Abstract
Purpose
Kidney transplantation is the treatment of choice for end stage renal disease, with long term allograft loss being the major obstacle, and for which potential treatments are based on a histological diagnosis. The problem is that markers for predicting graft rejection are limited in number and invasive and quite non-specific. We have hypothesized that protein biomarkers might be discovered in the urine of patients when acute or chronic rejection might be occurring.
Experimental design
We have established a workflow in which initial screening for candidate biomarkers is first performed using urine samples on large scale antibody microarrays. This approach generated several dozen candidates. The next step is to qualify some of the strongest signals using the high throughput Reverse Capture Protein Microarray platform.
Results
Four top candidates including ANXA11, Integrin α3 and Integrin β3, and TNFα initially identified by the antibody microarray platform were all qualified using Reverse Capture Protein Microarrays. We also used Receiver Operating Condition (ROC) curves to independently quantify the specificity and sensitivity of these four analytes.
Conclusions and clinical relevance
The present data suggest that these novel four analytes in the urine, together or independently, may contribute to a robust and quantitative urine proteomic signature for diagnosing acute or chronic rejection of renal allografts.
Keywords: graft rejection, urine, proteomics, protein arrays
1. Introduction
Kidney transplantation is an increasingly successful surgical intervention, although nearly 50% of transplants eventually fail due to graft rejection [1]. The choices for potential treatments for graft rejection are presently based on histological examination of a transplant biopsy, and the problem therefore facing clinicians is how to know when a needle biopsy is warranted. The problem is that markers for predicting graft rejection are limited in number and quite non-specific. The initial clinical features of Acute Rejection (AR) involve fever, anemia, graft tenderness, decreased urine output and elevated serum creatinine [2]. However, the rise in serum creatinine has very poor specificity for graft AR injury, since increases in serum creatinine can also occur with infections, drug nephrotoxicity, pre-renal injury, obstruction and malignancies [3]. Furthermore, significant immune mediated injury has already been established in the graft by the time the serum creatinine has elevated past the conventional 20% increase criterion for triggering invasive biopsy [4]. In addition, increases in blood urea nitrogen (BUN) are also non-specific indicators of kidney dysfunction. Finally, attempts have been made to use genetic predictors to anticipate those patients most likely to reject a renal allograft [5-7]. However, despite substantial effort, no definitive and consistent genetic predictors have been identified.
An attractive alternative to clinical intuition, or serum creatinine elevation, would be specific diagnostic features found in serum or plasma [8], or, preferentially, in a proximal biological fluid such as urine [4, 9-15]. In most of these prior allograft rejection studies, the proteomics technologies depended on the SELDI mass spectrometry platform. This platform identifies specific analytic entities by molecular mass, but does not actually identify the analyte. However, more recent advances in clinical proteomics ranging from mass spectrometry, to large scale antibody microarrays and Reverse Capture Protein Microarrays, have permitted explicit identification and quantitation of the analyte, as well as enhanced the sensitivity and specificity of the assays [16, 17]. In fact, a proteomic-based quest for informative biomarkers in urine has been the goal in diseases as diverse as ovarian cancer [18] and bladder cancer [19].
The aim of the present discovery study has been to expand the search for biomarkers for rejection, to validate candidate proteins using high-throughput assays, and to determine their significance with respect to clinical outcomes. In the present work, we have studied unfractionated urine samples from patients with acute and chronic allograft rejection, and contrasted these with urine samples from stable transplant patients and healthy controls. The workflow has been to initially study the pooled urinary proteome on 507-feature antibody microarrays, and then to deploy Reverse Capture Protein Microarrays to validate selected “hits” independently for individual samples. Each of these platforms consumes only microliter volumes of urine, while returning quantitative information on low abundance proteins. The data show that discovery of candidate biomarkers for acute and chronic rejection of kidney allografts can be achieved using these two proteomics platforms, and that a unique and robust candidate signature for rejection can be identified.
2. Materials and Methods
2.1. Patients
Following transplant, patients were maintained on multiple immuno-suppressive strategies. These included non depletional(anti-CD25 antibody) and depletional induction with monoclonal or polyclonal antibodies (Alemtuzumab or Thymoglobulin) followed by tacrolimus and/or mycophenolate mofetil or sirolimus typically in a steroid free strategy. Stable function (SF, 11 patients) was defined as at least 6 months post-transplant without change in renal function and the absence of any significant histological or clinical abnormalities. Chronic graft injury (ChR, 11 patients) was defined by a rise in serum creatinine of at least 18% from baseline, with characteristic histologic changes showing at least Banff grade 2 chronic glomerulopathy, and at least grade I interstitial fibrosis and tubular atrophy [20]. Acute rejection (AR, 10 patients) was defined based on renal biopsies that histologically satisfied the Banff criteria (Borderline, IA, IB, IIA or IIB) [20]. Healthy donors with no medical problems, without any medications, and with normal renal function constituted the controls (NK, 8 patients). All patients were enrolled in Institutional Review Board approved clinical trials at the National Institutes of Health after informed concent prior to being investigated in this study.
2.2. Urine Specimens
Urine from all patients was collected over a 24 hour period. All urine samples were handled within the first 4 hours after collection was complete, using standard biosafety precautions. Urine was centrifuged at 3000 rpm at 4°C for 10 minutes, and the supernatant aliquoted into siliconized tubes and stored at −70°C. Urine samples belonging to these patients were collected at NIDDK, National Institutes of Health, following Institutional Review Board requirements.
2.3. Protein Profiling Using Antibody Microarrays
2.3.1. Labeling of urine proteins
Urine from patients in each annotated set were pooled on the basis of equal volumes and labeled with Cy3. Samples of FDA-certified, pooled normal control male serum was labeled with Cy5 and multiplexed with the Cy3-labeled patient samples as an “internal standard”. Cy3-labeled pooled patient urine was mixed with Cy5-labeled control on equal volume basis. Each sample was incubated with a 507 feature antibody microarray (Clontech, Mountain View, CA), in a medium containing a detergent-based reagent to minimize protein-protein interactions. The methods were as described in our recent paper [16].
2.3.2. Fluorescence detection
The fluorescence at each spot on the antibody microarray was measured on a GenePix array reader (New Milton, New Hampshire, U.K.), and downloaded to an EXCEL spread sheet.
2.3.3. Data Quality Control
Each sample of urine provides 4 replicate data points on the array for the analysis. Sample selection therefore begins by rejection from calculations of all spots with intensities below the local background, as well as all spots with Signal-to-Noise-Ratio < 3. We then calculate the standard deviation (SD) for each protein. Outliers were rejected if their deviations are larger than 2SD’s from the average of the respective protein. The averages were recalculated by omitting outliers. If a given protein is still too noisy, the specific protein is excluded from the analysis. We quantitated volume-normalized protein levels by ratio’ing Cy3-labeled proteins in patient urine samples to the same protein, labeled with Cy5, in the normal control sera. Normal sera labeled with both Cy3 and Cy5 were ratio’ed to one another to calculate a labeling efficiency difference specific to each protein. Normalization according to either total protein or creatinine was tested and excluded on the basis of profoundly noisy outcomes. Therefore the protein levels defined by these assays are concentrations found in a 24 hour urine sample.
2.3.4. Statistical Methods
Two approaches were used. In the first approach, the differences between disease samples and normal controls were determined based on t-tests of the samples in each group. Statistical significance is defined as p< 0.05, or P < 0.01 for correlation analysis. P-values in Table 3 are calculated from 2-tailed t-tests. In the second approach, which we presently prefer, we applied the SAM (Statistical Analysis of Microarrays: www.stat.stanford.edu/~tibs/SAM) package to determine the false Discovery Rate. An FDR < 10% will be taken to be statistically significant.
Table 3. Statistical Discriminators for Acute vs Chronic Rejection.
Calculations are based on Reverse Capture Microarray data from Figure 4. 2-tailed t-tests are used to calculate p-values.
| TNFα | Integrin α3 | Integrin β3 | ANXA11 | ||
|---|---|---|---|---|---|
| Acute vs SF | p-value | 0.026 | 0.055 | 0.280 | 0.021 |
| Ratio | 2.9 | 7.8 | 4.0 | 32.1 | |
| AUC | 0.890 | 0.891 | 0.800 | 0.991 | |
| Chronic vs SF | p-value | 0.052 | 0.020 | 0.286 | 0.001 |
| Ratio | 2.3 | 5.0 | 2.1 | 8.0 | |
| AUC | 0.805 | 0.855 | 0.813 | 0.963 | |
|
Acute vs
Chronic |
p-value | 0.537 | 0.427 | 0.424 | 0.069 |
| Ratio | 1.3 | 1.6 | 1.9 | 4.0 | |
| AUC | 0.614 | 0.591 | 0.591 | 0.832 | |
| Rejection vs SF | p-value | 0.030 | 0.043 | 0.297 | 0.055 |
| Ratio | 2.6 | 6.4 | 3.0 | 20.0 | |
| AUC | 0.845 | 0.872 | 0.807 | 0.976 | |
2.4. Qualification using Reverse Capture Protein Microarray
This method, also termed “Reverse Phase Protein Microarray” or “lysate microarray”, is essentially a Western blot analysis performed on dot-blots of serially diluted samples [21, 22]. Antibodies for testing on this platform were chosen from those identified by the antibody microarray platform. Clontech (BD, Biosciences/Transduction Labs) supplies the exact same antibodies in soluble form as are printed on the microarrays. Urine from each individual archival patient sample was printed using an AUSHON printer (Waltham, MA) in serially diluted fashion (Janus Liquid Handling Workstation,) on a slide in triplicate. Patient urine samples were printed on multiple single slides, and the entire dataset was probed with given antibodies[16, 23].
2.4.1. Total levels of antigens
The total level of a given antigen in the urine was calculated by extrapolating the log of the measured intensities of the dilution series back to the y-axis (i.e., no dilution). The theoretical curve is linear with a slope of −1, with deviations occurring at the high end (due to saturation) and at the low end (due to noise). A slope of −1 indicates that there is a 1:1 relationship between printed antigen and bound antibody. Outliers and low signal to noise spots were excluded from the curve fitting [23].
2.5. Statistics
The significance of the differences between patient and normal control urine were determined based on ANOVA of quadruplicate samples, and significance validated at the P ≤ 0.05 levels. Statistical significance is defined as P < 0.05 or P < 0.01 for correlation analyses. P values in Table 3 are calculated from 2-tailed t-tests.
2.6 Ingenuity Pathways Analysis
To discriminate the molecular pathways responsible for stable function effects versus graft rejection, we used IPA software (www.ingenuity.com, Ingenuity Systems, Redwood City, CA). An average expression ratio of R > 2 in stable function versus graft rejection comparisons was used as a threshold. The reports with outlier proteins from antibody microarray analysis were uploaded and mapped to corresponding objects (genes/proteins) in IPA’s database.
3. Results
3.1. Discovery of candidate protein biomarkers for graft rejection on a large scale antibody microarray platform
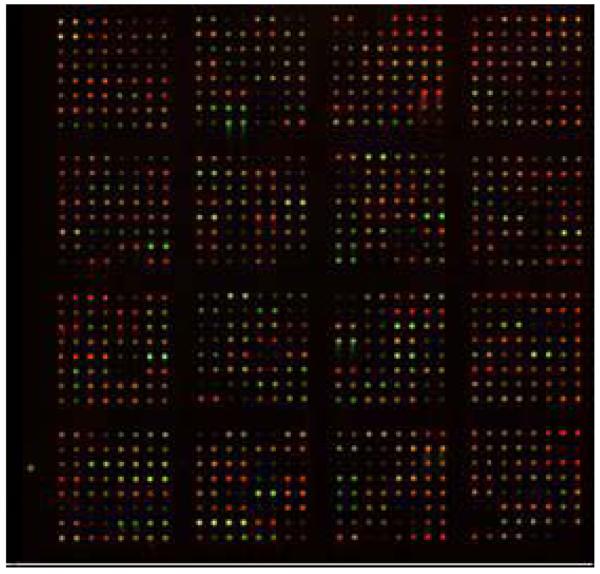
Individual pools of labeled urine samples from 21 patients with acute and chronic rejection, and urine samples from 11 gender-matched patients with stable function, were compared in parallel with 5 normal controls on large scale antibody microarrays. Figure 1 shows an example of one of these arrays, where the green pseudo color indicates spots where patient urine has a protein in greater concentration than in the internal standard, while the red pseudo color shows the reverse. Equal levels are imaged as a yellow (green = red) pseudo color. All urines analyzed on these antibody microarrays were scanned to provide independent values for intensity of either Cy3 or Cy5 labeled proteins.
Figure 1. Array image of urine from kidney transplant patient differing in graft survival vs. normal controls.
Proteins from kidney transplant urine (experimental) vs. normal kidney urine (control) were labeled with Cy3 or Cy5 dyes, respectively, and analyzed on an antibody microarray platform. Green features or red features indicate net increases for either condition. Yellow features indicate equivalent amounts of protein in both samples.
3.2. Scatter-plots of antibody microarray data for protein expression in urine from kidney transplant patients
Figure 2 shows graphs of protein expression in urine from different types of transplant function, compared to the same protein expression levels in urine from patients with normal kidneys. The statistics for each of these plots are summarized in Table 1. Figure 2a compares urines from 11 transplant patients in which the kidney function is stable, with urines from 8 patients with normal kidneys. The slope of this scatter-plot is 1.01, indicating that the two conditions are very similar. However, the R2 value (0.64) indicates that there are limits to this similarity, which we presume might be related to real differences in kidney function.
Figure 2. Scatter-plots for proteins found in transplant patient urines as a function of protein occurrence in urine from normal kidney urine.
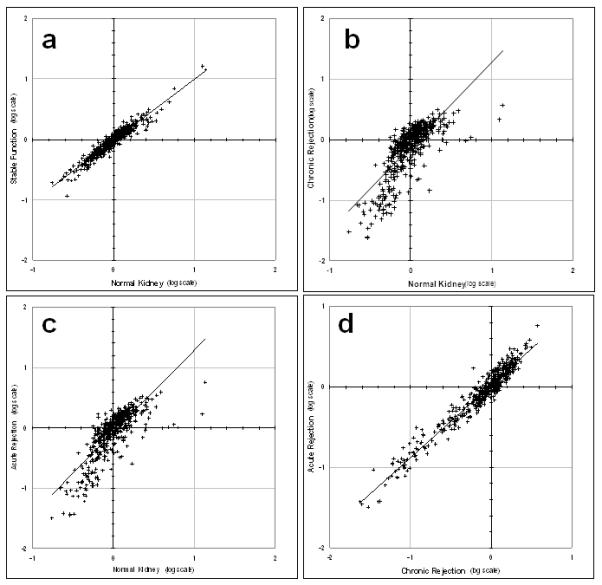
(a) Transplant patient with stable function vs normal kidney (slope = 1.01; R2=0.64). (b) Transplant patient with chronic rejection vs normal kidney (slope = 1.40; R2=0.59). (c) Transplant patient with acute rejection vs normal kidney (slope = 1.36; R2=0.64). (d) Transplant patient with chronic rejection vs transplant patient with acute rejection(slope = 0.90; R2=0.94). Data are normalized to the median of each array, and plotted on a log scale.
Table 1. Statistical analysis of proteomic relationships for kidney transplants*.
Calculations are based on antibody microarray data from Figure 2.
| Compare | slope | R2 |
|---|---|---|
| a. Stable Function vs Normal Kidney |
1.01 | 0.64 |
| b. Chronic Rejection vs Normal Kidney |
1.40 | 0.59 |
| c. Acute Rejection vs Normal Kidney |
1.36 | 0.64 |
| d. Chronic Rejection vs Acute Rejection |
0.90 | 0.94 |
Values are calculated from data in Figure 2.
Figure 2b compares protein expression in urines from 11 patients with chronic kidney transplant rejection, with urine proteins the same 8 controls. The slope of this scatter-plot is 1.40, indicating a substantial difference from the comparison of stable function vs control (cf, slope = 1.01, Figure 2a). In this case the R2 value is also a little lower (R2 = 0.59) indicating somewhat more variance. Figure 2c compares urine proteins from 10 patients with acute kidney transplant rejection, with urine proteins the same 8 controls as above. The slope of this scatter plot is 1.36, and the R2 value is 0.64. Finally, Figure 2d compares protein expression in urine of 11 chronic rejection patients with protein expression in urine from 10 patients with acute kidney transplant rejection. In this case the slope is 0.90, and the R2 value is 0.94. Clearly, these two classes of rejection have more in common than does the stable function kidney and the normal kidney (see Figure 2a and Table 1).
3.3. Identification of specific proteins as candidate biomarkers for transplant rejection
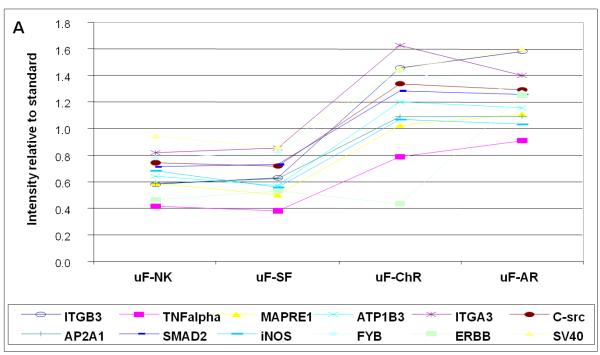
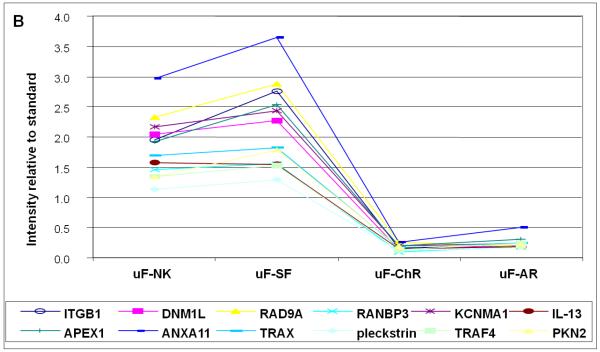
Figure 3 shows examples of candidate biomarkers that discriminate between patients with acute and chronic rejections, and patients that either have stable function, or are normal controls. Figure 3a shows data for 12 proteins which are relatively elevated in the two rejection stages. Among the most discriminating are TNFα (TNFA), Integrin α3 (ITGA3) and Integrin β3 (ITGB3). The differences are between 2- and 3-fold. Figure 3b shows data for proteins whose expression levels are reduced in the two rejection stages. Among the most discriminating are annexin A 11 (ANXA11) and Integrin beta 1 (ITGB1). In this case the differences are up to ten-fold. Table 2 contains all of the proteins in Figure 3a and Figure 3b, in addition to validating values for t-tests, calculations of local False Discovery rates, and q statistics.
Figure 3A. Proteins with increased excretion in urine with acute (AR) and chronic (ChR) kidney allograft rejection.
X-axis represents intensity relative to standard. Normal renal function (NK); stable function (SF); acute rejection (AR) and chronic rejection (ChR) are represented in the Y-axis.
Figure 3B. Proteins with decreased excretion in urine with acute (AR) and chronic (ChR) kidney allograft rejection.
X-axis represents intensity relative to standard. Normal renal function (NK); stable function (SF); acute rejection (AR) and chronic rejection (ChR) are represented in the Y-axis.
Table 2. Top markers for rejection in urine samples on Antibody Microarray using the average of normal kidney (NK) and stable function (SF), ratio’ed to chronic (ChrR) or acute (AR) rejection.
Table 1a is sorted by the ratio in urine showing markers most decreased in rejection. Table 1b below is sorted by the ratio in urine showing markers most increased in rejection. Table 1c below is sorted by the ratio in urine showing no change in markers in either state.
| ID | (NK+SF) vs (Chr+AR) | |
|---|---|---|
| Ratio | t-test | |
| a: Decreased in rejection state | ||
| integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
13.20 | 0.00009 |
| dynamin 1-like | 12.78 | 1.E-07 |
| RAD9 homolog (S. pombe) | 12.01 | 0.00001 |
| RAN binding protein 3 | 11.55 | 4.E-08 |
| potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 10.64 | 3.E-07 |
| interleukin 13 | 9.31 | 2.E-10 |
| APEX nuclease (multifunctional DNA repair enzyme) 1 | 8.85 | 0.00003 |
| annexin A11 | 8.75 | 0.00001 |
| translin-associated factor X | 8.60 | 2.E-06 |
| pleckstrin | 8.35 | 6.E-07 |
| TNF receptor-associated factor 4 | 7.90 | 1.E-06 |
| protein kinase C-like 2 | 7.76 | 0.00004 |
| b: Increased in rejection state | ||
| integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 2.51 | 2.E-06 |
| tumor necrosis factor (TNF superfamily, member 2) | 2.13 | 0.00007 |
| microtubule-associated protein, RP/EB family, member 1 | 1.96 | 0.00001 |
| ATPase, Na+/K+ transporting, beta 3 polypeptide | 1.94 | 5.E-06 |
| integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | 1.80 | 0.00006 |
| Cas-Br-M (murine) ecotropic retroviral transforming sequence | 1.80 | 6.E-08 |
| adaptor-related protein complex 2, alpha 1 subunit | 1.79 | 7.E-08 |
| MAD, mothers against decapentaplegic homolog 2 (Drosophila) | 1.76 | 2.E-08 |
| nitric oxide synthase 2A (inducible, hepatocytes) | 1.69 | 0.00004 |
| c: Not Changing in rejection state | ||
| nuclear autoantigenic sperm protein (histone-binding) | 1.02 | 0.92431 |
| Rho GTPase activating protein 1 | −1.02 | 0.06305 |
| caspase 9, apoptosis-related cysteine protease | −1.03 | 0.01703 |
| myogenic factor 3 | 1.07 | 0.00299 |
| nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 1.06 | 0.67978 |
| TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | 1.07 | 0.21566 |
| nuclear receptor subfamily 4, group A, member 1 | −1.01 | 0.87732 |
| protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1) | −1.02 | 0.32573 |
However, caution must be used when evaluating these tables because microarrays are as susceptible as any other microarray platform to false positives or false negatives. For that reason we have been careful to term these identified proteins as “candidates”, and now turn to a second platform, the Reverse Capture Protein Microarray, to qualify these identities.
3.4. Qualification of candidate urine protein biomarkers using the Reverse Phase Protein Microarray platform
Based on the most prominent hits from the antibody microarray data, we decided to use the Reverse Capture Protein Microarray platform to further qualify TNF alpha, Integrin alpha 3 (ITGA3), Integrin beta 3 (ITGB3) and annexin A 11 (ANXA11). In this case, samples of urine from each and every patient and control were serially diluted and printed on multiple slides. The slides were then probed with antibodies against the chosen antigens, and titers calculated. As shown in Figure 4 and Table 3 we were able to qualify all four of these candidate biomarkers. In addition, we calculated Receiver Operating Condition (ROC) curves, for which an area-under-the-curve (AUC) value of 1.0 would mark a perfect discrimination between rejection state and control. The values of AUC are independent of variance. By contrast (see Table 3), t-test calculations are dependant on variance. Therefore, for each condition, we also calculated significance using a 2-tailed t-test, and p values were calculated accordingly.
Figure 4. Reverse capture qualification of candidate urine protein biomarkers for graft rejection.
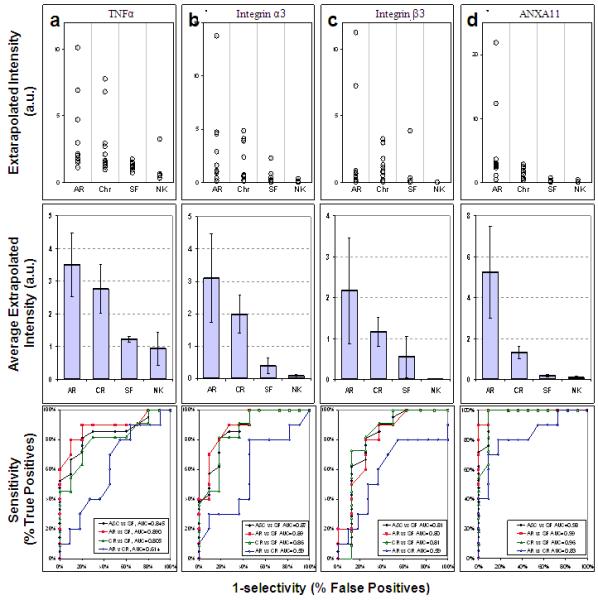
The levels of 4 biomarkers, a: TNFα, b: integrin α3, c: integrin β3, and d: annexin XI were evaluated using reverse capture microarrays. Top Panels: The distributions of immunoreactive activities in urine are given for all patients with documented acute rejection (AR), chronic rejection (CR), or stable function (Stable), and from normal (Normal) volunteers. Middle panels: the averages (± SEM) for the levels of the specific protein in urine of patients. Bottom panels: four different ROC curves are shown for each biomarker: A&C vs SF: Rejection (acute or chronic) vs. Stable in black; AR vs SF: Acute vs. Stable in red; CR vs SF: Chronic vs. Stable in green and AR v CR: Acute vs. Chronic Rejection in blue. The corresponding Area Under the Curve (AUC) values are shown in the respective insets.
Figure 4a shows qualification data for TNF alpha (TNFA, TNFα) in bar graph and dot-array forms, and the accompanying ROC curves. Table 3 summarizes the statistical information. The data clearly show that TNFα levels, on average, are 2.5-3-fold higher in the urine from acute and chronic rejection patients, compared to urine from either stable or control patients. Discrimination between urine from chronic rejection patients and stable patients is marginally significant (AUC = 0.81; p=0.052), while urines from acute vs stable patients show significant discrimination (AUC = 0.89; p=0.026). However, good discrimination is also achieved when comparing urines from both acute and chronic rejection patients (“R” for reject) with urine from stable transplant (“S” for stable function) patients (AUC = 0.845; p=0.030). The ROC curves for TNFα indicate that the discrimination between acute and chronic patients is limited (AUC = 0.61; p=0.537).
Figure 4b and Table 3 show qualification data for integrin-alpha-3 (ITGA3). The data clearly show that ITGA3 levels are in the range of ca. 5-7-fold higher in urine from either acute or chronic rejection patients, when compared to urine from stable patients (AUC = 0.87; p=0.043). In addition, the ROC curves for chronic vs stable and acute vs stable indicate AUC values of 0.85 (p=0.02) and 0.89 (p=0.055), respectively. However, this candidate biomarker fails to discriminate between urines from acute vs chronic rejection patients (AUC = 0.59; p=0.427).
Figure 4c and Table 3 show qualification data for integrin beta 3 (ITGB3). However, even though the average levels of ITGB3 are higher in urines from chronic and acute rejection patients, this biomarker actually fails all measures of significance. For example, the ROC curve comparing acute rejection with stable function data, has only a modest AUC value (0.59; p=0.427). The other comparisons are similarly marginal. Comparisons with the stable patients indicate that acute rejection (AUC = 0.80; p=0.280), chronic rejection (AUC = 0.81; p=0.286) fail because of problems with variance. When both acute and chronic rejection are compared with stable function, discrimination is remains marginal. (AUC = 0.81; p=0.297).
Figure 4d and Table 3 show qualification data for annexin A 11(ANXA11). In this case, urines from both acute and chronic rejection patients discriminate acute rejection well from chronic rejection patients. The ROC curve for this comparison has an AUC value of 0.98 (p=0.055). However, the signal for acute rejection, albeit with some variance, is much greater than the signal for chronic rejection. The ROC curve reflects this difference (AUC = 0.83; p=0.069). Nonetheless, the discrimination between urines from acute rejection, compared to stable function patients, is the best in our series (AUC = 0.99; p=0.021). By contrast, even with less variance, the difference between urine from chronic rejection vs stable function patients has an AUC value of 0.96 (p=0.001).
4. Discussion
Data in this paper support the concept that elevated expression levels of ANXA11, Integrin β3, Integrin α3, and TNFα can contribute to a candidate proteomic signature in urine for kidney allograft rejection. Receiver Operating Condition (ROC) calculations and stringent 2-tailed t-tests indicate that TNFα, a marker for inflammation, discriminates significantly between acute rejection and chronic rejection, and also clearly differentiates rejecting patients from stable function patients. By contrast, ANXA11 and integrin α3 levels are equivalently and significantly elevated in urine from patients with either acute or chronic kidney rejection. In addition, ANXA11 and integrin α3 also clearly discriminate rejecting patients from stable function patients. However, while Integrin α3 differs in terms of average ratio for all classes of transplant patient, this candidate biomarker only marginally passes the ROC test (AUC >> 0.5), but fails the t-test for significance. Thus at this discovery level of investigation, our patient set is sufficiently powered for ANXA11, Integrin α3, and TNFα, but not yet for Integrin β3. None of these proteins have been identified previously in the urine proteome from patients whose transplanted kidney is being rejected.
4.1. Integrins as biomarkers for kidney allograft rejection
Integrins are heterodimeric surface adhesion receptors, which mediate the interaction of cells to each other, and with the extracellular matrix [24]. The human genome contains a total of eighteen α subunits and eight β subunits, which occur as up to 24 different heterodimeric α/β pairs in different cellular systems [25]. The extracellular domain of the α subunit confers the binding specificity for the heterodimer [26]. By contrast, the intracellular domain of the β subunit is responsible for interacting with downstream signal transduction molecules [27], as well as cytoskeletal proteins such as talin and a-actinin [28, 29]. Some integrins are involved in leukocyte extravasation and inflammation [30] and integrin β3, a marker for leukocytes, has been reported to be elevated in circulating monocytes, T, B and NK lymphocytes of patients with ongoing allograft rejection [25].
However, of the three original candidate integrin biomarkers for allograft rejection in urine, both integrin β1 and integrin α3 have substantial histories of association with kidney physiology, structure, and development. For example, in human mature fetal and adult glomeruli, integrin β1 proteins are localized to the basal surfaces of endothelial cells, and to the podocytes abutting the glomerular basement membrane [31]. A more diffuse immunoreactivity is described for distal tubules and collecting ducts. Integrin α3 has also been shown to distribute similarly along basolateral aspects of endothelial cells in kidney, and the heterodimeric α3/β1 integrin has been shown to play an important role in kidney organogenesis [32]. The question then is the mechanism by which different integrins might be elevated in urine from patients with acute and chronic rejection.
In the case of integrin α3, we can conjecture that since the rejected kidney is losing its intrinsic structural integrity during the rejection process, the crucial molecules such as integrin α3, which tie kidney epithelial cells to each other, and to connective tissue, might be passively lost in the process. Integrin β1, the other part of the kidney-specific α3/β1 heteroduplex, was also identified by the antibody microarray as being elevated. However, qualification studies are ongoing on this candidate biomarker and others as part of our ongoing study. As for integrin β3, which, as mentioned earlier, has a history of association with kidney rejection, it is important to appreciate that this protein has been previously identified in circulating leukocytes from patients with allograft rejection [25]. Integrin β3 thus has little association with kidney structure, and has not previously been identified in the urine proteome. However, this novel finding does emphasize the potential contribution of proinflammatory cells to the rejection process, and may correlate with the elevated signal from TNFα, to be discussed below.
4.2. TNFα as a biomarker for allograft rejection
TNFα is a classical driver for inflammation and is readily detected in urine. It would therefore seem plausible for levels of TNFα to be elevated in fluid coming from a site of rejection. However, attempts to detect differences between TNFα levels in urine from patients undergoing renal allograft rejection have not previously met with success [33] Importantly, following biopsy-proven graft rejection, levels of TNFα have been shown to rise significantly in the plasma [34]. The rise in plasma concentration of TNFα has been reported to precede the diagnosis of clinical rejection by three days [35]. In the rejecting kidney itself, in situ hybridization has been used to detect TNFα mRNA and TNFα protein in severely rejected kidney grafts [36]. In this case, both TNFα mRNA and TNFα protein were found to be restricted to infiltrating leukocytes, defined only as “monomorphic”. In a study with type 1 diabetic children, kidney damage to renal proximal tubules was associated with elevated serum TNFα [37]. However, an analysis of urinary TNFα was not included in the study. On the other hand, IL-1α has been shown to induce cultured proximal tubular epithelial cells to produce TNFα [38]. With respect to the present study, we suggest that the elevated levels of TNFα and Integrin β3 in urine from acute and chronic rejecting allograft patients may represent contributions from infiltrating inflammatory cells, as well as from the damaged kidney tissue itself.
4.3. Annexins and cell function in the rejecting kidney
In the kidney, members of the annexin gene family are responsible for epithelial polarity, function as ion channels, and can act as extracellular autocrine regulators [39]. In experiments with acute renal failure in the rat, changes in ANXA2 were noted, although ANXA11 did not change [40]. However, the function of ANXA11 presently appears to be principally on the formation and maintenance of the chromosomal midbody, and ANXA11-depleted cells fail to complete cytokinesis and die by apoptosis [41]. We suggest that as ANXA11 seems to have a general role in cell maintenance, release of ANXA11 from acutely and chronically rejecting kidney into urine may represent an aspect of cell damage. How ANXA11 specifically relates to the rejection process, and from which cells may be contributing ANXA11 to the urine, must remain for future study.
We conclude that the current preliminary analyses suggest at least four qualified candidate biomarkers in urine for different types of kidney allograft rejection. The data presented here support the existence of a highly accurate and distinct multiplex proteomic set that can accurately distinguish between normal and stable function patients from acute and chronic rejection patients with AUC values ranging from 0.98 to 0.72.
4.4. Interaction analysis for Candidate Rejection Biomarker function in kidney
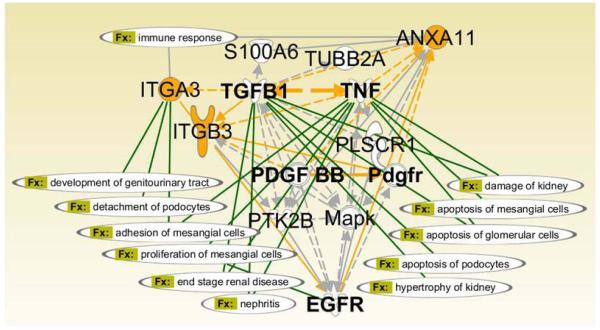
We have used the Ingenuity Pathways Analysis (IPA) platform to investigate possible functional relationships among the qualified candidate biomarkers for kidney allograft rejection. As shown in Figure 5, TNFα is a hub for pathways involved in major cytokine and cell survival regulation. These pathways include TGFB (Transforming Growth Factor beta), PDGF (Platelet Derived Growth Factor) and EGFR (Epidermal Growth Factor Receptor). This hub and its spokes have the potential to affect the Immune Response Biofunction, which is shared by ANXA11 and ITGA3. ITGA3 has several kidney-specific functions (see Section 4.1), and specifically includes Detachment of Podocytes and Adhesion of Mesangial Cells. Thus IPA analysis substantiates the selection of rejection biomarkers for their diagnostic and prognostic potential by connecting them to kidney-specific functions and positioning them within rejection-associated signaling pathways.
Figure 5. Interaction analysis for candidate rejection biomarker function in kidney.
The network was created using Path Designer (Ingenuity Pathways Analysis, IPA). The identified biomarkers are highlighted in yellow. The Function (Fx:) related connections in the network are highlighted in green.
5. Conclusions
The two antibody platforms used in this discovery study have the principal advantages of being able to identify and quantitate very low abundance proteins. This emphasizes advantages over more conventional proteomics platforms such as 2D gel electrophoresis, either alone or coupled to mass spectrometry, and even to newly developed mass spectrometry platforms such as MRM (Multiple Reaction Monitoring), which function independently. However, having identified candidate biomarker proteins in the antibody microarray discovery platform, qualification is necessary by at least one other method. For these reasons, we suggest that the two stage proteomic workflow, which we have employed here, constitutes a compelling model for discovery studies in other disease entities. Stage #1 is the use of a large scale antibody microarray to identify possible biomarkers. Stage #2 is a high capacity Reverse Capture Protein Microarray platform to simultaneously interrogate many patient and control samples. We conclude that the discovery/qualification phase of this project can now be upgraded to verification with a larger patient cohort. Furthermore, given the flexibility of the Reverse Capture platform, we will be able to further mine the antibody microarray data, and to add other candidate biomarkers for kidney allograft rejection, as they become known to the community.
Acknowledgements
The authors express appreciation to Ms. Wei Huang for outstanding technical support for this project, and gratefully acknowledge financial support from the NIH [RO1DK053051 (HBP); NO1HV28187 (HBP)], and the Cystic Fibrosis Foundation [HBP].
ABBREVIATIONS
- NK
normal renal function
- SF
stable function
- AR
acute rejection
- ChR
chronic rejection
- IPA
Ingenuity Pathways Analysis
References
- 1.Xue JL, Daniels F, Star RA, Kimmel PL, et al. Incidence and mortality of acute renal failure in medicare beneficiaries, 1992 to 2001. J. Am. Soc. Nephrol. 2006;17:1135–1142. doi: 10.1681/ASN.2005060668. [DOI] [PubMed] [Google Scholar]
- 2.Racusen LC. The Banff schema and differential diagnosis of allograft dysfunction. Transplant. Proc. 2004;36:753–754. doi: 10.1016/j.transproceed.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 3.Berl T. American Society of Nephrology Renal Research Report. J. Am. Soc. Nephrol. 2005;16:1886–1903. doi: 10.1681/ASN.2005030285. [DOI] [PubMed] [Google Scholar]
- 4.Schaub S, Rush D, Wilkins J, Gibson IW, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J. Am. Soc. Nephrol. 2004;1:219–227. doi: 10.1097/01.asn.0000101031.52826.be. [DOI] [PubMed] [Google Scholar]
- 5.Goldfarb-Rumyantzev AS, Naiman N. Genetic predictors of acute renal transplant rejection. Nephrol. Dial. Transplant. 2010 doi: 10.1093/ndt/gfp782. in press. (doi: 10.1093/ndt/gfp782) [DOI] [PubMed] [Google Scholar]
- 6.Hahn AB, Kasten-Jolly JC, Constantino DM, Graffunder E, et al. TNFalpha, IL-6. IFN-gamma, and IL-10 gene expression polymorphisms and the IL-4 receptor chain vatiant Q576R: effects on renal allograft outcome. Transplantation. 2001;72:660–665. doi: 10.1097/00007890-200108270-00017. [DOI] [PubMed] [Google Scholar]
- 7.Dimitrienko S, Hoar DI, Balshaw NA, Keown PA. Immune response gene polymorphisms in renal transplant recipients. Transplantation. 2005;80:1773–1782. doi: 10.1097/01.tp.0000184624.54005.9f. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Sigdel TK, Li Li., Kambham N, et al. Differentially Expressed RNA from Public Microarray Data Identifies Serum Protein Biomarkers for Cross-Organ Transplant Rejection and Other Conditions. PLOSComp.Biol. 2010;6(9):pii, e1000940. doi: 10.1371/journal.pcbi.1000940. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke W, Silverman BC, Zhang Z, Chan DW, et al. Characterization of Renal Allograft Rejection by Urinary Proteomic Analysis. Ann. Surg. 2003;237:660–665. doi: 10.1097/01.SLA.0000064293.57770.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Riordan E, Goligorsky MS. Emerging studies of the urinary proteome: the end of the beginning? Curr. Opin. Nephrol. Hypertens. 2005;6:579–585. doi: 10.1097/01.mnh.0000168425.60729.36. [DOI] [PubMed] [Google Scholar]
- 11.Parikh CR, Jani A, Mishra J, Ma Q, et al. Urine NGAL and IL-18 are predictive biomarkers for delayed graft function following kidney transplantation. Am. J. Transplant. 2006;6:1639–1645. doi: 10.1111/j.1600-6143.2006.01352.x. [DOI] [PubMed] [Google Scholar]
- 12.Barratt J, Topham P. Urine proteomics: the present and future of measuring urinary protein components in disease. CMAJ. 2007;177:361–368. doi: 10.1503/cmaj.061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bramham K, Mistry HD, Poston L, Chappell LC, et al. Thompson AJ. The non-invasive biopsy--will urinary proteomics make the renal tissue biopsy redundant? QJM. 2009;102:523–538. doi: 10.1093/qjmed/hcp071. [DOI] [PubMed] [Google Scholar]
- 14.Thomas CE, Sexton W, Benson K, Sutphen R, et al. Urine collection and processing for protein biomarker discovery and quantification. Cancer Epidemiol. Biomarkers Prev. 2010;19:953–959. doi: 10.1158/1055-9965.EPI-10-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quintana LF, Campistol JM, Alcolea MP, Bañon-Maneus E, et al. Application of label-free quantitative peptidomics for the identification of urinary biomarkers of kidney chronic allograft dysfunction. Mol. Cell Proteomics. 2009;8:1658–1673. doi: 10.1074/mcp.M900059-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava M, Eidelman o, Jozwik C, Paweletz C, et al. Serum proteomic signature for cystic fibrosis using an antibody microarray platform. Mol. Genet Metab. 2006;87:303–310. doi: 10.1016/j.ymgme.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 17.Pollard HB, Eidelman O, Jozwik C, Huang W, et al. De novo biosynthetic profiling of high abundance proteins in cystic fibrosis lung epithelial cells. Mol Cell Proteomics. 2006;5:1628–1637. doi: 10.1074/mcp.M600091-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Ye B, Skates S, Mok SC, Horick NK, et al. Proteomic-based discovery and characterization of glycosylated eosinophil-derived neurotoxin and COOH-terminal osteopontin fragments for ovarian cancer in urine. Clin. Cancer Res. 2006;12:432–441. doi: 10.1158/1078-0432.CCR-05-0461. [DOI] [PubMed] [Google Scholar]
- 19.Eiján AM, Sandes E, Puricelli L, Bal De Kier Joffé E, et al. Cathepsin B levels in urine from bladder cancer patients. Oncol. Rep. 2000;7:1395–1399. doi: 10.3892/or.7.6.1395. [DOI] [PubMed] [Google Scholar]
- 20.Girlanda R, Kleiner DE, Duan Z, Ford EAS, Wright EC, Mannon RB, Kirk AD. Monocyte infiltration and kidney allograft dysfunction during acute rejection. Am.J. Transplant. 2008;8:600–607. doi: 10.1111/j.1600-6143.2007.02109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paweletz CP, Charboneau L, Bichsel VE, Simone NL, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava M, Eidelman O, Zhang J, Paweletz C, et al. Digitoxin mimics CFTR-Gene Therapy, and Suppresses Hypersecretion of Proinflammatory Interleukin-8 (IL-8) from Cystic Fibrosis Lung Epithelial Cells. Proc.Nat.Acad.Sci.(USA) 2004;C101:7693–7698. doi: 10.1073/pnas.0402030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyorgy AB, Walker J, Wingo D, Eidelman O, et al. Reverse Phase protein microarray technology in traumatic brain injury. J. Neurosci. Meth. 2010;192:96–101. doi: 10.1016/j.jneumeth.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 24.Sugimori T, Griffith DL, Arnaout MA. Emerging paradigms of integrin ligand binding and activation. Kidney International. 1997;51:1454–1462. doi: 10.1038/ki.1997.199. [DOI] [PubMed] [Google Scholar]
- 25.Chandrakanton A, McDermott DH, Tran HTB, Jurewicz M, et al. Role of β3 integrin in acute renal allograft rejection in humans. Clin.J.Am Soc.Nephrol. 2007;2:1268–1273. doi: 10.2215/CJN.01380307. [DOI] [PubMed] [Google Scholar]
- 26.Kawaguchi S, Hemler ME. The role of the alpha subunit cytoplasmic domain in regulation of adhesive activity mediated by the integrin VLA-2. J.Biol.Chem. 1993;268:16279–16285. [PubMed] [Google Scholar]
- 27.Hannigan GE, Leung HC, Fitz GL, Coppolino MG, et al. Regulation of cell adhesion and anchorage-dependenjt growth by a new beta 1-integrinh linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 28.Otey CA, Vasquez GB, Burridge K, Erickson BW. Mapping of the alpha-actinin binding site within the beta 1 integrin cytoplasmic domain. J.Biol.Chem. 1993;268:21193–21197. [PubMed] [Google Scholar]
- 29.Chen HC, AQppeddu PA, Parsons JT, Hildebrand JD, et al. Interaction of focal adhesion kinase with cytoskeletal protein talin. J.Biol.Chem. 1995;267:18908–18914. doi: 10.1074/jbc.270.28.16995. [DOI] [PubMed] [Google Scholar]
- 30.Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 31.Korhonen M, Yianne J, Laitinen L, Virtanen I. Distribution of beta 1 and beta 3 integrins in human fetal and adult kidney. Lab.Invest. 1990;62:616–625. [PubMed] [Google Scholar]
- 32.Kreidberg JA, Donovan MJ, Goldstein SL, Rennke H, et al. Alpha 3 beta 1 integrin has a crucial role in kidney and lung organogenesis. Development. 1996;122:3537–3547. doi: 10.1242/dev.122.11.3537. [DOI] [PubMed] [Google Scholar]
- 33.Newstead CG, Lamb WR, Brenchley PE, Short CD. Serum and urine IL-6 and TNF-alpha in renal transplant recipients with graft dysfunction. Transplantation. 1993;56:831–835. doi: 10.1097/00007890-199310000-00010. [DOI] [PubMed] [Google Scholar]
- 34.Darge SE. Plasma levels of tumor necrosis factor (TNF) and soluble TNF receptors in kidney transplant recipients. Transplantation. 1994;58:1000–1008. doi: 10.1097/00007890-199411150-00005. [DOI] [PubMed] [Google Scholar]
- 35.Kutukculer N, Shenton BK, Clark K, Rigg KM, et al. Renal allograft rejection: the temporal relationship and predictive value of plasma TNF (alpha and beta), IFN-gamma and soluble ICAM-1. Transpl. Int. 1995;8:45–50. doi: 10.1007/BF00366710. [DOI] [PubMed] [Google Scholar]
- 36.Morel D, Normand E, Lemoine C, Merlio J-P, et al. Tumor necrosis factor alpha in human kidney transplant rejection-analysis by in situ hybridization. Transplantation. 1993;55:773–777. doi: 10.1097/00007890-199304000-00017. [DOI] [PubMed] [Google Scholar]
- 37.Myśliwiec M, Balcerska A, Zorena K, Myśliwska J, et al. Serum and urinary cytokine homeostasis and renal tubular function in children with type 1 diabetes mellitus. J Pediatr Endocrinol Metab. 2006;19:1421–1427. doi: 10.1515/jpem.2006.19.12.1421. [DOI] [PubMed] [Google Scholar]
- 38.Yard BA, Daha MR, Kooymans-Couthino M, Bruijn JA, et al. IL-1a – stimulated TNFa production by cultured human proximal tubular epithelial cells. Kidney. Internat. 1992;42:383–389. doi: 10.1038/ki.1992.299. [DOI] [PubMed] [Google Scholar]
- 39.Markoff A, Gerke V. Expression and functions of annexins in the kidney. Am J Physiol Renal Physiol. 2005;289:F949–F956. doi: 10.1152/ajprenal.00089.2005. [DOI] [PubMed] [Google Scholar]
- 40.Cheng C-W, Rifai A, Ka S-M, Shui H-A, et al. Calcium-binding proteins annexin A2 and S100A6 are sensors of tubular injury and recovery in acute renal failure. Kidney International. 2005;68:2694–2703. doi: 10.1111/j.1523-1755.2005.00740.x. [DOI] [PubMed] [Google Scholar]
- 41.Tomas A, Futter C, Moss SE. Annexin 11 is required for midbody formation and completion in the terminal phase of cytokinesis. J. Cell Biol. 2004;165:813–822. doi: 10.1083/jcb.200311054. [DOI] [PMC free article] [PubMed] [Google Scholar]