Abstract
We devised a noninvasive genetic selection strategy to identify positive regulators of bacterial virulence genes during actual infection of an intact animal host. This strategy combines random mutagenesis with a switch-like reporter of transcription that confers antibiotic resistance in the off state and sensitivity in the on state. Application of this technology to the human intestinal pathogen Vibrio cholerae identified several regulators of cholera toxin and a central virulence gene regulator that are operative during infection. These regulators function in chemotaxis, signaling pathways, transport across the cell envelope, biosynthesis, and adherence. We show that phenotypes that appear genetically independent in cell culture become interrelated in the host milieu.
Vibrio cholerae, the causative agent of the epidemic disease cholera, requires the coordinated expression of multiple virulence factors. Two of the most well studied factors are cholera toxin (CT) and toxin-coregulated pilus (TCP). CT is an ADP-ribosylating toxin largely responsible for eliciting the profuse diarrhea characteristic of this disease, and TCP is a type IV bundle-forming pilus that is essential for intestinal colonization in humans and in animal models of cholera (1, 2). Regulation of these factors involves the concerted actions of three proteins, ToxR, TcpP, and ToxT, which together form the V. cholerae virulence gene regulatory cascade (3–5). ToxR and TcpP are inner-membrane-associated transcriptional regulators that act cooperatively to induce the expression of ToxT in response to particular environmental signals. ToxT is a cytoplasmic transcriptional activator responsible for directly activating the transcription of the structural genes for CT, TCP, and other virulence factors (3). Investigation of the regulatory networks coordinating expression of these genes has been predominately limited to in vitro culture conditions largely because of the inability to monitor regulatory processes during infection of an intact host. Therefore, the true nature of the in vivo regulation of these virulence factors within a host remains unclear.
To extend our knowledge of virulence gene regulation during infection, we developed a noninvasive genetic selection that incorporates the use of the recombination-based in vivo expression technology (RIVET) and transposon mutagenesis to identify positive regulators of virulence genes during infection. RIVET relies on the construction of transcriptional fusions to a promoterless reporter gene, tnpR, encoding a site-specific DNA recombinase. After it is produced, TnpR functions in trans to permanently excise from the bacterial genome a tetracycline resistance (TcR) marker (tet) that is flanked by recombinase recognition sequences (res). The subsequent conversion to tetracycline (Tc) sensitivity serves as an ex post facto indicator of increased transcription of the gene fusion. RIVET has been used as a promoter-trap method to identify bacterial and fungal genes induced during infection (6–8) and as a tool to analyze the spatiotemporal patterns of induction of such genes (9). Using RIVET, we recently found that the patterns of transcriptional induction of genes encoding CT and TCP fundamentally differ during in vitro growth vs. during infection with respect to timing and interdependence (9). During infection, tcpA, encoding the pilin subunit of TCP, was transcriptionally induced in a biphasic pattern with an early induction occurring at 1 h after inoculation and a second more robust induction occurring at 4 h after inoculation. Meanwhile, ctxA, encoding the enzymatic subunit of CT, was not induced until 4 h after inoculation and was dependent on previous expression of TCP. To learn more about the regulatory pathways controlling these complex patterns of expression, we used a RIVET selection strategy to identify in vivo regulators of toxT and of ctxA. A number of these regulators are described, some of which affect virulence gene induction only during infection. The roles in gene regulation and colonization of infant mice of one major class of regulators controlling chemotaxis signaling were further investigated by defined mutational analysis.
Materials and Methods
Bacterial Strains and in Vitro Growth Conditions.
All of the V. cholerae strains used in this study are isogenic derivatives of El Tor biotype strain, C6709–1, containing the res-tet-res substrate for TnpR resolvase integrated within the endogenous lacZ gene (strain AC-V66) (10). Bacterial strains were either grown in Luria-Bertani (LB) broth with aeration at 37°C (virulence gene-noninducing condition) or AKI broth grown statically for 4 h followed by growth with aeration at 37°C (inducing condition) (11). Antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; streptomycin, 100 μg/ml; kanamycin, 50 μg/ml; and Tc, 3 μg/ml.
Transposon Mutagenesis and RIVET Selection.
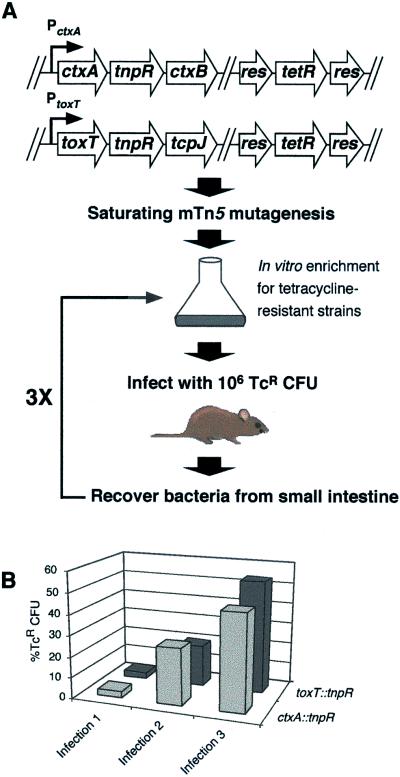
A promoterless recombinase gene, tnpR, was inserted in a nonpolar manner downstream of ctxA or toxT by allelic exchange into the strain AC-V66. In each fusion strain, random mutagenesis was performed with mini-Tn5Kn2 (mTn5) to generate 15,000 independent strains in 10 pools of 1,500 each. Each pool was grown in the presence of Tc and 106 colony-forming units (cfu) were intragastrically inoculated into 5-day-old CD-1 mice (2 mice per pool). After 7 h, the small intestines were removed and homogenized. The bacteria within each homogenate were grown overnight in the presence of Tc to enrich for TcR mutant strains. The animal passage and Tc enrichment were repeated three times. After each animal passage, but before Tc enrichment, a portion of the recovered bacteria was examined for previous induction of the toxT∷tnpR or ctxA∷tnpR fusion during infection by replica plating colonies onto LB agar containing Tc. The percentage of TcR cfu was calculated by dividing the number of TcR cfu by the total cfu.
Mutant Strain Construction.
The desired point and deletion mutations were constructed by using PCR, and the mutant alleles were cloned into the suicide shuttle vector pCVD442 (12). The deletion mutations of motY and flaA were of the entire coding region fusing the putative ATG start codon with the putative stop codon for each gene. For construction of the motAB deletion, the putative ATG start codon of motA was fused to the putative stop codon of motB. Escherichia coli strain, SM10λpir carrying the plasmids was mated with V. cholerae. The desired mutation was introduced into the genome by allelic exchange after appropriate counterselection for loss of the integrated plasmid. PCR, Southern blot, and DNA sequence analyses were used to confirm the correct mutations. The mTn5 insertion junctions for the isolated mutants were determined by arbitrary-primed PCR and DNA sequencing as described (13). The genes disrupted and sites of mTn5 insertion relative to the first base of the predicted coding regions are: sltA (1048), prpB (519), vps70/capK (816), rtxB (1817), cheA-2 (1977), cheY-3 (178), vieS (22), mfrha (239), cheZ (659), and mcpX (1774).
Swarm Assay and Electron Microscopy.
Chemotactic ability of the mutants were determined by stabbing cells into swarm agar (1% tryptone/0.5% NaCl/0.3% agar) and incubating overnight at 30°C. Production of the polar flagellum was determined by electron microscopy after negative staining with uranyl acetate.
In Vitro and in Vivo Transcription Assays.
Strains were grown either in LB broth or in AKI broth for 7 h at 37°C, and ≈150–200 cfu were plated on LB agar containing streptomycin. After growth overnight at 37°C, these plates were then replica plated onto LB agar containing Tc and the extent of toxT∷tnpR or ctxA∷tnpR induction was measured as the percent Tc-sensitive (TcS) cfu (TcS cfu divided by the total cfu). For the in vivo temporal gene fusion induction experiments, 105 cfu were intragastrically inoculated into 5-day-old CD-1 mice. At each hour after inoculation, the small intestines were removed and homogenized, and the percent TcS cfu was determined as described above.
Competition Assays of Mutant Strains and Population Dynamics Study.
The competitive index was determined by mixing a LacZ− mutant strain with a LacZ+ wild-type strain at approximately a 1:1 ratio, and 105 cfu were intragastrically inoculated into 5-day-old CD-1 mice and also into 2 ml of LB broth to be grown overnight at 37°C. Five mice were used per mutant strain tested. At 24 h after inoculation, the small intestines were removed, homogenized, and then plated on LB agar supplemented with streptomycin and 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside. Blue (LacZ+) and white (LacZ−) colonies were enumerated. The competitive index was calculated as the ratio of mutant to wild type recovered after 24 h of infection divided by the ratio of mutant to wild-type after 24 h of growth in LB broth, with both ratios corrected for deviations of the input from a value of 1:1. The population dynamics study was performed as follows: 106 cfu of wild-type or cheY-3D52N mutant were intragastrically inoculated into 5-day-old CD-1 mice. After 5 h, the stomach and small intestine were removed and the latter was cut into six equal-length segments (≈2 cm per segment). Each segment was homogenized, and the total cfu from each was determined. The cfu from each segment was divided by the inoculum (106) to calculate the percentage of input cfu recovered.
Results
RIVET Selection Strategy and Isolation of in Vivo Regulators.
During infection of a host, V. cholerae induce the expression of ToxT and CT to regulate the expression of virulence factors and manifest disease, respectively. To identify genes required for induction of toxT and ctxA during infection, we constructed V. cholerae strains bearing tnpR transcriptional fusions to toxT and ctxA in a strain background containing res-tet-res. These strains were then subjected to saturating mutagenesis by using mTn5, and the mutant libraries were used to seed infection in an infant mouse model of cholera (Fig. 1A). Bacteria were subsequently recovered from infected tissue and were subjected to Tc enrichment to select for mutants that failed to induce expression of the gene fusion within the host (such strains remain TcR). Importantly, bacteria were recovered from the small intestine at 7 h after inoculation when toxT and ctxA are known to be induced (9). Because recombination is not 100% efficient during the time course of this experiment, three rounds of enrichment and infection were necessary to isolate a pool of strains from the animals in which a large fraction exhibited reduced induction of the toxT∷tnpR or ctxA∷tnpR fusion during infection (Fig. 1B). After the third round of selection, 100 TcR strains per fusion were colony purified. The nucleotide sequence of the mTn5 junctions in 64 nonsibling strains was determined as described in Materials and Methods, and 14 of these were chosen for further analysis. The mutant phenotypes of these 14 were retested by first transducing the mutations into fresh backgrounds by using the generalized transducing phage CP-T1ts (18). These newly constructed strains were used for all subsequent experiments, including assaying for decreased induction of toxT∷tnpR and ctxA∷tnpR during infection. Of these 14 strains, 10 were shown to exhibit delayed or reduced induction of one or both fusions during infection (Figs. 3B and 5B and data not shown).
Figure 1.
RIVET-based selection for positive regulators of virulence gene expression during infection. (A) A schematic diagram of RIVET-based selection strategy is shown. Transposon mutagenesis was performed on derivatives of the El Tor biotype V. cholerae strain, C6709–1, containing the res-tet-res cassette and either a ctxA∷tnpR or toxT∷tnpR transcriptional fusion. Three rounds of in vitro enrichment and animal passage were done as described in Materials and Methods. (B) After each animal passage, but before Tc-enrichment, a portion of the recovered bacteria was examined for previous induction of the toxT∷tnpR or ctxA∷tnpR fusion during infection by replica plating colonies onto LB agar containing Tc. An increase in the percentage of TcR cfu after each animal passage indicates an overall enrichment of mutant strains that fail to induce expression of the toxT∷tnpR or ctxA∷tnpR fusion during the period of infection.
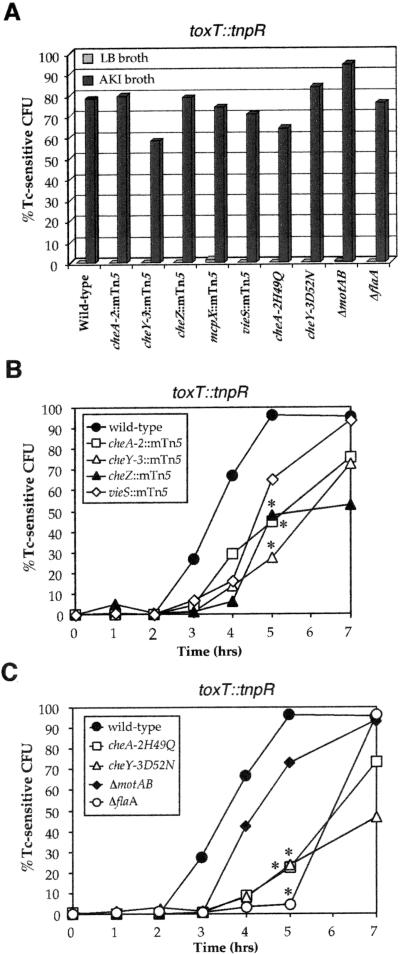
Figure 3.
Induction patterns of the toxT∷tnpR fusion in various strain backgrounds in vitro and during infection. (A) Induction profile of toxT∷tnpR fusion strains grown for 7 h either in LB broth (a normally noninducing condition), or in AKI broth (an inducing condition) was measured as percent Tc-sensitive cfu. The genetic backgrounds are denoted on the x axis. The data shown are the means from multiple independent experiments. (B and C) Induction kinetics of V. cholerae strains in infant mice. The data shown are the means from multiple independent experiments. Data at 5 h were tested for statistical significance by using the Student's two-tailed t test: Strains with reduced induction of the gene fusion compared with wild type (P < 0.1) are denoted by asterisks.
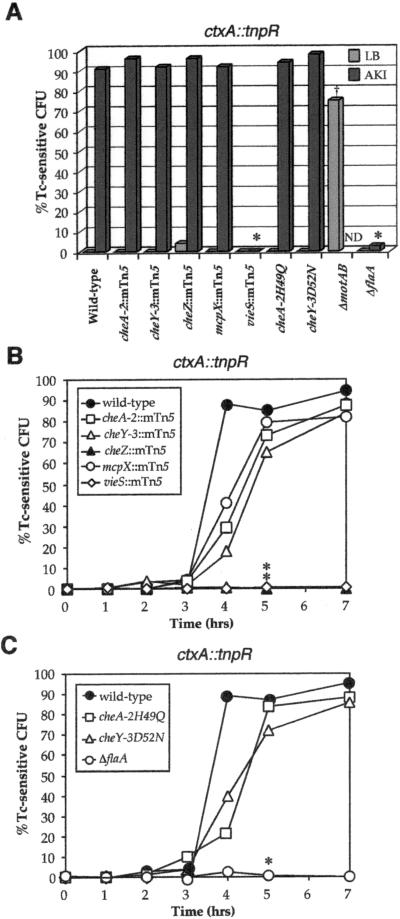
Figure 5.
Induction patterns of the ctxA∷tnpR fusion in various strain backgrounds in vitro and during infection. (A) The induction of ctxA∷tnpR was determined as described in the Fig. 3 legend. †, Percentage of Tc-sensitive cfu for the ΔmotAB strain was determined by testing 16 independently constructed strains for Tc sensitivity. ND, induction with the ΔmotAB strain in AKI broth was not tested because of constitutive activity of the ctxA∷tnpR fusion in this strain background. Strains with reduced induction of the gene fusion compared with wild-type (P < 0.01) are denoted by asterisks. (B and C) The induction kinetics of ctxA∷tnpR during infection were determined and statistically analyzed as described in the Fig. 3 legend. Strains with reduced induction of the gene fusion compared with wild type (P < 0.02) are denoted by asterisks.
The 10 genes identified as regulators of ctxA or toxT are shown in Table 1. These genes have putative functions in metabolism, biosynthesis or transport of extracellular factors, adherence, or signal transduction. Within this latter class is vieS, which encodes a putative sensor kinase of the BvgS family that may phosphorylate either or both of two putative response regulators, VieA and VieB. Previously, vieB was shown to be induced during infection but not during growth in vitro (10). Also of the latter class are four genes encoding putative bacterial chemotaxis components, cheA-2 and cheY-3, which encode the sensor kinase and response regulator pair that controls rotation of the polar flagellum, cheZ, which encodes the CheY phosphatase, and mcpX (C6.1), which encodes one of 43 putative methyl-accepting chemotaxis proteins of V. cholerae (14, 15). Methyl-accepting chemotaxis proteins bind attractants and repellents and control the phosphorylation state of CheA (14).
Table 1.
V. cholerae regulatory factors and their contributions to colonization of the infant mouse small intestine
| Strain* | Gene† | Putative protein function | Colonization fitness, CI‡ |
|---|---|---|---|
| T1.1 | sltA (VC 0866) | Soluble lytic transglycosylase | 0.18§ |
| T1.5 | prpB (VC 1336) | Carboxyphosphonoenolpyruvate phosphonomutase | 0.49 |
| T2.5 | vps70/capK (VC 0924) | Biofilm-associated exopolysaccharide biosynthesis | 0.58 |
| T2.9 | rtxB (VC 1448) | ABC transporter for RTX toxin | 0.55 |
| T8.5 | cheA-2 (VC 2063) | Chemotaxis sensor kinase | 5.3¶ |
| T10.6 | cheY-3 (VC 2065) | Chemotaxis response regulator | 4.5¶ |
| C2.3 | vieS (VC 1653) | Three-component sensor kinase | 1.32 |
| C3.4 | mfrha (VC 0443) | Mannose/fucose-resistant hemagglutinin | 0.58 |
| C3.5 | cheZ (VC 2064) | CheY phosphatase | 3.5¶ |
| C6.1 | mcpX (VC 2161) | Methyl-accepting chemotaxis protein | 2.2 |
| Site-specific mutations generated | |||
| — | cheA-2H49Q | Autophosphorylation mutant | 92¶ |
| — | cheY-3D52N | Phosphoreceiver mutant | 69¶ |
| — | ΔmotAB | Flagellar motor protein | 0.09§ |
| — | ΔmotY | Flagellar motor protein | 0.03§ |
| — | ΔflaA | Flagellar subunit | 0.07§ |
A strain designation starting with a T or C indicates isolation from the toxT∷tnpR or ctxA∷tnpR fusion strain selection, respectively. The cheA-2 gene was identified by both selections.
The gene name and locus designation (in parentheses) are from the Institute for Genomic Research database of the V. cholerae genome.
The competitive index (CI) was calculated as the mean of the ratios of mutant to wild-type cfu recovered from the small intestines of 5 mice after 24 h of infection. Mutant strains significantly less fit for colonization than wild type (
, P < 0.05), or more fit than wild type (
, P < 0.05) by the Wilcoxon signed ranked sum test are indicated.
Characterization of in Vivo Regulators and Their Roles in Chemotaxis and Bacterial Motility.
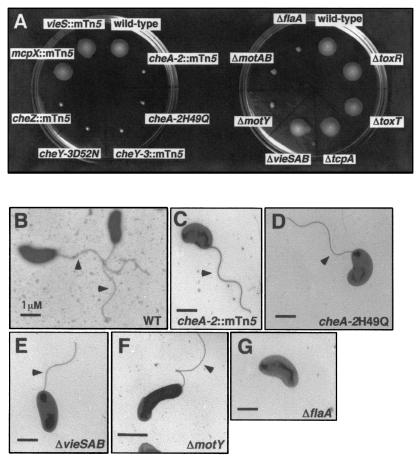
The identification of previously unknown regulators of ctxA and toxT, most of which are dispensable for regulation in vitro (Figs. 3A and 5A), indicates that the RIVET selection strategy is capable of identifying factors that perform regulatory roles within the context of an infection. Many of the identified regulators are hypothesized to function in chemotaxis. However, the roles for chemotaxis and the associated (but separable) properties of motility and flagellation in the virulence of V. cholerae are unclear (16–18). We therefore investigated the roles that chemotaxis, motility, flagellation, and vieS locus each play in the regulation of ctxA and toxT transcription and in virulence. Despite the fact that there are multiple cheA and cheY genes in V. cholerae, the cheA-2 and cheY-3 as well as the cheZ transposon insertion mutants identified in the selection were nonchemotactic, whereas the mcpX and vieS mutants exhibited wild-type chemotaxis (Fig. 2A). Strains engineered to contain point mutations in cheA-2 and cheY-3 that change the conserved sites of phosphorylation to nonphosphorylatable residues (cheA-2H49Q and cheY-3D52N) (19, 20) were nonchemotactic (Fig. 2A). Although lacking chemotaxis, the cheA-2 and cheY-3 mutants remain flagellated and motile (Fig. 2D and data not shown). Two additional mutant classes (nonmotile but flagellated, and aflagellate) were generated by in-frame deletion of the flagellar motor genes motAB or motY, and the major flagellin gene flaA, respectively. The expected in vitro phenotypes of each of these mutant strains were confirmed (Fig. 2 A, F and G and data not shown).
Figure 2.
Chemotaxis in semisolid agar and production of flagella. (A) Chemotactic ability of each mutant was determined by stabbing cells into swarm plates. Strain genetic backgrounds are indicated above each swarm location. The ΔvieSAB, ΔtoxR, ΔtoxT, and ΔtcpA strains were described (9, 10). (B–G) Production of the polar flagellum was determined by electron microscopy. The genetic background of each strain is indicated at bottom right corner.
In Vivo Regulation of toxT Transcription.
The effects of these mutations on the induction of toxT∷tnpR during growth in vitro and during infection were measured. For El Tor biotype V. cholerae, growth in LB broth does not activate transcription of toxT or ctxA, whereas microaerophilic growth in a bicarbonate-containing rich broth, AKI, induces both (Figs. 3 A and 5A) (21). None of the mutations affected induction of the toxT∷tnpR fusion during growth under these in vitro conditions by 7 h, which is the same time frame examined in the in vivo experiments (Fig. 3A). These results suggest that loss of chemotaxis, motility, or the flagellum do not severely affect expression of toxT in vitro. However, loss of these properties caused marked delays and reductions in toxT∷tnpR induction during infection. In the wild-type strain, toxT∷tnpR induction was detected by 3 h after inoculation and was induced in >95% of the bacteria by 5 h; whereas, in both the chemotaxis and vieS mutants, induction of toxT∷tnpR was delayed by 1–3 h and the former only reached 45–75% induction by 7 h (Fig. 3 B and C). The delay in the induction of toxT did not translate into a lower level of colonization of the small intestine. On the contrary, the chemotaxis mutants, in particular both point mutants, were hypercolonizers (Table 1), consistent with data previously reported for undefined nonchemotactic mutants (22).
Population Dynamics of Chemotaxis and Motility Mutants.
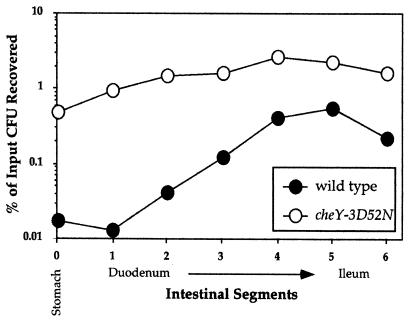
Despite the enhanced colonization exhibited by the nonchemotactic but motile V. cholerae cheY-3D52N mutant, this strain exhibited an aberrant distribution within the intestine. Whereas the wild-type strain was localized primarily within the distal half of the small intestine (Fig. 4 and ref. 23), the mutant strain was distributed throughout the stomach and small intestine. The observed hypercolonization within all sections of the small intestine might be because of any number of factors. It is consistent with the hypothesis (22) that there exists an antibacterial mechanism in the deeper layers of the intestinal epithelium of infant mice to which chemotactic V. cholerae are attracted and are accordingly reduced in number, but where nonchemotactic V. cholerae fail to go. In light of the additional observation of reduced levels of toxT∷tnpR induction in the nonchemotactic strains, these results suggest that chemotaxis is guiding the bacteria to a “preferred” location in the small intestine with respect to virulence gene induction, where it is optimal for expression but not for general survival. Thus, we speculate that the diminished survival of the wild-type strain is offset by a greater pathogenic potential of the properly colonized survivors.
Figure 4.
Colonization of the stomach and small intestine of infant mice by wild-type and nonchemotactic V. cholerae strains. Population dynamics of V. cholerae isogenic strains, AC-V66 (wild-type) and a nonchemotactic strain, cheY-3D52N, were determined during infection of infant mice. The stomach and small intestine were removed at 5 h after inoculation, and the latter was cut into six equal length segments as designated on the x axis. The number of bacteria recovered from each segment relative to the input inoculum is shown on the y axis. The values shown are the means from four mice per V. cholerae strain.
In contrast to the enhanced colonization exhibited by the motile but nonchemotactic strain, the ΔflaA and ΔmotAB strains are severely attenuated for colonization (Table 1); the latter is consistent with a previous report showing that a ΔmotB strain was avirulent (17). It is likely that motility, even in the absence of chemotaxis, is required for efficient penetration of the mucus gel layer that covers the intestinal epithelium, otherwise, cells would be quickly propelled through to the large intestine by peristalsis. The equivalent decrease in colonization potential of the flagellated and aflagellated nonmotile strains (ΔmotAB and ΔflaA, respectively) suggests that the presence of the polar flagellum per se, i.e., in the absence of rotation, is not required for colonization.
In Vivo Regulation of ctxA Transcription.
ToxT is a transcriptional activator of both CT and TCP, and thus the effects of the above mutations on induction of ctxA∷tnpR could mirror the effects already observed on induction of toxT∷tnpR. On the other hand, CT has been shown to be differentially expressed compared with TCP during infection (9), supporting the existing model that regulation of CT expression is more complex and requires the integration of multiple levels of control in addition to ToxT (3). Any differences in the effects of mutations in the regulators found in this study on ctxA∷tnpR vs. toxT∷tnpR induction might reveal new levels of regulation of ctxA. Such differences were indeed observed both in vitro and during infection. The vieS and ΔflaA mutant strains exhibited little or no induction of ctxA∷tnpR in AKI broth (an inducing condition) (Fig. 5A), whereas the ΔmotAB strain exhibited an elevation of ctxA∷tnpR induction during growth in LB broth (normally a noninducing condition) consistent with a previous report (17). This lack of induction of ctxA∷tnpR in the vieS mutant background correlates with reduced levels of ctxA mRNA by ribonuclease protection assay (A. Tischler and A.C., unpublished data). During infection, the vieS and ΔflaA mutant strains failed to induce ctxA∷tnpR over the time course examined (Fig. 5 B and C). These results suggest that VieS and flagellar rotation are factors that regulate ctxA transcription during infection. The ability of the vieS∷mTn5 strain to colonize the small intestine at wild-type levels (Table 1) despite having a defect in ctxA induction, is not contradictory in light of the fact that CT is not required for colonization (1, 24). As such, the vieS∷mTn5 mutation did not alter the kinetics of tcpA induction during infection (data not shown). This may be due to ToxT possibly having a higher affinity for the tcpA promoter than the ctxA promoter, and therefore the basal level of ToxT is sufficient for tcpA induction but not ctxA induction at early times of infection. The cheZ mutation also resulted in lack of measurable induction of the ctxA∷tnpR fusion during infection. This suggests that during infection CheZ is required for a pattern or mode of flagellar rotation necessary for ctxA∷tnpR induction that is not required during growth in vitro. Such an effect could be manifested at a macroscopic level, i.e., a difference in the microenvironments to which the bacteria swim, or could be completely internal to V. cholerae, e.g., a change in membrane potential via altered sodium movement through the flagellar motor (16). The finding that the VieS- and chemotaxis-signaling systems regulate virulence gene expression during infection, but play no role as such during in vitro cell culture, reinforces the importance of studying virulence factors in host environments.
Discussion
The roles that chemotaxis and motility play in virulence of V. cholerae are not fully understood. Richardson et al. (25) showed that flagellated and aflagellated nonmotile El Tor strains adhered less well and induced less fluid accumulation than the wild-type strain in rabbit ileal loop studies. However, in competition experiments using suckling mice, the nonmotile mutants colonized to similar levels compared with wild-type (25). Gardel et al. (17) showed that certain hyperswarmer strains were drastically reduced in their ability to colonize the small intestine of suckling mice. Both groups observed that some nonmotile mutants exhibited altered profiles of virulence factor expression (CT and TCP) during growth in normally noninducing in vitro conditions. Recently, it was shown that affecting flagellar rotation by increasing the viscosity of the medium elevated the expression of ToxT, the major transcriptional activator of virulence genes (16). These results suggest that there is an intimate relationship between motility and virulence gene expression. However, the exact nature of that relationship during an infection is unclear because of the fact that most nonmotile mutants used in the above studies contained undefined mutations and the virulence gene expression assays were performed under in vitro conditions. It is also unclear how chemotaxis and motility affect virulence during infection because of the contradictory results from different animal models.
A complete understanding of the pathogenicity of a microbe must include detailed knowledge of the patterns and regulation of virulence gene expression during the course of infection in a host. We have described in this study a selection strategy to identify positive regulators of virulence genes during infection. The selection strategy incorporates the use of transposon mutagenesis and an in vivo expression technology (RIVET) previously used to identify infection-induced genes (6). Through this technique, we have identified several previously unknown regulators of the V. cholerae virulence genes ctxA and toxT.
The newly identified in vivo regulators have diverse roles including those in chemotaxis, bacterial motility, signal transduction, biosynthesis of extracellular proteins, and toxin transport. Although many of the factors identified are not essential for colonization of infant mice, they are clearly involved in the proper kinetics of expression of V. cholerae virulence genes during infection. Specifically, factors involved in chemotaxis (CheY-3, CheA-2, and CheZ) and motility (FlaA) cause a delay in the induction of toxT during infection. This delay may be because of the inability of the nonchemotactic strains to swim toward a preferred niche optimal for virulence gene induction, whereas the nonmotile strains may fail to efficiently penetrate the mucus gel lining of the intestinal epithelium. In addition, our results indicate that full induction of CT, which occurs after colonization has taken place (9), requires additional signal inputs from the sensor kinase (VieS) of a three-component signal transduction system and from components of chemotaxis (CheZ). The delay and reduction in ctxA transcription observed in the mutant strains could not be solely attributed to reduced levels of toxT because both cheY-3D52N and cheA-2H49Q point mutations caused reduced levels of toxT, but they fully induced ctxA with proper kinetics. The added regulation afforded by CheZ and VieS may be needed to tightly control the expression of CT with respect to timing and location of expression.
Our results support a model whereby V. cholerae chemotaxis and motility are required early during the infection process to properly initiate virulence gene expression. Specifically, a subpopulation of the inoculated bacteria is directed by chemotaxis to a preferred location in the small intestine in which optimal expression of virulence factors occurs. Although this wild-type pathway of infection leads to damage and/or killing of a significant portion of the infecting bacteria, similar to that observed by Freter and O'Brien (22), the final result of these interactions is correct colonization and coordinated induction of CT. By identifying and analyzing how different systems such as chemotaxis, motility and signal transduction pathways affect the expression of key virulence factors in the mouse model of cholera, we gain a new appreciation for the complex interplay of bacterial regulatory factors within a host environment.
Acknowledgments
We thank M. Waldor, J. Mecsas, and D. Merrell for critical readings of the manuscript, M. Angelichio for statistical analyses, and C. Linsenmayer for electron microscopy work. This work was supported by National Institutes of Health (AI40262, AI45746), a Pew Scholar Award in the Biomedical Sciences, and the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928).
Abbreviations
- CT
cholera toxin
- TCP
toxin-coregulated pilus
- RIVET
recombination-based in vivo expression technology
- Tc
tetracycline
- TcR
Tc resistance
- cfu
colony-forming units
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skorupski K, Taylor R K. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 3.DiRita V J, Parsot C, Jander G, Mekalanos J J. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hase C C, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 6.Camilli A, Mekalanos J J. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe A M, Beattie D T, Deresiewicz R L. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 8.Staib P, Kretschmar M, Nichterlein T, Kohler G, Michel S, Hof H, Hacker J, Morschhauser J. Mol Microbiol. 1999;32:533–546. doi: 10.1046/j.1365-2958.1999.01367.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee S H, Hava D L, Waldor M K, Camilli A. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee S H, Angelichio M J, Mekalanos J J, Camilli A. J Bacteriol. 1998;180:2298–2305. doi: 10.1128/jb.180.9.2298-2305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwanga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 12.Donnenberg M S, Kaper J B. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Toole G A, Kolter R. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 14.Eisenbach M. Mol Microbiol. 1996;20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 15.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature (London) 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hase C C, Mekalanos J J. Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardel C L, Mekalanos J J. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hava, D. L. & Camilli, A. (2001) J. Microbiol. Methods, in press. [DOI] [PubMed]
- 19.Appleby J L, Bourret R B. Mol Microbiol. 1999;34:915–925. doi: 10.1046/j.1365-2958.1999.01653.x. [DOI] [PubMed] [Google Scholar]
- 20.Ellefson D D, Weber U, Wolfe A J. J Bacteriol. 1997;179:825–830. doi: 10.1128/jb.179.3.825-830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medrano A I, DiRita V J, Castillo G, Sanchez J. Infect Immun. 1999;67:2178–2183. doi: 10.1128/iai.67.5.2178-2183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freter R, O'Brien P C. Infect Immun. 1981;34:222–233. doi: 10.1128/iai.34.1.222-233.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelichio M J, Spector J, Waldor M K, Camilli A. Infect Immun. 1999;67:3733–3739. doi: 10.1128/iai.67.8.3733-3739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson K. Infect Immun. 1991;59:2727–2736. doi: 10.1128/iai.59.8.2727-2736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]