differ between the cryptic invader, Phragmites australis, and its native conspecific. Biological Invasions. 2012 10.1007/s10530-012-0246-5. [Google Scholar]
- Kettenring K, McCormick MK, Baron HM, Whigham DF. Mechanisms of Phragmites australis invasion: feedbacks among genetic diversity, nutrients, and sexual reproduction. Journal of Applied Ecology. 2011;48:1305–1313. [Google Scholar]
- Kulmatiski A, Beard KH, Meyerson LA, Gibson JC, Mock KE. Reconstruction of a cryptic invasion of Phragmites australis reveals potential for native haplotype extinction. Western North American Naturalist. 2010;70:541–552. [Google Scholar]
- Lambertini C, Gustafsson MHG, Frydenberg J, Lissner J, Speranza M, Brix H. A phylogeographic study of the cosmopolitan genus Phragmites (Poaceae) based on AFLPs. Plant Systematics and Evolution. 2006;258:161–182. [Google Scholar]
- Lambertini C, Mendelsshon I, Gustafsson MGH, Olesen B, Riis T, Sorrell BK, Brix H. Tracing the origin of Gulf Coast Phragmites (Poaceae)—a story of long distance dispersal and hybridization. American Journal of Botany. 2012a;99:538–551. doi: 10.3732/ajb.1100396. [DOI] [PubMed] [Google Scholar]
- Lambertini C, Sorrell BK, Riis T, Olesen B, Brix H. Exploring the borders of European Phragmites within a cosmopolitan genus. AoB PLANTS. 2012b doi: 10.1093/aobpla/pls020. pls020; doi:10.1093/aobpla/pls020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Guo W, Wang B, Li X, Chen H, Wei L, Wand Y, Wu J, Long H. Instability of chromosome number and DNA methylation variation induced by hybridization and amphidiploid formation between Raphanus sativus L. and Brassica alboglabra Bailey. BMC Plant Biology. 2010;10:207. doi: 10.1186/1471-2229-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WJ, Zhu BR, Zeng YF, Zhang DY. TETRA: an improved program for population genetic analysis of allotetraploid microsatellite data. Molecular Ecology Resources. 2008;8:1260–1262. doi: 10.1111/j.1755-0998.2008.02198.x. [DOI] [PubMed] [Google Scholar]
- Markwith SH, Stewart DJ, Dyer JL. TETRASET: a program for the population analysis of allotetraploid microsatellite data. Molecular Ecology Notes. 2006;6:586–589. [Google Scholar]
- Mayr E. Systematics and the origin of species. New York, USA: Columbia University Press; 1942. [Google Scholar]
- McCormick MK, Kettenring KM, Baron HM, Whigham DF. Extent and mechanisms of Phragmites australis spread in the Rhode River subestuary of the Chesapeake Bay, Maryland (USA) Wetlands. 2010a;30:67–74. [Google Scholar]
- McCormick MK, Kettenring KM, Baron HM, Whigham DF. Spread of invasive Phragmites australis in estuaries with differing degrees of development: genetic patterns, Allee effects and interpretation. Journal of Ecology. 2010b;98:1369–1378. [Google Scholar]
- Meadows RE, Saltonstall K. Distribution of native and introduced Phragmites australis in freshwater and oligohaline tidal marshes of the Delmarva Peninsula and southern New Jersey. Journal of the Torrey Botanical Society. 2007;134:99–107. [Google Scholar]
- Meudt HM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends in Plant Science. 2007;12:106–117. doi: 10.1016/j.tplants.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Meyerson LA, Chambers RM, Vogt KA. The effects of Phragmites removal on nutrient pools in a freshwater tidal marsh ecosystem. Biological Invasions. 1999;1:129–136. [Google Scholar]
- Meyerson LA, Saltonstall K, Windham L, Kiviat E, Findlay S. A comparison of Phragmites australis in freshwater and brackish marsh environments in North America. Wetlands Ecology and Management. 2000a;8:89–103. [Google Scholar]
- Meyerson LA, Vogt KA, Chambers RM. Linking the success of Phragmites australis to the decoupling of ecosystem nutrient cycles. In: Weinstein M, Kreeger D, editors. Concepts and controversies of tidal marsh ecology. Kluwer, Dordrecht; 2000b. pp. 817–834. [Google Scholar]
- Meyerson LA, Viola D, Brown RN. Hybridization of invasive Phragmites australis with a native subspecies in North America. Biological Invasions. 2010a;12:103–111. [Google Scholar]
- Meyerson LA, Lambert A, Saltonstall K. A tale of three lineages: expansion of common reed (Phragmites australis) in the U.S. Southwest and Gulf Coast. Invasive Plant Science and Management. 2010b;3:489–494. [Google Scholar]
- Orson R. A paleoecological assessment of Phragmites australis in New England tidal marshes: changes in plant community structure during the last millennium. Biological Invasions. 1999;1:149–158. [Google Scholar]
- Otto SP, Whitton J. Polyploid evidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Paul J, Vachon N, Garroway CJ, Freeland JR. Molecular data provide strong evidence of natural hybridization between native and introduced lineages of Phragmites australis in North America. Biological Invasions. 2010;12:2967–2973. [Google Scholar]
- Saltonstall K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proceedings of the National Academy of Sciences of the USA. 2002;99:2445–2449. doi: 10.1073/pnas.032477999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall K. Microsatellite variation within and among North American lineages of Phragmites australis. Molecular Ecology. 2003;12:1689–1702. doi: 10.1046/j.1365-294x.2003.01849.x. [DOI] [PubMed] [Google Scholar]
- Saltonstall K. Remnant native Phragmites australis maintains genetic diversity despite multiple threats. Conservation Genetics. 2011;12:1027–1033. [Google Scholar]
- Saltonstall K, Lambertini C. The value of repetitive sequences in chloroplast DNA for phylogeographic inference: a comment on Vachon and Freeland, 2011. Molecular Ecology Resources. 2012;12:581–585. doi: 10.1111/j.1755-0998.2012.03146.x. [DOI] [PubMed] [Google Scholar]
- Saltonstall K, Lambert A, Meyerson LA. Genetics and reproduction of common (Phragmites australis) and giant reed (Arundo donax) Invasive Plant Science and Management. 2010;3:495–505. [Google Scholar]
- Swearingen J, Saltonstall K. 2010. Phragmites field guide: distinguishing native and exotic forms of common reed (Phragmites australis) in the United States. Plant Conservation Alliance, Weeds Gone Wild. http://www.nps.gov/plants/alien/pubs/index.htm .
- Symonds VV, Soltis PS, Soltis ES. Dynamics of polyploid formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution. 2010;64:1984–2003. doi: 10.1111/j.1558-5646.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux J, 2, Richardson DM, Brysting AK, Suda J, Kubesova M, Pysek P. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall PH, Young A. AUTOTET: a program for analysis of autotetraploid genotypic data. The Journal of Heredity. 2000;91:348–349. doi: 10.1093/jhered/91.4.348. [DOI] [PubMed] [Google Scholar]
- van der Putten WH. Die-back of Phragmites australis in European wetlands: an overview of the European Research Programme on Reed Die-back and Progression (1993–1994) Aquatic Botany. 1997;59:263–275. [Google Scholar]
- van Puyvelde K, van Geert A, Triest L. ATETRA, a new software program to analyse tetraploid microsatellite data: comparison with TETRA and TETRASAT. Molecular Ecology Resources. 2010;10:331–334. doi: 10.1111/j.1755-0998.2009.02748.x. [DOI] [PubMed] [Google Scholar]
- van Tienderen PH, Meirmans PG. The analysis of inheritance, diversity and population structure in polyploids: old problems and new solutions. International Proceedings of Conference on Polyploidy, Hybridization and Biodiversity; Pruhonice, Czech Republic: 2012. 7–10 May 2012. [Google Scholar]
- Vachon N, Freeland JR. Phylogeographic inferences from chloroplast DNA: quantifying the effects of mutations in repetitive and non-repetitive sequences. Molecular Ecology Resources. 2011;11:279–285. doi: 10.1111/j.1755-0998.2010.02921.x. [DOI] [PubMed] [Google Scholar]
- Vela D, Guerreiro PGG, Fontdevila A. Adaptations of the AFLP technique as a new tool to detect genetic instability and transposition in interspecific hybrids. BioTechniques. 2011;50:247–250. doi: 10.2144/000113655. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]