Abstract
A mismatch amplification mutation assay (MAMA) was developed for identification of point mutations in quinolone resistance-determining region (QRDR) of gyrA at codons 91 and 95. MAMA PCR was used to detect mutations at codons 91 and 95 of gyrA in 117 Neisseria gonorrhoeae isolates (with ciprofloxacin MICs of 0.004 to >32 μg/ml) from Bangladesh during 1997 to 2001. The QRDR regions of the gyrA genes from 31 randomly selected isolates were sequenced, and the results were compared with those of MAMA PCR. Using mismatch PCR, a mutation at Ser91 could be detected in all 27 (resistant and intermediate) isolates, and an Asp95-to-Gly95 mutation could be detected in all 15 isolates, as detected by sequencing. MAMA PCR offers a simple, inexpensive, rapid, and easier alternative for detection of point mutations in fluoroquinolone resistance in N. gonorrhoeae.
Ciprofloxacin, a fluoroquinolone with excellent in vitro activity against Neisseria gonorrhoeae, has been demonstrated to be highly effective against gonococcal infection (7). It has been recommended by the World Health Organization and the Centers for Disease Control and Prevention as the first line of treatment against gonorrhea (2). However; in recent years there have been reports of increases in the prevalence of ciprofloxacin-resistant N. gonorrhoeae from many parts of the world, including Bangladesh (9, 10, 13, 18, 20, 23).
Quinolones have a bactericidal effect when they bind their target enzymes, DNA gyrase and topoisomerase IV, both of which are essential for DNA replication. DNA gyrase consists of two GyrA subunits and two GyrB subunits, encoded by the genes gyrA and gyrB, respectively. Topoisomerase IV, which is highly homologous to gyrase, consists of two ParC subunits and two ParE subunits, encoded by the genes parC and parE, respectively (9). The principal mechanisms of resistance are alterations in the target enzymes and a reduced cytoplasmic quinolone concentration due to overexpression of multidrug efflux systems (7). The most prevalent mechanism contributing to fluoroquinolone resistance in the gonococcus involves mutations in the quinolone resistance-determining region (QRDR) of gyrA and possibly in the analogous region of the parC locus on the chromosome (12).
In laboratory mutants and in clinical isolates of fluoroquinolone-resistant N. gonorrhoeae, amino acid changes have been identified in GyrA and GyrB of DNA gyrase and in ParC and ParE of topoisomerase IV (1, 12). Both clinical isolates and laboratory mutants showed identical mutations in gyrA. An amino acid change in GyrB has been found to be responsible for low-level resistance to nalidixic acid (6). These mechanisms are analogous to those observed in Escherichia coli and other bacteria; however, unlike in E. coli, mutations in the gyrB gene do not appear to have a significant impact on fluoroquinolone resistance in N. gonorrhoeae.
Earlier studies have shown that all clinical isolates with decreased susceptibility to quinolones have amino acid changes at Ser91 and/or Asp95 in the GyrA subunit. These findings have indicated that DNA gyrase is a primary target of fluoroquinolones in N. gonorrhoeae and that alterations in the GyrA subunit, particularly Ser91 and Asp95, are significantly associated with decreased susceptibilities to quinolones (4, 5).
Mutation in gyrA is detected mainly by direct sequencing of the QRDR region of the gyrA gene (5), PCR amplification using primers with specific restriction sites and subsequent restriction digestion of the amplicon (4), and the single-strand conformation polymorphism method (3). Zirnstein et al. have described a method for detection of point mutations, whereby mutation in gyrA was detected by mismatch amplification mutation assay (MAMA) PCR for Campylobacter jejuni (24). We have adapted the same concept of MAMA PCR for detection of mutations in the QRDR region of gyrA in ciprofloxacin-resistant or -intermediate N. gonorrhoeae isolates and have compared it with sequencing.
MATERIALS AND METHODS
Bacterial isolates.
A total of 117 isolates of N. gonorrhoeae isolated from female sex workers during 1997 to 2001 were studied. After isolation, the identity of the organism was confirmed by colony morphology, Gram staining, and oxidase, catalase, and sugar oxidation tests. Isolates were further confirmed by PCR with primers which amplify a 390-bp region of the gonococcal cryptic plasmid (12). Isolates were stored at −70°C in Trypticase soy broth with 20% glycerol until further use.
MIC of ciprofloxacin.
The MIC of ciprofloxacin was determined by the agar dilution method as described elsewhere (14). Briefly, GC agar base (BBL Microbiology System) supplemented with 1% IsoVitaleX and twofold serial dilutions of ciprofloxacin (Bayer, Hampshire, United Kingdom) were used. Plates were inoculated with 104 CFU of bacteria and incubated at 37°C in 5% CO2 for 24 h. The end point was determined as the lowest concentration of ciprofloxacin giving complete inhibition of growth. Five N. gonorrhoeae reference strains, WHO A to WHO E, for which the MICs were known were included for quality control in the test. Each test was repeated three times. The antimicrobial susceptibility was judged by breakpoint criteria defined by the National Committee for Clinical Laboratory Standards (15). The twofold serial dilutions of ciprofloxacin were at concentrations of 0.004 to 32 μg/ml.
DNA extraction.
N. gonorrhoeae isolates were subcultured from −70°C stock on modified Thayer-Martin medium. After 24 h of incubation at 37°C with 5% CO2, chromosomal DNA was extracted as described elsewhere (22).
Primer design for mismatch PCR.
Three sets of primers were used in this study: (i) GYRA-1 and GYRA-2 primers for PCR amplification of the QRDR of gyrA for sequencing, (ii) GYRA-1 and SF-91 primers for detection of mutation (Ser to Phe) at codon 91, and (iii) D-95 and DG-95 primers for detection of mutation (Asp to Gly) at codon 95 (Table 1). The GYRA-1 and GYRA-2 primers were designed from the N. gonorrhoeae gyrA DNA sequence in GenBank (accession no. U08817). The SF-91, D-95, and DG-95 primers were designed by comparing DNA sequences near codons 91 and 95 in the QRDRs of the gyrA genes of fluoroquinolone-susceptible and -resistant N. gonorrhoeae by using the online Gene Fisher interactive primer design software (http://bibiserv.techfak.uni-bielefeld.de/genefisher/). The expected amplicon size for the presence of the Ser-to-Phe (TCC-to-TTC) mutation at codon 91 in the QRDR of the N. gonorrhoeae gyrA gene was 249 bp, and that for the presence of the Asp-to-Gly (GAC-to-GGC) mutation at codon 95 in the QRDR of N. gonorrhoeae gyrA was 230 bp.
TABLE 1.
Primers used in this study
| Method | Primer | Sequence (5′ to 3′) | Direction |
|---|---|---|---|
| Mismatch PCR | gyrA 1 | AACCCTGCCCGTCAGCCTTGA | Forward |
| SF-91 | ACGATGGTGTCGTAAACTGCAA | Reverse | |
| D-95 | ATGCGCAAAAGCTATCTCGACTAC | Forward | |
| DG-95 | CGCCATACGGACGATGGTAC | Reverse | |
| gyrA gene se- quencing | GYRA-1 | AACCCTGCCCGTCAGCCTTGA | Forward |
| GYRA-2 | GGACGAGCCGTTGACGACCAG | Reverse |
PCR.
PCR for detection of both mutations was done in a 50-μl reaction volume containing 5 μl of purified chromosomal DNA (∼75 ng of DNA in sterile Tris-EDTA-RNase), 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 0.2 μM primers, and 1 U of TaqDNA polymerase. Thirty cycles of PCR were carried out as follows: initial denaturation at 95°C for 5 min; 30 cycles of denaturation at 95°C for 1 min, annealing at 66°C for 20 s, and extension at 72°C for 20 s; and a final extension at 72°C for 5 min. Ten microliters of the PCR products was analyzed after electrophoresis in ethidium bromide-containing 2% agarose gels prepared in 0.5× Tris-borate-EDTA. DNAs from a known ciprofloxacin-susceptible isolate and a known ciprofloxacin-resistant isolate were used as positive and negative controls in each PCR.
Sequencing of the gyrA region.
A total of 31 randomly selected isolates for which ciprofloxacin MICs were 0.004 to 32 μg/ml were used for sequencing. The QRDR region was amplified, and the amplified fragments were purified in PCR purification kits (Qiagen Inc., Valencia, Calif.) and run in a cycle sequencing with Big-Dye terminator as previously described (12). The extension products were sequenced by capillary electrophoresis in an ABI PRISM 310 genetic analyzer (Applied Biosystems). The sequences were analyzed by BioEdit software.
Nucleotide sequence accession numbers.
Sequences obtained during this study were assigned GenBank accession numbers AY443500 to AY443530 for the 450-bp gyrA genes of N. gonorrhoeae MD295, MA240, MD73, MA219, MA285, MA309, MA360, MA418, MA89, MA223, MA381, MA422, MA148, MA290, MA366, MA378, MA345, MA275, MA424, MA356, MA220, MA369, MA441, MA84, MA335, MA298, MA270, MA87, MD23, MA178 and MD12, respectively.
RESULTS
Antimicrobial susceptibility.
A total of 117 gonococcal isolates collected during 1997 to 2001 were analyzed in this study. Approximately two-thirds of the isolates were resistant to ciprofloxacin. Among the isolates, 27 were susceptible (MICs of <0.06), 10 were intermediate (MICs of 0.125 μg/ml [4 isolates] and 0.25 μg/ml [6 isolates]), and 80 were resistant (MICs of 1.0 μg/ml [9 isolates], 2.0 μg/ml [8 isolates], 4.0 μg/ml [25 isolates], 8.0 μg/ml [17 isolates], 16.0 μg/ml [10 isolates], and ≥32.0 μg/ml [11 isolates]) to ciprofloxacin.
Mutation at codons 91 and 95 determined by MAMA PCR.
Twenty-seven ciprofloxacin-susceptible isolates (MICs of <0.125 μg/ml), 10 ciprofloxacin-intermediate isolates (MICs of >0.125 and <0.5 μg/ml), and 80 ciprofloxacin-resistant isolates (MICs of ≥1.0 μg/ml) were analyzed for mutations at codon 91 (Ser to Phe) and codon 95 (Asp to Gly) by mismatch PCR (Fig. 1). All intermediate and resistant isolates had a mutation at codon 91 (Ser91 to Phe91). On the other hand, 50% (5 of 10) of the intermediate isolates and 38% (31 of 80) of the resistant isolates had a mutation at codon 95 (Asp95 to Gly95). None of the susceptible isolates had either of these two mutations.
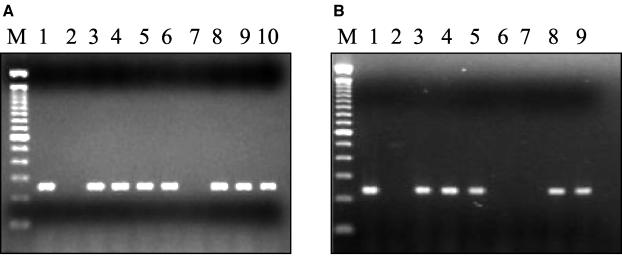
FIG. 1.
(a) Agarose gel electrophoresis of MAMA PCR products of representative strains of N. gonorrhoeae for the detection of mutation at codon 91 (Ser91 to Phe91). A 249-bp PCR product was obtained in strains having the Ser91-to-Phe91 mutation. Lane M, 100-bp DNA ladder; lane 1, N. gonorrhoeae MA87 (MIC, 4 μg/ml); lane 2, N. gonorrhoeae MA178 (MIC, 0.004 μg/ml); lane 3, N. gonorrhoeae MA270 (MIC, 4 μg/ml); lane 4, N. gonorrhoeae MA360 (MIC, 4 μg/ml); lane 5, N. gonorrhoeae MA89 (MIC, 16 μg/ml); lane 6, N. gonorrhoeae MA441 (MIC, 8 μg/ml); lane 7, N. gonorrhoeae MA240 (MIC, 0.004 μg/ml); lane 8, N. gonorrhoeae MA275 (MIC, >32 μg/ml); lane 9, N. gonorrhoeae MA290 (MIC, >32 μg/ml); lane 10, N. gonorrhoeae MA223 (MIC, 16 μg/ml). (b) Agarose gel electrophoresis of MAMA PCR products of representative strains of N. gonorrhoeae for detection of mutation at codon 95 (Asp91 to Gly95). A 230-bp PCR product was obtained in strains having the Asp95-to-Gly95 mutation. Lane M, 100-bp DNA ladder; lane 1, N. gonorrhoeae MA270 (MIC, 4 μg/ml); lane 2, N. gonorrhoeae MA178 (MIC, 0.004 μg/ml); lane 3, N. gonorrhoeae MA309 (MIC, 4 μg/ml); lane 4, N. gonorrhoeae MA335 (MIC, 4 μg/ml); lane 5, N. gonorrhoeae MA441 (MIC, 8 μg/ml); lane 6, N. gonorrhoeae MA240 (MIC, 0.004 μg/ml); lane 7, N. gonorrhoeae MA 366 (MIC, >32 μg/ml; lane 8, N. gonorrhoeae MA 378 (MIC, >32 μg/ml; lane 9, N. gonorrhoeae MA 275 (MIC, >32 μg/ml).
Mutation at codons 91 and 95 determined by sequencing.
Among the 117 isolates included in the study, the QRDRs of the gyrA genes of 31 isolates (4 susceptible, 2 intermediate, and 25 resistant) were sequenced. A mutation at codon 91 (Ser91 to Phe91) was detected in all of the 25 resistant isolates. A mutation at codon 95 from Asp to Gly was found in 15 isolates, and a mutation from Asp to Asn was detected in 10 isolate. No correlation was found between the MIC of ciprofloxacin and the number of mutations, and none of the isolates had a single mutation only at Asp95. A mutation at codon 91 (Ser91 to Phe91) was detected in both intermediate isolates, and no mutation was detected in susceptible isolates.
Correlation between mismatch PCR and sequencing.
By using mismatch PCR, a mutation at Ser91 could be detected in all 25 resistant isolates and an Asp95-to-Gly95 mutation could be detected in all 15 isolates, as detected by sequencing. However, in one isolate the Asp95 to Asn95 mutation, as detected by sequencing, was detected as Asp95 to Gly95 by MAMA PCR (Table 2). A mutation at codon 91 (Ser91 to Phe91) was detected in both intermediate isolates, as detected by sequencing.
TABLE 2.
MIC distribution and comparison of gyrA QRDR sequencing and MAMA PCR for 31 N. gonorrhoeae isolates from Bangladesh
| Isolate | Ciproflox- acin MIC (μg/ml) | Amino acid alteration in GyrA as determined by:
|
|||
|---|---|---|---|---|---|
| Sequence analysis
|
MAMA PCR
|
||||
| Position 91 | Position 95 | Position 91 (Ser 91 to Phe) | Position 95 (Asp 95 to Gly) | ||
| MD12 | 0.002 | No | No | ||
| MD295 | 0.002 | No | No | ||
| MA178 | 0.004 | No | No | ||
| MA240 | 0.004 | No | No | ||
| MD23 | 0.125 | Ser91 to Phe | Yes | No | |
| MD73 | 0.25 | Ser91 to Phe | Yes | No | |
| MA87 | 4 | Ser91 to Phe | Asp95 to Asn | Yes | Yes |
| MA219 | 4 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA270 | 4 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA285 | 4 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA298 | 4 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA309 | 4 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA335 | 4 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA360 | 4 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA84 | 8 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA418 | 8 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA441 | 8 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA89 | 16 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA220 | 16 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA223 | 16 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA369 | 16 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA381 | 16 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA356 | 32 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA422 | 32 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA424 | 32 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA148 | >32 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA275 | >32 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
| MA290 | >32 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA345 | 32 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA366 | >32 | Ser91 to Phe | Asp95 to Asn | Yes | No |
| MA378 | >32 | Ser91 to Phe | Asp95 to Gly | Yes | Yes |
Correlation between mutation and MIC.
Among the sequenced isolates, the MIC for 14 isolates was 16 μg/ml or greater, and 7 of these (50%) had the Asp95-to-Asn95 mutation, compared to 2 (28%) out of 11 of the isolates for which the MIC was less then 8 μg/ml.
DISCUSSION
The principal mechanisms of resistance are alterations in the target enzymes and reduced cytoplasmic quinolone concentration due to overexpression of multidrug efflux systems (9). There are several reports on the association between mutations in the QRDRs of the corresponding genes gyrA and parC and resistance to fluoroquinolones in other gram-negative bacteria and in gonococci (8, 11, 16, 17, 19, 21). Mutations in gyrB have been evaluated for several gram-negative bacteria, including N. gonorrhoeae (8, 12).
Fluoroquinolone resistance in N. gonorrhoeae has been shown to be due to point mutation of Ser91 to Phe91 and of Asp95 to either Asn95 or Gly95 in the gyrA gene. Besides this, parC and parE mutation has also been implicated in resistance of N. gonorrhoeae (5, 12). We have designed and used two PCR primers for the detection of Ser91-to-Phe91 and Asp95-to-Gly95 mutations of gyrA, which contribute to quinolone resistance of N. gonorrhoeae. The same concept can be adapted for the detection of Asp95-to-Asn95 mutation in the gyrA gene. Since many laboratories do not have the equipment, time, or expertise to sequence the gene for investigation of mutations relevant to antibiotic resistance and since there are easy ways to detect such mutations by MAMA PCR, we felt that it was worthwhile to adapt the MAMA PCR for N. gonorrhoeae. This is an easy and less expensive method then sequencing, as many laboratories do not have sequencing facilities or expertise to do sequencing.
The 3′-terminal nucleotides of primer SF-91 for detection of mutation (Ser to Phe) at codon 91 and of primer DG-95 for detection of mutation (Asp to Gly) at codon 95 pair correctly with the codon for Phe in codon 91 and Gly in codon 95 of ciprofloxacin-resistant N. gonorrhoeae strains. Neither the nucleotide at the 3′ end nor the nucleotide immediately 5′ to it paired with the fluoroquinolone-susceptible N. gonorrhoeae gyrA gene sequence, and thus no amplicon was obtained.
Using MAMA PCR, we were able to identify the Ser-to-Phe mutation at codon 91 in all resistant isolates and the Asp-to-Gly mutation at codon 95 in all isolates. An Asp95-to-Asn95 mutation in one isolate was detected as Asp95 to Gly95 by MAMA PCR, which might be due to mixed population of bacteria with both Asp-to-Gly and Asp-to-Asn mutations at codon 95 or to failure of the MAMA PCR.
By using primers specific for detection of the Asp95-to-Gly95 mutation, 50% of the intermediate and 38% of the resistant isolates were found to have the Asp95-to-Gly95 mutation. The remaining isolates might have the Asp95-to-Asn95 mutation, which could not be detected by this specific set of primers. Primers specific for detecting mutation at codon 95 from Asp to Asn could be used to detect such a mutation.
Although the MAMA PCR described here is relatively simple and easy to perform, it has certain limitations: (i) it can identify only the known mutations, and (ii) for detection of each of the mutations, a specific primer is needed. However, it can be used as a quick screening method for public health laboratories that are interested in studying resistance profiles of outbreak isolates but that have little access to sequencing facilities. It might also be interesting to screen intermediate isolates or isolates for which the MIC is low for identification and in-depth studies of other mechanisms for fluoroquinolone resistance.
We believe that the MAMA PCR method is a simple, specific, rapid, inexpensive, and portable alternative to nonradioisotopic single-strand conformation polymorphism methods and DNA sequencing for detection of this important fluoroquinolone resistance mutation.
Acknowledgments
This study was conducted at the ICDDR,B Centre for Health and Population Research with the support of cooperative agreement HRN-A-00-96-90005-00 from USAID. The ICDDR,B acknowledges with gratitude the commitment of USAID to the center's research effort.
REFERENCES
- 1.Belland, R. J., S. G. Morrison, C. Ison, and W. M. Huang. 1994. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 14:371-380. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2002. Sexually transmitted diseases treatment guidelines 2002. Morb. Mortal. Wkly. Rep. 51:1-80. [Google Scholar]
- 3.Charvalos, E., E. Peteinaki, I. Spyridaki, S. Manetas, and Y. Tselentis. 1996. Detection of ciprofloxacin resistance mutations in Campylobacter jejuni gyrA by nonradioisotopic single-strand conformation polymorphism and direct DNA sequencing. J. Clin. Lab Anal. 10:129-133. [DOI] [PubMed] [Google Scholar]
- 4.Deguchi, T., M. Yasuda, M. Nakano, S. Ozeki, T. Ezaki, S. Maeda, I. Saito, and Y. Kawada. 1996. Rapid detection of point mutations of the Neisseria gonorrhoeae gyrA gene associated with decreased susceptibilities to quinolones. J. Clin. Microbiol. 34:2255-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deguchi, T., M. Yasuda, M. Nakano, S. Ozeki, T. Ezaki, I. Saito, and Y. Kawada. 1996. Quinolone-resistant Neisseria gonorrhoeae: correlation of alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV with antimicrobial susceptibility profiles. Antimicrob. Agents Chemother. 40:1020-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deguchi, T., M. Yasuda, M. Nakano, S. Ozeki, E. Kanematsu, Y. Kawada, T. Ezaki, and I. Saito. 1996. Uncommon occurrence of mutations in the gyrB gene associated with quinolone resistance in clinical isolates of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 40:2437-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.el Amin, N. A., S. Jalal, and B. Wretlind. 1999. Alterations in GyrA and ParC associated with fluoroquinolone resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 43:947-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorito, S., P. Galarza, I. Pagano, C. Oviedo, A. Lanza, J. Smayevsky, G. Weltman, L. Buscemi, and E. Sanjuan. 2001. Emergence of high level ciprofloxacin resistant Neisseria gonorrhoeae strain in Buenos Aires, Argentina. Sex. Transm. Infect. 77:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivens, D., I. Martin, and C. Ison. 2000. Neisseria gonorrhoeae in a London sexually transmitted infection clinic not fully sensitive to quinolones: are isolates imported and how effective is ciprofloxacin as a first-line therapy? Int. J. STD AIDS 11:774-776. [DOI] [PubMed] [Google Scholar]
- 11.Jalal, S., and B. Wretlind. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb. Drug Resist. 4:257-261. [DOI] [PubMed] [Google Scholar]
- 12.Lindback, E., M. Rahman, S. Jalal, and B. Wretlind. 2002. Mutations in gyrA, gyrB, parC, and parE in quinolone-resistant strains of Neisseria gonorrhoeae. APMIS 110:651-657. [DOI] [PubMed] [Google Scholar]
- 13.Llanes, R., J. Sosa, D. Guzman, Y. Gutierrez, A. Llop, and O. Ricardo. 2001. Neisseria gonorrhoeae resistant to ciprofloxacin: first report in Cuba. Sex. Transm. Dis. 28:82-83. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1993. Approved standards M7-A3. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 15.National Committee for Clinical Laboratory Standards. 1994. Performance standards for antimicrobial susceptibility testing 14. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 16.Oppegaard, H., and H. Sorum. 1994. gyrA mutations in quinolone-resistant isolates of the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 38:2460-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oram, M., and L. M. Fisher. 1991. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob. Agents Chemother. 35:387-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otero, L., B. Alcala, J. A. Varela, M. D. Miguel, J. A. Vazquez, and F. Vazquez. 2001. First isolate of a Neisseria gonorrhoeae strain associated with an ofloxacin treatment failure in Spain: case report. Sex. Transm. Dis. 28:576-578. [DOI] [PubMed] [Google Scholar]
- 19.Rahman, M., G. Mauff, J. Levy, M. Couturier, G. Pulverer, N. Glasdorff, and J. P. Butzler. 1994. Detection of 4-quinolone resistance mutation in gyrA gene of Shigella dysenteriae type 1 by PCR. Antimicrob. Agents Chemother. 38:2488-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman, M., Z. Sultan, S. Monira, A. Alam, K. Nessa, S. Islam, S. Nahar, A. W. Shama, S. Alam Khan, J. Bogaerts, N. Islam, and J. Albert. 2002. Antimicrobial susceptibility of Neisseria gonorrhoeae isolated in Bangladesh (1997 to 1999): rapid shift to fluoroquinolone resistance. J. Clin. Microbiol. 40:2037-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reyna, F., M. Huesca, V. Gonzalez, and L. Y. Fuchs. 1995. Salmonella typhimurium gyrA mutations associated with fluoroquinolone resistance. Antimicrob. Agents Chemother. 39:1621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Tanaka, M., H. Nakayama, M. Haraoka, and T. Saika. 2000. Antimicrobial resistance of Neisseria gonorrhoeae and high prevalence of ciprofloxacin-resistant isolates in Japan, 1993 to 1998. J. Clin. Microbiol. 38:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zirnstein, G., Y. Li, B. Swaminathan, and F. Angulo. 1999. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J. Clin. Microbiol. 37:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]