Summary
Clinically useful diagnostic tests of dengue virus infection are lacking. We prospectively evaluated the performance of real-time reverse transcriptase (rRT)-PCR, NS-1 antigen and IgM antibody tests to confirm dengue virus infection in acute blood specimens from 162 patients presenting with undifferentiated febrile illness compatible with dengue infection. rRT-PCR was the most sensitive test (89%) and potentially could be used as a single test for confirmation of dengue infection. NS-1 antigen and IgM antibody were not sufficiently sensitive to be used as a single confirmatory test with sensitivities of 54% and 17% respectively. The specificities of rRT-PCR, NS-1 antigen and IgM antibody tests were 96%, 100% and 88% respectively. Combining NS-1 and rRT-PCR or the combination of all three tests resulted in the highest sensitivity (93%) but specificities dropped to 96% and 83% respectively. We conclude that at least the combination of two tests, either agent detection (rRT-PCR) or antigen detection (NS-1) plus IgM antibody detection should be used for laboratory confirmation of dengue infection.
Keywords: Dengue, PCR, Serology, Evaluation, Diagnosis, Thailand
1. Introduction
Dengue virus is the most important arboviral disease in humans, with an estimated 100 million cases of dengue fever (DF) and several hundred thousand cases of dengue haemorrhagic fever (DHF) each year.1 Cases of DF and DHF were increasingly reported in nine countries within the South East Asia region between 1985 and 2006, with Thailand reporting the highest number of cases in the region until 2003.2 In South East Asia, adults with dengue virus infection usually present with an acute, undifferentiated, febrile illness.3, 4, 5, 6, 7 Previous reports have documented the difficulty in clinically differentiating dengue from other causes of fever, including leptospirosis8 and scrub typhus.5 Given this difficulty, and the fact that delayed antimicrobial treatment for such infections may result in increased mortality, reliable and rapid dengue confirmatory tests are needed. Additionally, rapid confirmation of dengue infection would facilitate improved monitoring of confirmed cases for development of complications such as shock or haemorrhage.9
Accurate laboratory confirmation of dengue infection involves a combination of tests depending on timing of infection. During the acute phase of infection, virus culture, nucleic acid detection (RT-PCR)10, 11, 12 or antigen detection (for example, by NS-1 antigen ELISA13, 14, 15) may be used for diagnosis. Serology is also used to confirm infection and distinguish primary and secondary infections by determining the differences between IgM and IgG antibody response and is currently more widely used as one of the laboratory diagnostic methods.16 There are a variety of serological methods described, including ELISA and haemagglutination inhibition (HI) tests, some of which are commercially available.17, 18 However, serological diagnosis of dengue infection requires paired serum specimens, resulting in retrospective, rather than rapid and clinically useful, confirmation of infection.
In the present paper, we have prospectively evaluated three diagnostic methodologies (IgM antibody ELISA, NS-1 antigen ELISA, and real-time reverse transcriptase [rRT]-PCR) for the detection of dengue virus infection in patients presenting with undifferentiated fever in a dengue endemic area, with the aim of determining the optimal acute specimen testing strategy in a routine clinical setting. This is the first head-to-head assessment of PCR and a selection of widely-applied commercial serological assays (NS-1 antigen, IgM and IgG antibodies). It is also the first diagnostic assessment that has demonstrated the improved diagnostic accuracy when selections of PCR, NS-1 antigen, IgM and IgG antibody test results are combined.
2. Materials and Methods
2.1. Patients
Shoklo Malaria Research Unit (SMRU) undertakes surveillance for malaria and other infectious diseases, and provides general medical care for the migrant and refugee population, in five clinics in rural Thailand, on the Thailand-Myanmar (Burma) border. We conducted dengue surveillance from April to August 2008 in the SMRU clinics at Mawker Thai village (MKT) and Maela refugee camp (MLA), both located in rural Tak province approximately 500 km northwest of Bangkok. Adult patients (age ≥15 years old) presenting to the clinics with fever ≥38°C of less than seven days duration and any clinical symptoms or physical signs consistent with dengue (abnormal bleeding, eye redness, headache, myalgia or rash) were included in the surveillance and blood was drawn to inform clinical management (blood culture, complete blood count, and plasma for serology). Patients who had a clear alternative diagnosis such as malaria, urinary tract infection or pneumonia were excluded from the surveillance.
In addition to dengue virus, patients were serologically investigated for leptospirosis (ELISA with confirmation by the microscopic agglutination test) and rickettsial infection (ELISA with confirmation by indirect immunofluorescence assay), the other common causes of undifferentiated fever in SE Asia.
2.2. Plasma specimens
A 6 ml acute venous blood specimen was collected from each patient in a sterile EDTA tube (Becton Dickinson, Franklin Lakes, NJ, USA) for dengue rRT-PCR, NS-1 antigen detection, and dengue IgM and IgG antibody detection tests. The patients were reviewed at a 10–14 day follow-up visit and a repeat 6 ml convalescent venous blood specimen was collected for dengue IgM and IgG antibody tests. Plasma was separated by centrifugation and frozen at −80 °C prior to the assays.
2.3. RNA extraction and RT-PCR
Viral RNA was extracted from 150 μl of the acute plasma specimens using the NucleoSpin RNA virus extraction kit (Macherey Nagel, Düren, Germany) according to the manufacturer's instructions and was stored at −80 °C prior to testing. One-step SYBR Green-based rRT-PCR, modified from Shu et al., was performed using the RotorGene 6000 real-time PCR system (Corbett Research, San Francisco, CA, USA).11 The reaction volume was 25 μl, comprising 10 μl of extracted RNA, 1 μl of each primer (10 μM), 12.5 μl of 2X SYBR Green reaction mix (containing 0.4 mM of each dNTP and 6 mM MgSO4) and 0.5 μl of SuperScript III RT/Platinum Taq Mix (SuperScript III Platinum SYBR Green One-Step Quantitative RT-PCR Kit, Invitrogen, Carlsbad, CA, USA). The amplification conditions consisted of reverse transcription at 50 °C for 30 min, 95 °C for 5 min for Taq inhibitor inactivation, followed by 45 cycles of 95 °C for 10 s, 54 °C for 30 s and 72 °C for 30 s. Melting curve analysis was used to confirm the specificity of the amplicons. Positive (extracted RNA from control strains of DENV1-4) and negative controls were included in each PCR run and the run only accepted if all controls gave appropriate results.
The serotype of dengue virus was sought from the acute plasma specimen in all patients with serologically confirmed dengue infection using a nested RT-PCR assay,12 modified by the Armed Forces Research Institute of Medical Sciences (AFRIMS), Bangkok, Thailand.19
2.4. NS-1 antigen detection
The Panbio Dengue Early ELISA (cat. no. E-DEN01P, lot. no. 08140; Panbio, Brisbane, Queensland, Australia) was used to detect NS-1 antigen in the acute plasma specimens only, following the manufacturer's instructions. A positive specimen was defined as having >11 Panbio units; <9 Panbio units was defined as negative; and 9–11 Panbio units was equivocal and the specimen retested to confirm the result.
Panbio units were calculated by first determining the assay cut-off value: the lot specific calibration factor was multiplied by the average absorbance result of the kit calibrator (internal control, run in triplicate). Subsequently, an index value was calculated for each patient specimen by dividing the specimen absorbance result by the cut-off value. Finally the Panbio units were determined by multiplying the index value by 10.
2.5. Dengue IgM and IgG antibody capture ELISAs
We used the dengue IgM (Panbio: cat. no. E-DEN01 M, lot. no. 08316) and IgG (Panbio: cat. no. E-DEN02G, lot. no. 09080) antibody capture ELISAs to detect IgM and IgG antibodies in both the acute and convalescent plasma specimens, following the manufacturer's instructions. A positive specimen was defined as having >11 Panbio units for IgM and >22 for IgG antibodies; <9 and <18 Panbio units was defined as negative for IgM and IgG antibodies, respectively; 9–11 Panbio units was equivocal for IgM and 18–22 Panbio units was equivocal for IgG antibodies and the specimen retested to confirm the result. Panbio units were calculated as described above.
2.6. Classification of dengue infection status
Dengue infection was classified using the above described commercial serological assays as ‘confirmed’ acute dengue infection based on WHO dengue diagnostic criteria as defined in Table 1. ‘Confirmed’ acute dengue cases were those that demonstrated an IgM or IgG antibody sero-conversion based on paired serum collections.16 Patients with static IgM positivity (i.e., positive in both acute and convalescent specimens, but with no rise in Panbio units) were considered to have evidence of recent dengue infection.
Table 1.
Interpretation algorithm for dengue virus serology using the Panbio Dengue Early ELISA, IgM and IgG antibody capture ELISA kits
| Patients (n = 76) | Acute specimen |
Convalescent specimen |
Interpretations |
|||||
|---|---|---|---|---|---|---|---|---|
| [Median Panbio Units (IQR)] |
Justification for ‘acute dengue’ interpretation | WHO criteria interpretation | Reference interpretation | |||||
| NS1 Ag ≥ 11 Positive | IgM ≥ 11 Positive | IgG ≥ 22 Positive | IgM ≥ 11 Positive | IgG ≥ 22 Positive | ||||
| 4 | 1.5 (1.5-1.6) | 30.1(23.5-33.8) | 4.0 (3.2-6.8) | 24.4 (20.9-27.5) | 4.2 (3.2-7.0) | IgM positive but no increase | - | Recent Dengue Infection |
| 3 | 29.0 (24.9-29.4) | 2.1 (1.9-2.3) | 1.2 (1.1-1.3) | 47.7 (32.9-49.0) | 16.0 (12.1-16.9) | NS1 Ag positive; IgM sero-conversion | Confirmed | Confirmed Acute Dengue Infection |
| 23 | 2.4 (1.7-5.8) | 3.7 (2.8-5.2) | 2.1 (1.7-3.8) | 34.5 (24.4 -47.0) | 43.4 (38.0-49.7) | IgM & IgG sero-conversion | ||
| 6 | 3.1 (2.3-4.1) | 4.0 (3.6-4.1) | 6.0 (2.3-7.9) | 7.6 (6.8-8.4) | 45.3 (39.9-51.5) | IgG sero-conversion | ||
| 4 | 4.3 (4.0-5.4) | 21.8 (16.5-29.5) | 3.3 (1.3-5.3) | 56.9 (54.2-57.3) | 43.1 (42.6-44.4) | IgM pairs increase; IgG sero-conversion | ||
| 27 | 24.2 (16.1-37.2) | 4.1 (2.8-6.5) | 1.2 (1.0-1.8) | 45.5 (35.1-51.8) | 41.5 (39.8-46.8) | NS1 Ag positive; IgM & IgG sero-conversion | ||
| 1 | 41.4 | 4.8 | 23.7 | 12.1 | 40.5 | NS1 Ag positive; IgG pairs increase | ||
| 8 | 23.4 (14.1-37.1) | 21.8 (13.9-40.0) | 3.3 (2.5-4.7) | 46.0 (29.8-51.5) | 37.2 (34.1-41.3) | NS1 Ag positive; IgM pairs increase; IgG sero-conversion | ||
2.7. Data Analysis
All statistical analyses were performed by using STATA/SE for Macintosh, version 10.1 (Stata Corporation, College Station, TX, USA). Continuous variables were compared using Wilcoxon rank sum test (since non-normally distributed). Characteristics of the tests were analysed in 2 x 2 tables using the ‘diagt’ routine.20 The McNemar test was used to compare sensitivities of the tests.
3. Results
3.1. Patients
Two hundred and twenty nine patients were examined. All patients met the WHO clinical case definition for acute dengue infection.21 One hundred and sixty two patients (71%) returned for the follow-up specimen collection visit and were considered further.
Of the 162 patients included in the current analysis, 72 patients (44%) were given a laboratory confirmed diagnosis of dengue infection by paired serology (Table 1). Four patients were found to have evidence of recent dengue infection. The demographic and clinical data of the patients are shown in Table 2. Patients with confirmed dengue infection had lower white cell counts (4.8 vs 7.2, P<0.0001) and platelets (147 vs. 162, P=0.03) than patients with non-dengue infection.
Table 2.
Demographic and clinical data of patients (n = 162)
| Dengue cases (n = 72) | Non-dengue cases (n = 90) | |
|---|---|---|
| Sex (male) | 42/72 (58%) | 56/90 (62%) |
| Age (years) | 23 (range: 15–63) | 27 (range: 15–60) |
| Headache | 72/72 (100%) | 86/90 (96%) |
| Arthralgia | 55/72 (76%) | 67/90 (74%) |
| Myalgia | 44/72 (61%) | 57/90 (63%) |
| Retro-orbital pain | 46/72 (64%) | 52/90 (58%) |
| Rash | 4/72 (6%) | 4/90 (4%) |
| Fever duration (days) | 2 (range: 1–5) | 2 (range: 1–6) |
| Presenting temperature (°C) | 38.8 (IQR: 38.3–39.2) | 38.4 (IQR: 38.0–38.9) |
| Haemoglobin (g/dL) | 13.3 (IQR: 12.0–14.6) | 13.7 (IQR: 12.1–14.4) |
| White cell count (x103/mm3) | 4.8 (IQR: 3.6–6.4) | 7.2 (IQR: 5.2–9.7) |
| Platelets (x103/mm3) | 147 (IQR: 106–196) | 162 (IQR: 133–212) |
Twenty six patients (16%) were found to have non-dengue causes for their illness: four patients (2%) grew Salmonella typhi from blood cultures, by serology nine patients (6%) had murine typhus, seven (4%) had scrub typhus, and six (4%) had leptospirosis. Only one patient had evidence of dual infection: acute secondary dengue and typhoid; this patient had positive NS-1 antigen and dengue rRT-PCR and also grew Salmonella typhi from a blood culture.
The serotype of dengue virus was sought by performing nested RT-PCR on the acute specimens from the 72 serologically confirmed cases. Seventy one of the cases (99%) were serotype 3, with the remaining case serotype 2. The four patients with serological evidence of recent dengue infection were negative in the nested RT-PCR.
3.2. Comparison of the characteristics of the individual diagnostic tests
The performance characteristics of NS-1 antigen detection, rRT-PCR, and IgM antibody detection in the acute plasma specimen against our ‘gold standard’ of paired serology are shown in Table 3. NS-1 antigen or IgM antibody test alone had low sensitivity compared with the paired serology result, 54% and 17% respectively. rRT-PCR sensitivity was 89%, significantly higher than both NS-1 antigen and IgM antibody tests (P<0.00001). Specificities of NS-1 antigen detection, rRT-PCR, and IgM antibody detection were 100%, 96% and 88% respectively.
Table 3.
Diagnostic accuracy of NS-1 antigen detection, rRT-PCR, and IgM antibody detection on acute plasma specimens
| Tests | Paired serology result |
Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % (95% CI) | NPV % (95% CI) | |||
|---|---|---|---|---|---|---|---|---|
| Dengue | Not dengue | |||||||
| Individual assays | NS-1 | + | 39 | 0 | 54.2 (42.0-66.0) | 100 (96.0-100) | 100 (91.0-100) | 73.2 (64.4-80.8) |
| − | 33 | 90 | ||||||
| rRT-PCR | + | 64 | 4 | 88.9 (79.3-95.1) | 95.6 (89.0-98.8) | 94.1 (85.6-98.4) | 91.5 (83.9-96.3) | |
| − | 8 | 86 | ||||||
| IgM | + | 12 | 11 | 16.7 (8.9-27.3) | 87.8 (79.2-93.7) | 52.2 (30.6-73.2) | 56.8 (48.2-65.2) | |
| − | 60 | 79 | ||||||
| Combined assays | NS-1+rRT-PCR | + | 67 | 4 | 93.1 (84.5-97.7) | 95.6 (89.0-98.8) | 94.4 (86.2-98.4) | 94.5 (87.6-98.2) |
| − | 5 | 86 | ||||||
| NS-1+IgM | + | 43 | 11 | 59.7 (47.5-71.1) | 87.8 (79.2-93.7) | 79.6 (66.5-89.4) | 73.1 (63.8-81.2) | |
| − | 29 | 79 | ||||||
| rRT-PCR+IgM | + | 66 | 15 | 91.7 (82.7-96.9) | 83.3 (74.0-90.4) | 81.5 (71.3-89.2) | 92.6 (84.6-97.2) | |
| − | 6 | 75 | ||||||
| NS-1+rRT-PCR+IgM | + | 67 | 15 | 93.1 (84.5-97.7) | 83.3 (74.0-90.4) | 81.7 (71.6-89.4) | 93.8 (86.0-97.9) | |
| − | 5 | 75 | ||||||
rRT: reverse transcriptase real-time; PPV: positive predictive value; NPV: negative predictive value.
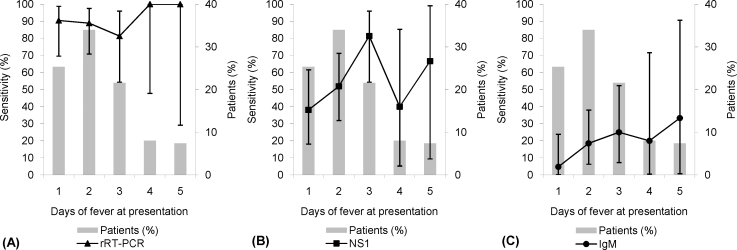
The effect of fever duration at presentation on assay sensitivity is shown in Figure 1. NS-1 antigen sensitivity peaked in the early stages of fever (three days of fever at presentation). IgM antibody sensitivity rose later, peaking in patients presenting with five days of fever. The sensitivity of rRT-PCR remained high throughout. However, as a result of the relatively small number of specimens on each day, particularly for four and five days of fever, the 95% confidence intervals around the sensitivities are wide.
Figure 1.
The effect of fever duration on test sensitivity: (A) rRT-PCR, (B) NS-1 antigen and (C) IgM antibody.
3.3. Comparison of the characteristics of diagnostic test combinations
The performance characteristics of combinations of the acute specimen tests are shown in Table 3 (combined by an ‘OR’ operator). The overall sensitivity of rRT-PCR combined with IgM antibody detection (92%) was significantly better than IgM antibody detection alone (P<0.00001), but not significantly better than rRT-PCR alone (P=0.5). This was also true for the combination of IgM+NS-1+rRT-PCR (93%), which was significantly better than NS-1 antigen or IgM antibody detection alone (P<0.00001), but not better than rRT-PCR alone (P=0.2). The combination of NS-1+rRT-PCR was significantly more sensitive than NS-1 antigen detection alone (P<0.00001), but was not significantly better than rRT-PCR alone (P=0.2). NS-1+IgM was significantly more sensitive than IgM antibody alone (P<0.0001), but not significantly better than NS-1 antigen detection alone (P=0.1). Combining tests resulted in a fall in specificity. Combining rRT-PCR with IgM antibody alone or with IgM antibody and NS-1 antigen resulted in a specificity of 83%, whereas combining NS-1 antigen with rRT-PCR or IgM antibody, the specificities were 96% and 88% respectively.
4. Discussion
An accurate and rapid method for the diagnosis of dengue virus infection would facilitate optimal patient management. In resource-poor settings, diagnosis based on clinical features is the norm but, as shown in the current study, clinical diagnosis of dengue using WHO criteria is non-specific: only 72/162 (44%) of patients meeting the clinical case definition in the current study had laboratory confirmed dengue infection. Twenty six patients (16%) meeting the dengue clinical case definition had an alternative, and antibiotic-treatable, cause for their illness. Unfortunately, for a definitive laboratory diagnosis of dengue using current techniques often a combination of tests are required.22 In spite of this, many clinical laboratories continue to rely on a single assay to confirm dengue infection, which, as we have demonstrated, may lead to diagnostic inaccuracy. Additionally, many currently available rapid immunochromatographic tests (ICT), used in the field for clinical decision-making, suffer from poor performance characteristics.19
Antigen or molecular-based assays are attractive options for rapid diagnosis of dengue infection because they can potentially detect infection before an antibody response develops. Indeed, detection of dengue NS-1 antigen by ELISA allowed detection of infection prior to sero-conversion and could be detected in serum from the first day after onset of fever up to day nine of fever.23 Shu et al. confirmed the ability of PCR to detect dengue virus RNA between day one and day seven of fever.11
In the current study, NS-1 antigen and acute IgM antibody detection did not detect the majority of confirmed dengue cases. The positive predictive value (PPV) for NS-1 antigen detection was excellent (100%) but the negative predictive value (NPV) was poor at 73%, resulting in many patients with confirmed dengue being missed if the test were to be used alone. Acute IgM antibody detection had poor PPV and NPV (52% and 57% respectively), resulting in many misdiagnosed cases. rRT-PCR had the best operating characteristics (sensitivity 89%, specificity 96%, PPV 94%, NPV 92%) and would be potentially sufficient as a single assay for confirmation of dengue infection, since it allows for accurate confirmation or refuting of infection. The combinations of NS-1+rRT-PCR or NS-1+IgM+rRT-PCR resulted in the highest sensitivity (93%), although this was associated with an inevitable fall in specificity (96% and 83% respectively).
Compared to previous studies on NS-1 antigen ELISA we report a slightly lower sensitivity. Dussart et al. found the Panbio NS-1 antigen ELISA to have a sensitivity of 60% when used on stored serum specimens from French Guiana14 and, in a similar study from Puerto Rico, Bessoff et al. reported a sensitivity of 65%.13 On prospectively collected specimens from clinically suspected dengue cases in Laos, Blacksell et al. reported a sensitivity of 63%.24 The sensitivity of rRT-PCR was slightly better than reported by the original authors who found that PCR detected viral RNA 83% of acute specimens from patients with confirmed dengue.11 Comparing operating characteristics of assays between studies can be difficult, since there are many potential confounding factors. Firstly, in the current study, specimens were collected prospectively on patients with illness broadly compatible with dengue whereas several of the previous evaluations of NS-1 antigen ELISA have been retrospective, using well characterised serum specimen collections. We feel that the results presented here are likely to more accurately reflect the operating characteristics of the tests in a routine clinical setting. Secondly, infections due to dengue serotype 3 predominated in our study, and previous work has noted that the Panbio NS-1 antigen ELISA may miss infections caused by this serotype.24 Thirdly, timing of presentation and specimen collection may affect assay performance: in our study, most patients presented very early in the course of their infection. Although we demonstrated trends in the sensitivity of each assay, the small number of patients presenting with more than three days of fever limited our ability to perform statistical analysis. Previous studies have demonstrated the effect of timing of presentation on NS-1 antigen and IgM antibody24 or PCR11 assays, but no comparison between antigen detection, PCR, and serology on the same patient population has been described. Finally, infection status (primary infection versus secondary infection) may also make study-to-study comparisons difficult. We identified very few patients with acute primary infection (3/72, using Panbio kit criteria), resulting in an inability to determine potential differences in test characteristics between primary and secondary infections. We plan to perform further work to delineate the optimum sampling ‘window’ for each assay for patients with primary and secondary dengue infection.
This work suggests that, in a clinical setting, combined tests (IgM antibody ELISA, NS-1 antigen ELISA and/or rRT-PCR) are still required to confidently diagnose acute dengue infection from a single blood specimen: at least a combination of two tests, either agent detection (rRT-PCR) or antigen detection (NS-1) plus IgM antibody should be used. Unfortunately, this results in slow laboratory confirmation of dengue infection. Given that rRT-PCR was the most accurate single assay in our assessment, prospective evaluations of new field-deployable rapid diagnostic molecular tests, for example LAMP-based assays, are planned. These evaluations will include comparisons with the newer combined NS-1 antigen and IgM antibody ICT rapid test kits. Although cheap and/or field-deployable PCR systems are some way off at the current time, progress has been made in this area and development and refinement of current techniques may result in nucleic acid detection becoming the standard for rapid dengue diagnosis even in resource-poor settings.25
Funding
This work was supported by the Wellcome Trust (Grant no. 077166/Z/05).
Conflicts of interest
None declared.
Ethical approval
Not required.
Authors’ statement: The opinions or assertions contained herein are the private views of the authors, and not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
Authors’ contributions
FHN, CLT and PT conceived the work; CLT was responsible for the clinical work and specimen collection; AT and WW conducted and interpreted the laboratory work (serology and real-time PCR) under the supervision of SDB and PT; RGJ interpreted and analysed the dengue serotyping PCR results; SDB, PT and WW performed the final data analysis. WW prepared the first draft and all authors contributed to the revision of the manuscript and read and approved the final version. WW and FHN are guarantors of the paper.
Acknowledgements
We are very grateful to all the patients who participated in this surveillance, the doctors, medical students (Cherry Alviani and Thomas Van den Bussche), nurses, and staff of the SMRU clinics and Microbiology Laboratory, and the AFRIMS staff for technical advice.
References
- 1.Monath T.P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994 Mar 29;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Dengue/DHF. Reported cases of DF/DHF in selected countries in SEA Region (1985 – 2005). http://www.searo.who.int/en/Section10/Section332_1101.htm [accessed 9 September 2009].
- 3.Shu P.Y., Su C.L., Liao T.L., Yang C.F., Chang S.F., Lin C.C. Molecular characterization of dengue viruses imported into Taiwan during 2003-2007: geographic distribution and genotype shift. Am J Trop Med Hyg. 2009 Jun;80:1039–1046. [PubMed] [Google Scholar]
- 4.Leelarasamee A., Chupaprawan C., Chenchittikul M., Udompanthurat S. Etiologies of acute undifferentiated febrile illness in Thailand. J Med Assoc Thai. 2004;87:9. [PubMed] [Google Scholar]
- 5.Watt G., Jongsakul K., Chouriyagune C., Paris R. Differentiating dengue virus infection from scrub typhus in Thai adults with fever. Am J Trop Med Hyg. 2003;68:3. doi: 10.4269/ajtmh.2003.68.536. [DOI] [PubMed] [Google Scholar]
- 6.Solomon T., Dung N.M., Vaughn D.W., Kneen R., Thao L.T., Raengsakulrach B. Neurological manifestations of dengue infection. Lancet. 2000;355:1053–1059. doi: 10.1016/S0140-6736(00)02036-5. [DOI] [PubMed] [Google Scholar]
- 7.Suttinont C., Losuwanaluk K., Niwatayakul K., Hoontrakul S., Intaranongpai W., Silpasakorn S. Causes of acute, undifferentiated, febrile illness in rural Thailand: results of a prospective observational study. Ann Trop Med Parasitol. 2006;100:8. doi: 10.1179/136485906X112158. [DOI] [PubMed] [Google Scholar]
- 8.Brown M.G., Vickers I.E., Salas R.A., Smikle M.F. Leptospirosis in suspected cases of dengue in Jamaica, 2002-2007. Trop Doct. 2010;40:92–94. doi: 10.1258/td.2009.090340. [DOI] [PubMed] [Google Scholar]
- 9.Potts J.A., Rothman A.L. Clinical and laboratory features that distinguish dengue from other febrile illnesses in endemic populations. Trop Med Int Health. 2008;13:1328–1340. doi: 10.1111/j.1365-3156.2008.02151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chutinimitkul S., Payungporn S., Theamboonlers A., Poovorawan Y. Dengue typing assay based on real-time PCR using SYBR Green I. J Virol Methods. 2005;129:8. doi: 10.1016/j.jviromet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Shu P.Y., Chang S.F., Kuo Y.C., Yuen Y.Y., Chien L.J., Sue C.L. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J Clin Microbiol. 2003;41:9. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanciotti R.S., Calisher C.H., Gubler D.J., Chang G.J., Vorndam A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992 Mar;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessoff K., Delorey M., Sun W., Hunsperger E. Comparison of two commercially available dengue virus (DENV) NS1 capture enzyme-linked immunosorbent assays using a single clinical sample for diagnosis of acute DENV infection. Clin Vaccine Immunol. 2008;15:1513–1518. doi: 10.1128/CVI.00140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dussart P., Petit L., Labeau B., Bremand L., Leduc A., Moua D. Evaluation of two new commercial tests for the diagnosis of acute dengue virus infection using NS1 antigen detection in human serum. PLoS Negl Trop Dis. 2008;2:e280. doi: 10.1371/journal.pntd.0000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapphra K., Sangcharaswichai A., Chokephaibulkit K., Tiengrim S., Piriyakarnsakul W., Chakorn T. Evaluation of an NS1 antigen detection for diagnosis of acute dengue infection in patients with acute febrile illness. Diagn Microbiol Infect Dis. 2008;60:387–391. doi: 10.1016/j.diagmicrobio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization. Dengue guidelines for diagnosis, treatment, prevention and control. Geneva, Switzerland: WHO Press; 2009. [PubMed]
- 17.Kuno G., Gomez I., Gubler D.J. An ELISA procedure for the diagnosis of dengue infections. J Virol Methods. 1991;33:101–113. doi: 10.1016/0166-0934(91)90011-n. [DOI] [PubMed] [Google Scholar]
- 18.David W.V., Nisalak A., Solomon T., Kalayanarooj S., Dung N.M., Kneen R. Rapid serologic diagnosis of dengue virus infection using a commercial capture ELISA that distinguished primary and secondary infections. Am J Trop Med Hyg. 1999;60:6. doi: 10.4269/ajtmh.1999.60.693. [DOI] [PubMed] [Google Scholar]
- 19.Blacksell S.D., Newton P.N., Bell D., Kelley J., Mammen M.P., Jr., Vaughn D.W. The comparative accuracy of 8 commercial rapid immunochromatographic assays for the diagnosis of acute dengue virus infection. Clin Infect Dis. 2006;42:1127–1134. doi: 10.1086/501358. [DOI] [PubMed] [Google Scholar]
- 20.Seed P. DIAGT: Stata module to report summary statistics for diagnostic tests compared to true disease status. Stata J. 2001;4:490. [Google Scholar]
- 21.WHO. WHO recommended surveillance standards. 2nd ed. Geneva: World Health Organization [date unknown].
- 22.Hang V.T., Nguyet N.M., Trung D.T., Tricou V., Yoksan S., Dung N.M. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl Trop Dis. 2009;3:e360. doi: 10.1371/journal.pntd.0000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alcon S., Talarmin A., Debruyne M., Falconar A., Deubel V., Flamand M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J Clin Microbiol. 2002 Feb;40:376–381. doi: 10.1128/JCM.40.2.376-381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blacksell S.D., Mammen M.P., Jr., Thongpaseuth S., Gibbons R.V., Jarman R.G., Jenjaroen K. Evaluation of the Panbio dengue virus nonstructural 1 antigen detection and immunoglobulin M antibody enzyme-linked immunosorbent assays for the diagnosis of acute dengue infections in Laos. Diagn Microbiol Infect Dis. 2008;60:43–49. doi: 10.1016/j.diagmicrobio.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Parida M., Horioke K., Ishida H., Dash P.K., Saxena P., Jana A.M. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]