Abstract
Mycoplasma genitalium is a leading cause of chlamydia-negative, nongonoccocal urethritis and has been directly implicated in numerous other genitourinary as well as extragenitourinary tract pathologies. Detection of M. genitalium has relied almost entirely on PCR amplification of clinical specimens and evidence of seroconversion since these mycoplasmas are highly fastidious and culture isolation by microbiological techniques is very rare. We have established a combinatorial strategy using confocal immunoanalysis (CIA) and real-time PCR to qualitatively and quantitatively assess patterns of M. genitalium infection in women attending a sexually transmitted disease-related health clinic in San Antonio, Tex. CIA allows spatial examination of mycoplasmas on surfaces and inside human target cells, plus the ability to evaluate cell-to-cell patterns and variances within samples. Real-time PCR permits determination of genome copy numbers of mycoplasmas and human cells by multiplex amplification using mycoplasma gyrA and human RNase P gene sequences, which indicates overall levels of mycoplasma infection and degree of parasitism. These assays are strongly correlated and, in combination, permit detection and elucidation of heretofore-unrecognized patterns of M. genitalium infections in clinical and experimental samples.
Mycoplasma genitalium is considered the smallest self-replicating cell, with a genome consisting of 580,074 bp, and is theorized to approximate the essential complement of genes necessary to sustain life (14, 20). M. genitalium was first isolated in 1981 from human male urethral cultures and identified as a causative agent of nongonococcal urethritis (31, 36). Since that time numerous reports have linked M. genitalium to additional genitourinary symptoms, including endometritis, salpingitis, cervicitis, and pelvic inflammatory disease as well as a range of other pathologies such as arthritis, pneumonia, AIDS progression, chronic fatigue, and autoimmune disorders (2, 5, 13, 17, 22, 24, 35, 37). Mycoplasmas are capable of invading human target cells and persisting and replicating for extended periods intracellularly (6, 9). Two pathways by which M. genitalium and other mycoplasmas accomplish this parasitism are tip organelle-mediated attachment and translocation of mycoplasma cytoplasmic enzymes to mycoplasma membrane surfaces to facilitate tissue colonization. Tip organelle-mediated attachment requires numerous adhesins and cytadherence accessory proteins (3, 7, 21), while translocation of mycoplasma cytoplasmic enzymes to its membrane surface permits adherence to extracellular matrix molecules, such as mucin and fibronectin (1, 10). These examples of pathogenic specialization, along with other attributes, including complete dependence on sterols and a wide range of biosynthetic precursors (amino acids, nucleotides, and fatty acids) for growth, serve to illustrate how mycoplasmas have evolved with genetically streamlined genomes and a total parasitic lifestyle. Since mycoplasmas have no parallel among other prokaryotes, they have attracted the interests of microbiologists and infectious disease specialists, as well as geneticists and cell biologists, in pursuit of understanding the “minimalist” cell or pathogen. However, the identification of biological properties of M. genitalium that correlate with virulence determinants has been hampered by its highly fastidious nature, its use of UGA as a tryptophan codon, and other genetic and biochemical characteristics not encountered in other bacteria (19, 28, 29).
Furthermore, distinct biological features of M. genitalium, which include its small size and lack of cell wall, prevent the use of traditional and relatively simple histopathological staining and light microscopy methods to assess relationships between M. genitalium and tissue colonization and disease. An experimental animal model using chimpanzees as a host for M. genitalium infection has provided direct evidence for its pathogenic capabilities, based upon colonization and shedding of mycoplasmas over many months, genitourinary tract-associated inflammatory pathologies, and >4-fold seroconversion (26, 32). However, other subhuman primates are much less susceptible, if at all, and alternate animal species are even less relevant in providing a model that parallels human M. genitalium disease. Since M. genitalium is emerging as an important infectious agent (8, 13, 30), it is becoming increasingly important to correlate the in situ presence of M. genitalium with human disease. Unfortunately, direct isolation and cultivation of M. genitalium from clinical genitourinary samples in cell-free broth or agar medium are a very rare event (4, 36). In the absence of consistent “gold standard” culture methods to establish the existence of M. genitalium in clinical samples, recent associations of M. genitalium with clinical signs and symptoms have been predominantly based upon PCR and evidence for seroconversion (11, 12, 27), both of which have substantial limitations in evaluating the role of M. genitalium in human diseases. For example, in the case of serological assessment, sequential serum samples are a prerequisite for establishing active infection, and these are seldom available. For PCR assessment, assay sensitivity along with other test variables, such as quality and preparation of DNA samples and presence of inhibitory factors in clinical specimens, can confound data interpretation (4).
In this study we use confocal immunoanalysis (CIA) and real-time PCR to visually and analytically monitor frequency and infectious load of M. genitalium in vaginal specimens among a population of Mexican-American and African-American women attending a specialized clinic for sexually transmitted diseases (STDs), who were culture positive for M. genitalium (4; J. E. Korte, J. B. Baseman, J. M. Piper, M. P. Cagle, A. E. C. Holden, S. T. Perdue, J. D. Champion, and R. N. Shain, unpublished data) This combinatorial approach enables detection and evaluation of infectious patterns and trends within individual vaginal cells infected with M. genitalium and reinforces the emergence of M. genitalium as an important cause of STDs.
MATERIALS AND METHODS
Clinical sample preparation.
Vaginal samples consisting of epithelium and endometrial cells and traces of blood were obtained by swab from an 18-patient subset of high-risk minority women enrolled in an ongoing longitudinal STD investigation (NIAID-sponsored STD Cooperative Research Center to The University of Texas Health Science Center at San Antonio [UTHSCSA]). This study was approved by the institutional review boards of UTHSCSA and the San Antonio Metropolitan Health District. This patient subset was selected based upon M. genitalium culture-positive, PCR, and enzyme-linked immunosorbent assay results (4). Individual swabs were stored at 4°C in 5 to 10 ml of sterile phosphate-buffered saline (PBS) prior to transporting on ice to UTHSCSA. Clinical specimens were centrifuged at 2,500 × g at 4°C for 10 min, washed, and pelleted three times in ice-cold PBS prior to resuspension in 2 ml of PBS. Processing vaginal epithelial and endometrial cell specimens in this manner markedly reduced exogenous, nonadhering bacteria and eliminated swab-associated fibers. One milliliter of each sample was used for DNA isolation, and the other milliliter was used for CIA.
Human and mycoplasma cell cultures.
HEp-2 cell (human larynx; American Type Culture Collection) and M. genitalium strain G37 cultures were used to standardize measurements of confocal and real-time PCR protocols. HEp-2 cells were grown to 80% confluence in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum using T-75 vented culture flasks incubated in air-7% CO2 at 37°C. Cell monolayers were trypsin treated, split into multiple 25-cm2 vented culture flasks, and grown for 48 h to ∼75% confluence. M. genitalium strain G37 was grown in 100 ml of SP-4 medium in T-150 culture flasks for 72 h at 37°C, and surface-attached mycoplasmas were washed in PBS and harvested (5, 36).
DNA and RNA sample preparation.
Genomic DNAs of M. genitalium and 13 other common bacterial, fungal, or parasitic species, which were obtained from the Department of Microbiology and Immunology laboratories at UTHSCSA, were isolated using Easy-DNA kit (Invitrogen), and DNA concentrations were determined by optical densities at 260 nm. The 13 distinct DNA samples were derived from Bacillus subtilis, Candida albicans, Chlamydia trachomatis, Escherichia coli 6824, Gardnerella vaginalis, Mycoplasma fermentans, Mycoplasma orale, Mycoplasma penetrans, Mycoplasma pirum, Mycoplasma pneumoniae, Neisseria gonorrhoeae, Streptococcus pyogenes, and Trichomonas vaginalis. These DNAs served as controls to determine M. genitalium primer and probe sequence specificities. Human DNA samples obtained from HEp-2 cell cultures (Easy-DNA kit; Invitrogen) and Applied Biosystems (source of human RNase P gene) were used as standards for defining human vaginal cell genomic copy numbers. DNA samples from clinical vaginal swabs were prepared as described above.
M. genitalium total RNA from strain G37 and three clinical specimens was obtained using Tri Reagent as recommended by the manufacturer (Sigma) and quantified by absorbance at 260 nm. Two micrograms of RNA was subjected to DNase I treatment (Invitrogen) to remove traces of genomic DNA and used in reverse transcription PCR.
Confocal immunoanalysis of M. genitalium infection of human cells.
Multiplicities of infection (MOI) (mycoplasmas/HEp-2 cells) of 5:1, 2:1, 1:1, 1:2, and 1:10 were established in 10-ml volumes of Dulbecco's modified Eagle's medium with 10% fetal bovine serum, and incubation continued for 24 to 48 h at 37°C. CIA was employed to visually monitor the extent of mycoplasma infection of HEp-2 cells, which served as the basis for assessing M. genitalium infectious load in clinical vaginal cell specimens (6). CIA permits the generation of optical z-sections (sequential serial sections of 0.1- to 1.0-μm thickness through the z axis of individual cells) without invasive manipulation. Infected HEp-2 cell samples and controls (HEp-2 and M. genitalium alone) were processed by harvesting and dispersing cells through a 21-gauge needle until no visible cell clumps were evident. Triton X-100 (0.1%) and neutral buffered formalin (10%) were added to samples, which were incubated at 37°C for 45 min to permeabilize and fix cells. Subsequently, cell preparations were washed, centrifuged, and resuspended once in PBS; blocked with 1% bovine serum albumin (Gibco BRL) for 30 min; and washed, centrifuged, and resuspended two more times. Polyclonal rabbit anti-M. genitalium strain G37 antiserum at a 1/1,000 dilution in a volume of 1 ml was added to each of the fixed cell samples, and incubation continued at 37°C for 45 min with rocking. Individual samples were washed, centrifuged, and resuspended by gentle agitation in 1 ml of PBS to remove unbound antibodies. Samples were blocked with 1% bovine serum albumin (Gibco BRL) for 30 min and washed, centrifuged, and resuspended two more times. Secondary immunolabeling was performed with fluorescein isothiocyanate-labeled goat anti-rabbit antibody (Molecular Probes) at 1/100 dilution, along with propidium iodide (0.25 mM; Molecular Probes) to stain nucleic acids. Samples were again incubated for 45 min with rocking at 37°C, washed, centrifuged, and resuspended three times in 1 ml of PBS. After the final wash, pelleted cells were resuspended in 300 μl of Vecta-Shield mounting medium (Vector Laboratories Inc.) for slide preparation. Slides were viewed with an Olympus IX70 microscope with a UPLANAPO 60× objective and 1.3 numerical aperture, and an Olympus FV500 confocal system was used to capture and process images. Argon (emission 488) and helium-neon (emission 543) lasers were employed to visualize fluorescein isothiocyanate and propidium iodide labeling patterns, respectively. Once experimental conditions were established, which included the lack of cross-reactive fluorescence in parallel samples with M. pneumoniae-infected HEp-2 cells (data not shown), clinical vaginal specimens were processed as described above. All experiments were repeated in triplicate, and 20 fields of 8 to 10 cells per sample were examined to determine M. genitalium qualitative infectious patterns.
Electron microscopy.
Aliquots of clinical vaginal cell preparations were fixed in 4% formaldehyde and 1% glutaraldehyde, subjected to a series of alcohol dehydrations, and embedded in LR White epoxy resin (15). Embedded sections were cut into ultrathin sections, placed onto 150-mesh nickel grids, and labeled with whole organism anti-M. genitalium strain G37 primary antibodies (1/100 dilution) and 10-nm-diameter gold bead goat anti-rabbit immunoglobulin G secondary label. Samples were stained with a combination of uranyl acetate (saturated aqueous 7%) and Reynolds lead citrate. Specimens were viewed under a Philips 208S transmission electron microscope at an accelerating voltage of ∼60 kV.
Real-time PCR determinations.
Quantification of M. genitalium genome number was achieved by real-time PCR using TaqMan chemistry (Applied Biosystems). M. genitalium species-specific oligonucleotide primers and TaqMan probe were designed for detection of M. genitalium gene gyrA (MG003; single copy) using Primer Express software (version 2.0). Forward (FOR, 5′-AATCCTTGAACGCTTCATGTTAGAAA-3′) and reverse (REV, 5′-TTGTGAAAATCGAGTTCTTCGCTCT-3′) sequence detection primers along with probe 5′-6-FAM-CCCACAAGAAGCAAACGATATCATCAGAA-TAMRA-3′ (where FAM is 5′-fluorescein and TAMRA is 3′-rhodamine), which was localized within the amplicon, yielded an 86-bp amplicon. Human vaginal and HEp-2 cells were quantified similarly by detecting single-copy gene probe RNase P (TaqMan RNase P detection reagents [Applied Biosystems]; primer and probe sequences are proprietary) using VIC fluorophore (Applied Biosystems).
Standard curves were established by real-time PCR amplification for M. genitalium strain G37 DNA serially diluted from 107 to 1 copy per reaction using M. genitalium-specific MGgyrA primers and for human DNA diluted 108 to 1 copy per reaction using RNase P gene primers. All reactions were performed in triplicate (for exception, see below) using TaqMan Universal PCR Master Mix, a 0.5 μM concentration of M. genitalium primers, and a 0.2 μM concentration of mycoplasma probe, or RNase P gene primers and probe as suggested by the manufacturer (Applied Biosystems). We plotted threshold coefficients (CTs) against corresponding DNA concentrations, determined the linear dynamic range of DNA concentrations for both target DNAs (slope of −3.33 for mycoplasma DNA and −3.32 for human DNA assays), and observed a significant correlation coefficient (r = 0.997 for MGgyrA and r > 0.99 for RNase P gene). However, reactions containing a single M. genitalium genome yielded inconsistent results, and therefore, assays were performed 10 times. Repeatedly, 3 out of 10 reactions (30%) did not yield the expected amplicon based upon Poisson distribution of positives. Amplification conditions consisted of 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min.
Mycoplasma and human DNA templates were tested singly and in combination (multiplex reaction) with both MGgyrA and RNase P gene primers and TaqMan probes to confirm the absence of interference and cross-binding. Multiplex real-time PCR (ABI PRISM HP7900 SDS; Applied Biosystems) permitted measurements of the ratio of M. genitalium to human genomic DNA in order to quantify the dynamics of variations of mycoplasma-host cell interactions.
Reverse transcription-PCR.
Reverse transcription-PCR analysis was performed based upon detection of 16S rRNA of M. genitalium. Isolated RNA was reverse transcribed with primer MGSO-2 (5′-CACCACCTGTCACTCGGTTAACCTC-3′) and later, together with forward primer My-ins (5′-GTAATACATAGGTCGCAAGCGTTATC-3′), used to amplify the predicted 517-bp fragment. SuperScript First-Strand Synthesis System (Invitrogen) was used for all reverse transcription reactions. Amplified products were electrophoresed on 1% agarose alongside 100-bp DNA ladder standards (Invitrogen).
Statistical analyses.
For statistical analysis of results, the correlation coefficient between M. genitalium levels as determined by CIA and real-time PCR was calculated, with each modeled as a continuous variable. A nonparametric test for trend was used to determine whether real-time PCR measurements of M. genitalium load increased across CIA categories (negative, positive, and strongly positive). The test for trend was also used to determine whether the degree of parasitism of vaginal cells increased across CIA categories. All analyses were conducted using STATA (version 7.0).
RESULTS
Confocal detection and localization of M. genitalium in human cells.
We monitored trends and patterns of immunolabeled M. genitalium within infected cells by CIA. In preliminary experimental samples, a range of MOI of mycoplasma/HEp-2 cells allowed us to measure the extent of mycoplasma parasitism and establish assay parameters, which served as the basis for assessing M. genitalium infectious load in clinical specimens. For example, high-MOI (5:1 mycoplasmas/HEp-2 cell) short-term infections revealed intense surface-localized fluorescent bodies with limited discrete intracellular fluorescence label while low MOI (1:2 and 1:10) showed only limited surface immunofluorescence and very little intracellular labeling. These observations were similar to our earlier report of experimental mycoplasma infectious trends involving WI-38 human lung cells (6). In contrast, clinical specimens of heavily parasitized vaginal cells demonstrated moderate surface and intense internal immunofluorescence (Fig. 1). Therefore, we utilized a qualitative visual scoring system to define the level of M. genitalium parasitism among individual vaginal cells within clinical samples (Fig. 1 and 2). Optical z-series revealed locations of immunolabeled mycoplasmas, which were readily determined by referencing x, y, and z axes, in conjunction with nuclear and outer cell membranes of individual vaginal cells (Fig. 1C, and D). For these measurements, 150 to 200 vaginal cells were screened microscopically per test specimen, and a representative group of 20 to 25 cells was selected for scoring of discrete immunofluorescence bodies. Although subjective, this scoring system permitted us to distinguish negative from intermediate to heavily parasitized cells and to define infectious trends and patterns. For negative controls, preimmune rabbit sera and anti-M. pneumoniae antibody reagents exhibited no fluorescence under similar experimental and clinical samples.
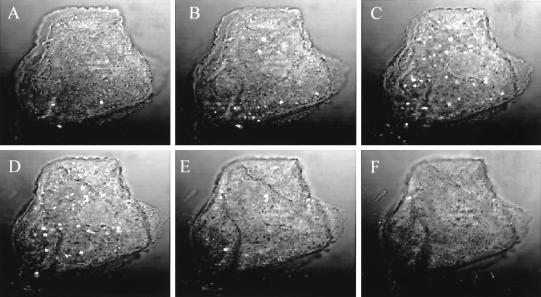
FIG. 1.
Sequential visualization of clinically derived vaginal cells using CIA. Both surface-associated and internalized immunolabeled M. genitalium cells are readily observed using 1-μm-diameter z-series optical sections. (A to F) Images are representative of a cell with a strong positive score. Mycoplasmas can be readily visualized as intense, discrete white fluorescing bodies, while the cell nucleus can be visualized using propidium iodide as detected (B to E).
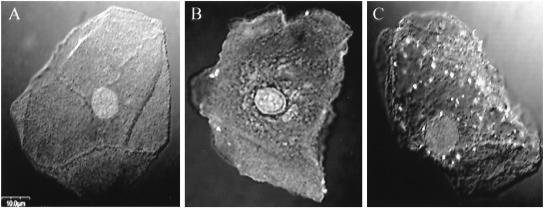
FIG. 2.
Representative clinically derived vaginal cells based upon immunolabeled patterns of M. genitalium as described in Fig. 1 legend. Mycoplasmas appear as distinct fluorescent bodies as in Fig. 1, and the vaginal cell nucleus is clearly observed in each panel. CIA scores: negative (A), positive (B), and strongly positive (C).
We scored gross levels of infection per clinical sample as follows: negative, less than 3% of vaginal cells exhibited any fluorescence and no internalized fluorescent bodies visualized in a complete z-series of individual cells (Fig. 2A); positive, approximately 3 to 30% of vaginal cells exhibited fluorescence with 5 to 15 fluorescent bodies detected on surfaces and within individual cells (Fig. 2B); and strongly positive, 30 to 100% of vaginal cells exhibited positive immunofluorescence with 16 or more fluorescent bodies detected on surfaces and within individual cells (Fig. 2C). In the last of these, we capped the maximal count of mycoplasmas per individual vaginal cell at 20 because of difficulties in establishing accurate CIA measurements beyond that number. As noted in Fig. 1 and 2, a single vaginal cell can be infected with 50 or more mycoplasmas. Although individual z-series cell-to-cell patterns varied within specific clinical samples, vaginal cells classified as positive usually exhibited immunofluorescence confined to the cell periphery, while cells categorized as strongly positive exhibited predominantly intracellular immunofluorescence. These scoring criteria were shown to be reproducible by blindly repeating confocal evaluations of independent samples from the same specimen.
Transmission electron microscopy of clinical samples:.
To further visualize and confirm the existence and location of mycoplasmas in clinical samples and reinforce confocal immunolabeling patterns within individual vaginal cells, we performed immunogold electron microscopy (Fig. 3) (15). Immunogold-labeled mycoplasma cells with intact classic mycoplasma unit membranes and size variations between ∼300 and ∼1,200 nm were readily observed in clinical vaginal cell samples (23), corroborating CIA antibody label specificity and predicted absence of cell wall structures. Typically, mycoplasmas stained darker by uranyl acetate and lead citrate than surrounding host cell components, which was verified by whole organism polyclonal anti-M. genitalium antibody coupled with 10-nm-diameter immunogold beads (Fig. 3). The predominant mycoplasma cell morphology was oblong, approximately 500 by 1,000 nm, and lacked tip structures observed in broth-grown cultures.
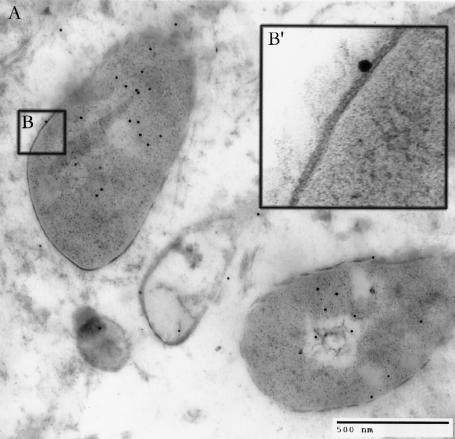
FIG. 3.
Electron micrograph of epoxy-embedded immunolabeled clinically derived vaginal cell sample. (A) Intracellular M. genitalium cells labeled with 10-nm-diameter immunogold beads appear as two nearly complete bisected bodies and two smaller ones, each surrounded by a unit membrane and associated with gold beads. (B′) Enlarged inset of panel B, revealing intact mycoplasma unit membrane. Bar = 500 nm.
Real-time PCR analysis.
To demonstrate specificity of M. genitalium MGgyrA primers in real-time PCR assays, we screened 13 distinct microbial DNA samples (104 copies/reaction mixture) for background and interference effects (16). Amplification of expected products was observed only with M. genitalium DNA, and no PCR products were detected when other genomic DNAs were used as templates. Using primers and probes of M. genitalium (MGgyrA) and human (RNase P gene), we established standard curves for genomes of M. genitalium and human DNA as outlined in Materials and Methods.
To evaluate possible interference of human DNA in M. genitalium TaqMan assays, we used a multiplex reaction, which allowed determination of copy numbers for both host tissues and M. genitalium simultaneously. This was implemented by the addition of genomic HEp-2 cell DNA to M. genitalium DNA amplification reactions. We observed no significant differences in CT of individual versus multiple simultaneous amplification reactions. In a final set of control experiments, 10-fold dilutions of M. genitalium DNA with parallel dilutions of HEp-2 cell DNA were compared in the multiplex TaqMan assay with simultaneous detection of MGgyrA and RNase P gene in order to determine whether amplification occurred more readily in combination for either primer set. No significant differences in CT were detected between pure or various dilutions of combined templates.
After establishing assay reliability and reproducibility of scoring criteria, we performed multiplex real-time PCR analysis on DNA samples from clinical specimens. Based upon standard curve quantification of M. genitalium and human cell DNA, we determined the ratio of M. genitalium to human cell genomes among negative, positive, and strongly positive samples. The median ratio of M. genitalium to vaginal cells as determined by PCR was 0.14 for the CIA negative group, while the M. genitalium/vaginal cell ratios were 0.65 and 3.21 for the CIA positive and strongly positive groups, respectively (Table 1). The test for trend in median M. genitalium/vaginal cell values across CIA categories was statistically significant (P < 0.001). The test for trend in prevalence of heavy parasitism (as defined by the percentage of evaluated cells per patient with ≥20 mycoplasmas) across CIA categories was also statistically significant (P < 0.001; Table 1). The quantitative real-time PCR ratios reflected general trends of infectious load throughout the entire clinical sample and correlated extremely well with CIA (r = 0.88) (Fig. 1 and 2).
TABLE 1.
Relationship between M. genitalium-based CIA scoring group, real-time PCR result, and degree of CIA-determined parasitisma
| Confocal score | No. of patients (n = 18) | Median ratio of mycoplasmas/host cell (range) | Median proportion (%) of cells that are heavily parasitizedb (range) |
|---|---|---|---|
| Negative | 5 | 0.14 (0.01-0.37) | 0 |
| Positive | 8 | 0.65 (0.4-1.17) | 8 (4-12) |
| Strongly positive | 5 | 3.21 (1.75-3.9) | 25 (10-45) |
| Test for trend Z-statistic (P) | 3.85 (<0.001) | 3.79 (<0.001) |
Median ratio reflects real-time PCR result, and median proportion reflects CIA-determined parasitism.
Individual vaginal cells with ≥20 mycoplasmas.
Reverse transcription-PCR of M. genitalium in vaginal cells.
Nonquantitative reverse transcription-PCR of M. genitalium 16S rRNA was performed to demonstrate metabolically active, and predictably viable, M. genitalium in clinical samples of vaginal cells (25, 38). RNA was harvested from clinical samples that tested positive and negative by CIA and real-time PCR (Table 1) and was amplified by reverse transcription-PCR. Reverse transcription analyses of positive vaginal samples resulted in amplification of M. genitalium rRNA (Fig. 4, lanes 4, 6, and 8) while a representative negative clinical specimen generated no amplified product (Fig. 4, lane 10). No product amplification occurred in the absence of reverse transcriptase in samples that scored positive or negative (Fig. 4, odd-numbered lanes), confirming the lack of genomic DNA contamination. These experimental results further served to reconfirm CIA, immunogold electron microscopy, and real-time PCR data, indicating the existence of intact and metabolically active (and, presumably, replicating) M. genitalium cells in vaginal specimens since reverse transcription-PCR utilized unique M. genitalium 16S rRNA gene sequences.
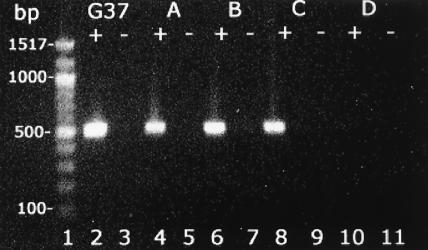
FIG. 4.
Reverse transcription-PCR of M. genitalium G37 control and individual clinical vaginal specimens. In lanes G37, A, B, and C, positive lanes (+) indicate M. genitalium 16S rRNA 517-bp fragment amplification with addition of reverse transcriptase; negative lanes (-) are without reverse transcriptase. Vaginal cells from patients A, B, and C scored positive using immunofluorescent confocal microscopy while negative sample D showed no amplification.
DISCUSSION
Valid and reliable detection procedures and clinical diagnostic criteria are essential for purposes of defining M. genitalium as an important and emerging bacterial pathogen. Other than the original isolations of M. genitalium from males with nongonococcal urethritis in 1981 and several subsequent successful but scarce reports (4, 5, 18, 34, 36), serological and PCR assessments comprise the major evaluative criteria for linking M. genitalium to STDs and other human pathologies (13, 24, 33). In separate studies we have described a population of minority women who attend an STD health clinic in San Antonio and who are culture-positive for M. genitalium (4; Korte et al., unpublished). This subgroup has been characterized over many months of clinical visits using M. genitalium-specific PCR, enzyme-linked immunosorbent assay, and immunoblot measurements. This study indicates that M. genitalium infects and persists in women for extended periods and is linked to a variety of genitourinary tract pathologies. However, understanding the infectious patterns of M. genitalium in clinical samples requires a dynamic assessment of mycoplasma-host cell interactions.
In the present study we applied CIA to assess infectious patterns of M. genitalium in clinical specimens. By examining vaginal cells of infected individuals, we could spatially localize M. genitalium on vaginal cell surfaces and intracellularly evaluate the extent of parasitism which, heretofore, had been totally undefined. Representative vaginal cell images from clinical specimens of M. genitalium culture-positive women allowed us to compare mycoplasma infectious levels within specimens and between individuals. As presented in Fig. 1 and 2, the extent of M. genitalium parasitism varied from strongly positive to negative in vaginal cells within the same sample field. Moderately infected samples, designated positive by CIA, possessed predominately extracellular mycoplasmas. Interestingly, heavily parasitized strong-positive cells revealed considerably greater numbers of intracellular than surface-associated mycoplasmas. This distribution pattern suggested extensive invasion of mycoplasmas, accompanied by intracellular replication and persistence as reported by us for experimentally infected human cell cultures (6, 9). Based upon CIA, we obtained a qualitative assessment of the range of infectious patterns displayed by M. genitalium in human cells, which indicated an unexpectedly high level of parasitism.
Transmission immunoelectron microscopy confirmed the intracellular presence and cell integrity of M. genitalium in vaginal samples (Fig. 3). The size and unit membrane structure of individual mycoplasmas, along with specific gold immunolabeling and staining patterns, reinforced the integrity and morphology of M. genitalium in a clinical setting. Interestingly, the stereotypical tip organelle, flask-shaped appearance of mycoplasmas visualized in broth-passaged cultures was difficult to discern in vaginal cells. Instead, an oblong morphology was more commonly observed. This could have been due to orientation of mycoplasmas within thin sections prepared for electron microscopic analysis or may truly reflect the in situ and intracellular morphological and physiological state of mycoplasmas, which may no longer require tip-mediated cytadherence for survival and replication. We have described alternate pathways of mycoplasma-host cell interactions that are not tip mediated (1, 10), which might be more advantageous for in vivo dissemination and persistence of mycoplasmas.
To provide additional insights into the extent of M. genitalium parasitism of vaginal cells, we performed real-time PCR, which permitted quantitative analysis of the ratios of mycoplasmas to vaginal cells. M. genitalium gyrA primers were used to consistently detect 10 copies/reaction mixture while 1 genome copy could be detected 70% of the time. The correlation between real-time PCR values of mycoplasma/human genome ratios and CIA determinations was very strong (r = 0.88), and we were able to show a statistically significant trend in this ratio across CIA categories (Table 1). This suggests that either CIA or real-time PCR might be useful in identifying potentially high transmitters, assessing response to antimicrobial therapy, and evaluating levels of mycoplasma persistence, chronicity of infection and immune competence in the host. We further demonstrated by reverse transcription-PCR that M. genitalium is transcriptionally active in vaginal specimens (Fig. 4), consistent with a physiologically dynamic and replicative state (25).
The experimental strategies outlined in this report enable both qualitative and quantitative approaches to assess M. genitalium infectious patterns. The system is robust and provides an in-depth appreciation of the dynamics of M. genitalium parasitism in human tissues. In the absence of reproducible culture techniques for the isolation of M. genitalium, this combinatorial approach provides additional insights as to how M. genitalium establishes subclinical to overt infections and long-term survival. In the latter case, M. genitalium appears to persist intracellularly, thereby circumventing mycoplasmacidal drugs and immune surveillance (6, 9). Thus, latent or persistent M. genitalium infections are likely to be common although the relationships between M. genitalium in the genitourinary tract and host immunity, nutrition, physiological conditions and disease progression are unclear. Furthermore, it is uncertain whether vaginal cells that were strong positives on the basis of CIA score represent a relatively early or intermediate stage of infection, or whether they are the result of long-term persistence. In either scenario, transition from negative to positive and strongly positive CIA-based designations may reflect an inability of the host to successfully eradicate M. genitalium. Alternatively, heavily parasitized vaginal cells may be in the process of elimination through cell death or apoptosis and cell shedding. Whatever the case, in situ replication of M. genitalium and cross talk between mycoplasmas and host cells are consistent with both possibilities, and high mycoplasma load, as observed in strong-positive M. genitalium-infected vaginal cells (Fig. 1 and 2), may directly reflect an infectious cycle of mycoplasma replication inside tissue target cells. This pattern is consistent with symptomatology and seroconversion observations described in M. genitalium culture-positive women (4; unpublished data).
Therefore, the infectious process and progression of disease associated with M. genitalium are expected to be influenced by pathogen load and degree of virulence, tissue tropism, immune competence and other host status variables, and antimicrobial interventions. Currently, we are examining fundamental biological properties of recent clinical isolates of M. genitalium to identify properties which can be linked to pathogenic and transmission mechanisms.
Acknowledgments
We thank Daniele Provenzano for his insightful discussions concerning the manuscript, Lauren Chestnut for preparing tissue sections and electron microscopic advice, and Mariana Cagle for patient selection and various microbiology and testing assistance. We also thank J. Alderete, J. Gunn, W. Haldenwang, K. Klose, B. Wickes, and G.-M. Zhong and their respective laboratories for providing organisms and DNA used as controls and J. Piper, W. Peairs, and staff at the Avanti Health Clinic for sample preparation and delivery. Images were generated in the Core Imaging Facility, UTHSCSA.
This work was supported by grants AI 45429 and AI 41010 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Alvarez, R. A., M. W. Blaylock, and J. B. Baseman. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol. Microbiol. 48:1417-1425. [DOI] [PubMed] [Google Scholar]
- 2.Anagrius, C., and B. Lore. 2002. Chlamydia-like symptoms can have another etiology. Mycoplasma genitalium-an important and common sexually transmitted disease. Lakartidningen 99:4854-4855, 4858-4859. (In Swedish.) [PubMed] [Google Scholar]
- 3.Baseman, J. B. 1993. The cytadhesins of Mycoplasma pneumoniae and Mycoplasma genitalium, p. 243-259. In I. Kahane (ed.), Subcellular biochemistry: mycoplasma cell membranes. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 4.Baseman, J. B., M. Cagle, J. E. Korte, C. Herrera, W. G. Rasmussen, J. G. Baseman, R. Shain, and J. Piper. 2004. Diagnostic assessment of Mycoplasma genitalium in culture-positive women. J. Clin. Microbiol. 203-211. 42: [DOI] [PMC free article] [PubMed]
- 5.Baseman, J. B., S. F. Dallo, J. G. Tully, and D. L. Rose. 1988. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J. Clin. Microbiol. 26:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baseman, J. B., M. Lange, N. L. Criscimagna, J. A. Giron, and C. A. Thomas. 1995. Interplay between mycoplasmas and host target cells. Microb. Pathog. 19:105-116. [DOI] [PubMed] [Google Scholar]
- 7.Baseman, J. B., S. P. Reddy, and S. F. Dallo. 1996. Interplay between mycoplasma surface proteins, airway cells, and the protean manifestations of mycoplasma-mediated human infections. Am. J. Respir. Crit. Care Med. 154:S137-S144. [DOI] [PubMed] [Google Scholar]
- 8.Baseman, J. B., and J. G. Tully. 1997. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 3:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dallo, S. F., and J. B. Baseman. 2000. Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb. Pathog. 29:301-309. [DOI] [PubMed] [Google Scholar]
- 10.Dallo, S. F., T. R. Kannan, M. W. Blaylock, and J. B. Baseman. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46:1041-1051. [DOI] [PubMed] [Google Scholar]
- 11.de Barbeyrac, B., C. Bernet-Poggi, F. Febrer, H. Renaudin, M. Dupon, and C. Bebear. 1993. Detection of Mycoplasma pneumoniae and Mycoplasma genitalium in clinical samples by polymerase chain reaction. Clin. Infect. Dis. 17(Suppl. 1):S83-S89. [DOI] [PubMed] [Google Scholar]
- 12.Deguchi, T., C. B. Gilroy, and D. Taylor-Robinson. 1995. Comparison of two PCR-based assays for detecting Mycoplasma genitalium in clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 14:629-631. [DOI] [PubMed] [Google Scholar]
- 13.Deguchi, T., and S. Maeda. 2002. Mycoplasma genitalium: another important pathogen of nongonococcal urethritis. J. Urol. 167:1210-1217. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths, G., J. M. Lucocq, and T. M. Mayhew. 2001. Electron microscopy applications for quantitative cellular microbiology. Cell. Microbiol. 3:659-668. [DOI] [PubMed] [Google Scholar]
- 16.Grondahl, B., W. Puppe, A. Hoppe, I. Kuhne, J. A. Weigl, and H. J. Schmitt. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 37:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay, P. E., and D. Taylor-Robinson. 1996. Defining bacterial vaginosis: to BV or not to BV, that is the question. Int. J. STD AIDS 7:233-235. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, J. S., H. T. Hansen, and K. Lind. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kannan, T. R., and J. B. Baseman. 2000. Expression of UGA-containing Mycoplasma genes in Bacillus subtilis. J. Bacteriol. 182:2664-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin, E. V. 2000. How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet. 1:99-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause, D. C. 1998. Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol. 6:15-18. [DOI] [PubMed] [Google Scholar]
- 22.Lind, K., and G. B. Kristensen. 1987. Significance of antibodies to Mycoplasma genitalium in salpingitis. Eur. J. Clin. Microbiol. 6:205-207. [DOI] [PubMed] [Google Scholar]
- 23.Luo, D., W. Xu, G. Liang, S. Wang, Z. Wang, Z. Bi, and W. Zhu. 1999. Isolation and identification of Mycoplasma genitalium from high risk populations of sexually transmitted diseases in China. Chin. Med. J. 112:489-492. [PubMed] [Google Scholar]
- 24.Manhart, L. E., C. W. Critchlow, K. K. Holmes, S. M. Dutro, D. A. Eschenbach, C. E. Stevens, and P. A. Totten. 2003. Mucopurulent cervicitis and Mycoplasma genitalium. J. Infect. Dis. 187:650-657. [DOI] [PubMed] [Google Scholar]
- 25.Marois, C., C. Savoye, M. Kobisch, and I. Kempf. 2002. A reverse transcription-PCR assay to detect viable Mycoplasma synoviae in poultry environmental samples. Vet. Microbiol. 89:17-28. [DOI] [PubMed] [Google Scholar]
- 26.Morrison-Plummer, J., D. H. Jones, K. Daly, J. G. Tully, D. Taylor-Robinson, and J. B. Baseman. 1987. Molecular characterization of Mycoplasma genitalium species-specific and cross-reactive determinants: identification of an immunodominant protein of M. genitalium. Isr. J. Med. Sci. 23:453-457. [PubMed] [Google Scholar]
- 27.Palmer, H. M., C. B. Gilroy, E. J. Claydon, and D. Taylor-Robinson. 1991. Detection of Mycoplasma genitalium in the genitourinary tract of women by the polymerase chain reaction. Int. J. STD AIDS 2:261-263. [DOI] [PubMed] [Google Scholar]
- 28.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy, S. P., W. G. Rasmussen, and J. B. Baseman. 1995. Molecular cloning and characterization of an adherence-related operon of Mycoplasma genitalium. J. Bacteriol. 177:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor-Robinson, D. 2002. Mycoplasma genitalium—an up-date. Int. J. STD AIDS 13:145-151. [DOI] [PubMed] [Google Scholar]
- 31.Taylor-Robinson, D. 1983. The role of mycoplasmas in non-gonococcal urethritis: a review. Yale J. Biol. Med. 56:537-543. [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor-Robinson, D., J. G. Tully, and M. F. Barile. 1985. Urethral infection in male chimpanzees produced experimentally by Mycoplasma genitalium. Br. J. Exp. Pathol. 66:95-101. [PMC free article] [PubMed] [Google Scholar]
- 33.Totten, P. A., M. A. Schwartz, K. E. Sjostrom, G. E. Kenny, H. H. Handsfield, J. B. Weiss, and W. L. Whittington. 2001. Association of Mycoplasma genitalium with nongonococcal urethritis in heterosexual men. J. Infect. Dis. 183:269-276. [DOI] [PubMed] [Google Scholar]
- 34.Tully, J. G., D. L. Rose, J. B. Baseman, S. F. Dallo, A. L. Lazzell, and C. P. Davis. 1995. Mycoplasma pneumoniae and Mycoplasma genitalium mixture in synovial fluid isolate. J. Clin. Microbiol. 33:1851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tully, J. G., J. W. Shih, R. H. Wang, D. L. Rose, and S. C. Lo. 1993. Titers of antibody to Mycoplasma in sera of patients infected with human immunodeficiency virus. Clin. Infect. Dis. 17(Suppl. 1):S254-S258. [DOI] [PubMed] [Google Scholar]
- 36.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288-1291. [DOI] [PubMed] [Google Scholar]
- 37.Wang, R. Y., T. Grandinetti, J. W. Shih, S. H. Weiss, C. L. Haley, M. M. Hayes, and S. C. Lo. 1997. Mycoplasma genitalium infection and host antibody immune response in patients infected by HIV, patients attending STD clinics and in healthy blood donors. FEMS Immunol. Med. Microbiol. 19:237-245. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, T., T. Deguchi, M. Ito, S. Maeda, M. Tamaki, and H. Ishiko. 2002. Quantitative detection of Mycoplasma genitalium from first-pass urine of men with urethritis and asymptomatic men by real-time PCR. J. Clin. Microbiol. 40:1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]