Abstract
Porphyromonas gingivalis is strongly associated with periodontal diseases and is regarded as one of the risk factors for periodontitis. Insertion sequence element IS1126-based PCR was used to investigate the genetic heterogeneity of P. gingivalis from periodontitis patients and to examine the frequency of the parent-child and spouse-spouse transmission. Two sets of IS1126-specific primers were used for the PCR. The inward primer set (PI1 and PI2), which amplifies the IS1126 fragment of approximately 690 bp, was used to identify P. gingivalis. The outward primer set (PI1RC and PI2RC), which is reverse complementary to PI1 and PI2, respectively, and amplifies the gene fragments between the adjacent IS1126 elements was used to characterize the genotypes of the P. gingivalis strains. PCR of P. gingivalis with PI1RC and PI2RC resulted in the production of two to seven amplicons, which showed a unique electrophoretic pattern in each strain (4 laboratory strains and 37 clinical isolates cultured from 12 patients with aggressive periodontitis). The usefulness of the method for transmission study was confirmed by detecting identical genotypes between the isolates and the plaque samples from which the isolates were cultured and between the plaque samples from different tooth sites in the same patient. Thirty probands with periodontal diseases and their thirty immediate family members were included in the transmission study. In 11 of 14 parent-child pairs (78.6%), P. gingivalis revealed an identical or similar band pattern, whereas 5 of 16 spouse pairs (31.25%) had this similarity. These results show that IS1126-based PCR for genotyping P. gingivalis has a highly discriminating potential with reproducible data and is a simple and reliable method for a transmission study.
Porphyromonas gingivalis is a gram-negative, anaerobic, nonmotile short rod-shaped, black-pigmented bacterium. Studies on the epidemiology, the virulence factors, the host response, and the pathogenicity in animal models have provided strong evidence implicating this bacterium in the etiology of periodontitis (9, 10, 12). P. gingivalis is known to be a risk factor for periodontitis (3). In addition, there is accumulating evidence showing that an infection with P. gingivalis may predispose an individual to cardiovascular disease and cause the delivery of preterm infants (4, 5, 24). This emphasizes the importance of preventing an infection by this bacterium. In this respect, information on the transmission route of P. gingivalis between individuals is important for determining the risk factors for periodontitis and for developing strategies for preventing the transmission of the pathogen to uninfected individuals.
Several studies on the transmission of periodontopathogens have been performed by detecting identical strains between related family members. The transmission of Actinobacillus actinomycetemcomitans and P. gingivalis has been reported by comparing either the serotypes (15, 32) or the genotypes. The transmission rate reported thus far varies from study to study. Information on the vertical transmission of P. gingivalis had been restricted due to the difficulties in detecting this bacterium in young children by using culture methods.
A recent study used PCR to detect and investigate the transmission of P. gingivalis within extended family members (30). In that study, the presence or absence of P. gingivalis was examined in samples taken from the mesial sulcus of four family generations. These authors found that contact with an infected family member substantially increased the relative risk of colonization for spouses, children and their parents, and adults and their mothers, as well as between siblings. A method that characterizes the strains directly from clinical samples and differentiate them would be valuable for transmission studies. The direct identification of P. gingivalis present in periodontitis patient samples was performed at the strain level by an arbitrarily primed PCR (AP-PCR) (1) or a heteroduplex analysis of the ribosomal DNA intergenic spacer region (ISR) (16, 18).
The insertion sequence (IS) elements in several bacterial species have been shown to be an excellent tools for molecular typing and an epidemiological study (6, 13, 28). IS1126 of P. gingivalis is an insertion sequence with 1,338 bp, which is flanked by 12-bp inverted repeats and generates a 5-bp target site duplication (21). It contains a single open reading frame encoding the transposase and belongs to the IS5 family of ISs based on the homology of the enzyme (20). The P. gingivalis genome harbors multiple copies of IS1126, and the value of this element as a rapid epidemiological tool for strain-specific identification of P. gingivalis has been suggested because Southern blot analysis of a number of P. gingivalis strains with IS1126 as a probe showed a unique hybridization pattern after digestion with the restriction enzymes (8, 21).
The present study describes a rapid and simple method that distinguishes the strains of P. gingivalis in subgingival plaque samples by PCR with the IS1126-specific primers that amplify the regions between the adjacent IS1126. The method was performed (i) to identify the P. gingivalis strains cultured from patient materials, (ii) to compare the multiple P. gingivalis strains cultured from the same patient, (iii) to compare the P. gingivalis isolates and plaque samples from which the isolates were obtained, and (iv) to compare the P. gingivalis strains in the different tooth sites from the same patient. Finally, the present study applied the method to a transmission study between parent-child pairs and between spouse pairs. Using this method, we were able to differentiate P. gingivalis in both forms of isolates and plaque samples at the strain-specific level. In addition, we successfully used this method to estimate the transmission rate by comparing the PCR band pattern of family members. Here we report that a PCR method with IS1126-specific primers is a very sensitive typing method and can potentially be useful for transmission studies of P. gingivalis.
MATERIALS AND METHODS
Bacterial strains
Four laboratory strains of P. gingivalis and 17 other oral bacterial strains were included to evaluate the specificity of the primers used for PCR (Table 1). A. actinomycetemcomitans, Prevotella intermedia, Prevotella nigrescens, and Fusobacterium nucleatum were grown in a brain heart infusion (BHI; Difco, Detroit, Mich.) broth in an anaerobic chamber (N2 at 85%, H2 at 5%, and CO2 at 10%) at 37°C for 2 to 3 days. The streptococcus species were grown aerobically in BHI broth at 37°C for 2 days. P. gingivalis was grown anaerobically in BHI broth supplemented with hemin (10 μg/ml) and vitamin K (1 μg/ml) for 3 days. The bacterial cells were harvested by centrifuging at 5,000 × g for 10 min at 4°C.
TABLE 1.
Reference strains used for determining primer specificity
| Bacterium | Strain | Sourcea |
|---|---|---|
| Actinobacillus actinomycetemcomitans | ATCC 43717 | ATCC |
| Actinobacillus actinomycetemcomitans | ATCC 33384 | ATCC |
| Prevotella intermedia | ATCC 49046 | ATCC |
| Prevotella nigrescens | NCTC 9336 | ATCC |
| Porphyromonas gingivalis | ATCC 33277 | ATCC |
| Porphyromonas gingivalis | HG564 | KCTC |
| Porphyromonas gingivalis | ATCC 53978 | ATCC |
| Porphyromonas gingivalis | A7A1-28 | KCTC |
| Fusobacterium nucleatum | ATCC 25586 | ATCC |
| Fusobacterium nucleatum | ATCC 10953 | ATCC |
| Fusobacterium nucleatum | 5119 | KCTC |
| Streptococcus gordonii | ATCC 49818 | ATCC |
| Streptococcus rattus | BHT | KCTC |
| Streptococcus sanguis | 4 | KCTC |
| Streptococcus mitior | 903 | ATCC |
| Streptococcus sobrinus | ATCC 27351 | ATCC |
| Streptococcus mutans | Ingbritt | KCTC |
ATCC, American Type Culture Collection (Manassas, Va.); KCTC, Korean Collection for Type Culture (Taejon, Korea).
Plaque samples and P. gingivalis isolates.
Subgingival plaque samples (31 samples) taken from 12 patients with aggressive periodontitis who had been previously analyzed in an epidemiology study for Koreans (17) were included in the present study (Table 2). All of these plaque samples tested P. gingivalis positive when detected by dot blot hybridization (17). P. gingivalis isolates (37 strains) were cultured from these plaque samples to compare the PCR band patterns between the isolates and between the plaque samples from which the isolates were cultured. The plaque samples were serially diluted up to 10−6. Aliquots of 100 μl were plated on BHI agar medium supplemented with 5% defibrinated sheep blood, hemin (10 μg/ml), and vitamin K (1 μg/ml) and then incubated in an anaerobic chamber for 10 days. Black-pigmented colonies were streaked to purity. The identification of P. gingivalis was performed based on Gram staining, trypsin-like enzyme activity, and dot blot hybridization with an oligonucleotide probe specific for the P. gingivalis 16S rRNA genes (7). Each colony identified as P. gingivalis was subjected to subculture and stored in 15% glycerol stock at −70°C until needed.
TABLE 2.
Plaque samples from 12 patients with aggressive periodontitis and their 37 P. gingivalis isolatesa
| Subject no. | Plaque sample(s)b |
|---|---|
| P1 | #16(3), #21, #36, #37 |
| P2 | #36(8) |
| P3 | #11(4), #21, #33, #34 |
| P4 | #28(1), #36, #46 |
| P5 | #13(2) |
| P6 | #11(8), #16, #26, #46 |
| P7 | #11(4), #16, #26, #33, #47 |
| P8 | #14, #16, #21(1), #35, #47 |
| P9 | #46(1) |
| P10 | #43(1) |
| P11 | #16(1) |
| P12 | #24(3) |
The 12 patients with aggressive periodontitis were originally described elsewhere (17).
The first number after the number symbol (#) indicates the maxillary (1 and 2) and mandibular (3 and 4) teeth. The second number after the number symbol refers to the tooth sites for the sampling (1 and 2, incisor; 3, canine; 4 and 5, premolar; 6 and 7, molar). The numbers of isolates cultured from the tooth sites are indicated in parentheses. Designations with no number in parentheses indicate there was no isolate.
The genotypes of some plaque samples collected from the different tooth sites in the same patient (Table 2) were also compared.
Subjects and sampling for the transmission study.
The subjects for the transmission study were Korean patients with periodontal disease who were referred to the Department of Periodontology at the Hospital in the College of Dentistry at Seoul National University, as well as their immediate family members. A total of 30 probands and their immediate family members were selected based on the presence of P. gingivalis: 14 parent-child pairs (10 adults as a proband and their mothers and 4 mothers as a proband and their adult children), and 16 spouse pairs. The probands were diagnosed with various types of periodontal diseases, as shown in Table 3. The criteria for selecting the family members of the patient probands were as follows: the child had to be natural and had to have lived with their parents for more than 2 years at the time of sampling. The spouses had to have been married for more than 5 years. The affected sites of the probands had a probing depth of >6 mm and an attachment loss of 3 mm.
TABLE 3.
Detection of the P. gingivalis genotypes in the parent-child pairs and in the spouse-spouse pairs
| Type of transmission and patient no. | Sex | Age (yr) | Diagnosis | Family member analyzed | Genotypea |
|---|---|---|---|---|---|
| Vertical (parent-child pair) | |||||
| 1 | Male | 27 | Chronic periodontitis | Mother | + |
| 2 | Female | 28 | Chronic periodontitis | Mother | + |
| 3 | Male | 19 | Chronic gingivitis | Mother | + |
| 4 | Female | 26 | Aggressive periodontitis | Mother | + |
| 5 | Male | 22 | Aggressive periodontitis | Mother | + |
| 6 | Male | 32 | Chronic periodontitis | Mother | − |
| 7 | Male | 20 | Gingival hyperplasia | Mother | + |
| 8 | Male | 37 | Chronic periodontitis | Mother | + |
| 9 | Male | 27 | Chronic periodontitis | Mother | + |
| 10 | Male | 32 | Chronic periodontitis | Mother | − |
| 11 | Female | 61 | Chronic periodontitis | Daughter | + |
| 12 | Female | 67 | Chronic periodontitis | Daughter | + |
| 13 | Female | 47 | Chronic periodontitis | Son | − |
| 14 | Female | 52 | Chronic periodontitis | Daughter | + |
| Horizontal (spouse-spouse pair) | |||||
| 1 | Female | 35 | Chronic periodontitis | Spouse | − |
| 2 | Male | 51 | Chronic periodontitis | Spouse | − |
| 3 | Male | 51 | Chronic periodontitis | Spouse | − |
| 4 | Male | 63 | Chronic periodontitis | Spouse | − |
| 5 | Female | 25 | Chronic periodontitis | Spouse | + |
| 6 | Male | 32 | Chronic periodontitis | Spouse | − |
| 7 | Female | 49 | Chronic periodontitis | Spouse | + |
| 8 | Male | 37 | Chronic periodontitis | Spouse | + |
| 9 | Female | 61 | Chronic periodontitis | Spouse | − |
| 10 | Female | 53 | Chronic periodontitis | Spouse | + |
| 11 | Male | 61 | Chronic periodontitis | Spouse | − |
| 12 | Male | 68 | Chronic periodontitis | Spouse | − |
| 13 | Male | 67 | Chronic periodontitis | Spouse | − |
| 14 | Male | 38 | Chronic periodontitis | Spouse | − |
| 15 | Male | 32 | Chronic periodontitis | Spouse | + |
| 16 | Male | 68 | Chronic periodontitis | Spouse | − |
+, Identical genotype between family members; −, different genotype between family members.
For subgingival plaque sampling, excess saliva was removed by using a cotton roll, and three sterile paper points were inserted into the mesial sulcus of diseased sites of the probands and the deepest sites of their family members for 30 s. The paper points were transferred to autoclaved 1.5-ml microfuge tube and stored at −20°C until needed. For the DNA isolation, the bacteria from the paper points were eluted by vigorously vortexing them after the addition of 1 ml of phosphate-buffered saline in a sample tubes.
DNA isolation and PCR with IS1126-specific primers.
The DNA of the reference strains, the clinical isolates of P. gingivalis, and the plaque samples was obtained by a phenol-chloroform extraction method according to standard protocols. The quantity of DNA dissolved in the TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA) was estimated by measuring the optical density at 260 nm.
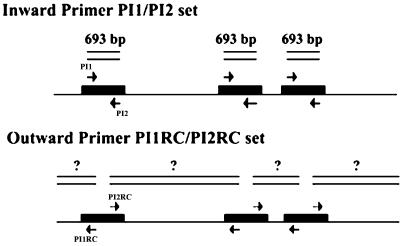
The specificity of the inward primers for IS1126 (Fig. 1), PI1 (5′-CCC GGC TTA TGA CGT GAT TTC TCT-3′) and PI2 (5′-CTG TTG CGT TTG TGC CCT TGT GC-3′), was tested with 4 laboratory strains of P. gingivalis, along with 17 other oral bacterial strains. Thirty-seven clinical P. gingivalis isolates were also included in order to verify the specificity of the primers. The clinical samples were then subjected to PCR with the same primers to detect the P. gingivalis. All of the samples haboring this bacterium were further analyzed for genotyping. The inward primers amplified the 693-bp fragment of IS1126 (Fig. 1).
FIG. 1.
IS1126-specific PCR primers for P. gingivalis. The inward primer pair (PI1 and PI2) was used to amplify the IS1126 fragment of 693 bp, and the outward primer pair (PI1RC and PI2RC) was used to amplify the gene fragments between the adjacent IS1126 elements. The nucleotide sequences of the outward primers, PI1RC and PI2RC, are reverse complementary to those of the inward primers (PI1 and PI2, respectively).
For genotyping the P. gingivalis, the following primers, which are reverse complementary to the inward primers, were used (Fig. 1): PI1RC (5′-AGA GAA ATC ACG TCA TAA GCC GGG-3′) and PI2RC (5′-GCA CAA GGG CAC AAA CGC AAC AG-3′).
PCR was carried out in a total volume of 50 μl containing 1× PCR buffer, 2.5 U of Taq polymerase (TaKaRa Shuzo, Shiga, Japan), deoxynucleoside triphosphates (0.2 μM), 0.3 μM concentrations of each primer, and 100 ng of DNA. The amplifications were performed in a thermal cycler (GeneAmp PCR system 9600; Perkin-Elmer). The temperature profile selected involved 30 cycles of denaturation at 94°C for 30 s, annealing at 45°C for 30 s, and extension at 72°C for 1 min, with an initial denaturation at 94°C for 5 min and a final extension at 72°C for 5 min. The PCR products were analyzed on ethidium bromide-stained agarose gels (1.0%) and visualized by using a Las-1000 Plus luminescent image analyzer (Fuji, Tokyo, Japan). The PCR was repeated three times for all samples.
RESULTS
Specificity of IS1126-based PCR
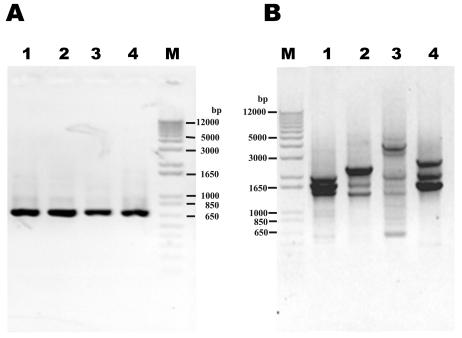
The specificity of the PCR primers for IS1126 was determined by using the different bacteria listed in Table 1. PCR with the IS1126-specific inward primers (PI1 and PI2) detected only the P. gingivalis reference strains and produced a single band with the expected size of ca. 690 bp (Fig. 2A). PCR with the outward primer pair (PI1RC and PI2RC) produced two to seven major bands in all of the P. gingivalis strains (Fig. 2B). No band was observed by PCR with both primer pairs in any of the other species tested. The PCR detection limit was 1 pg of P. gingivalis DNA, corresponding to the genomic equivalents of approximately 100 bacteria (data not shown).
FIG. 2.
PCR products of four laboratory P. gingivalis strains obtained with the primer pairs PI1-PI2 (A) and PI1RC-PI2RC (B). Lanes: 1, HG564; 2, ATCC 33277; 3, A7A1; 4, ATCC 53978; M, DNA size marker.
Genotypes of P. gingivalis.
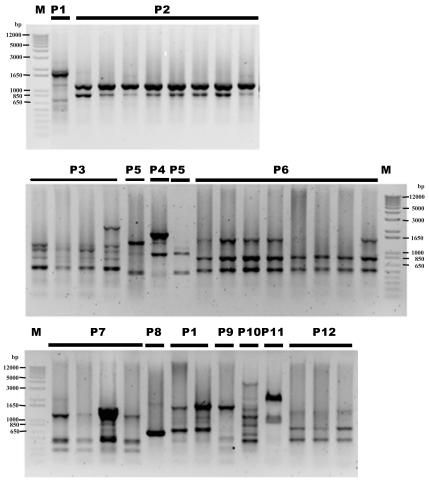
Figure 3 shows the band patterns of 37 clinical isolates cultured from 12 patients with aggressive periodontitis. The isolates showed their own characteristic band patterns, whereby the multiple isolates from the same patient revealed an identical band pattern with slight differences in some cases. Single-band gains or losses were observed in some isolates (Fig. 3, P3, P5, and P6).
FIG. 3.
Genotypes of the 37 P. gingivalis strains cultured from 12 patients with aggressive periodontitis (Table 2) and detected by IS1126-based PCR with the primers PI1RC and PI2RC. The numbers of patients correspond to the numbers indicated in Table 2.
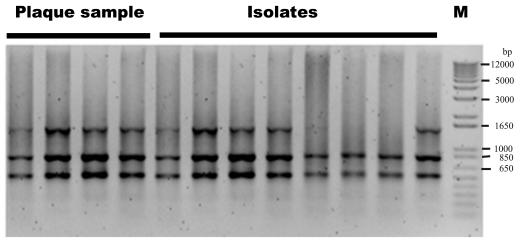
In order to determine whether or not the genotypes of the isolates were the same as those from the plaque samples from which the isolates were cultured, the band pattern of six plaque samples (Table 2) taken from the aggressive periodontitis patients and their 30 isolates (P1 to P3, P6, P7, and P12) were examined. There was no difference in the band pattern between the isolates and the plaque samples when we considered a one-band difference between the isolates from the same plaque samples, as shown in P3, P5, and P6. Figure 4 represents the genotypes of isolates and plaque samples from P6. Three of eight isolates showed one missing band compared to their plaque sample (P6#11). The plaque samples taken from the different tooth sites of the same patient were also analyzed, which revealed an identical band pattern (Fig. 4).
FIG. 4.
Representative electropherogram showing genotypes between the plaque samples from the different tooth sites and their isolates from the same patient (P6 #11 in Table 2). DNA from the plaque samples and the isolates was extracted, and PCR was performed with the primers PI1RC/PI2RC.
Intrafamilial transmission of P. gingivalis.
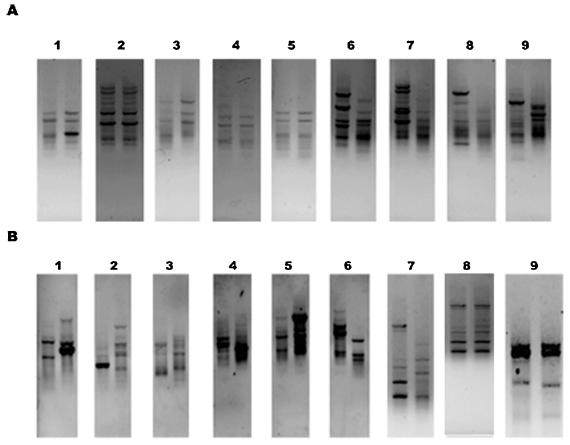
After evaluation of the IS1126-specific primers that were used to identify and characterize P. gingivalis at the strain level by PCR, this method was applied to the transmission study. We examined the frequency of the vertical (14 parent-child pairs) and horizontal (16 spouse pairs) transmission of P. gingivalis. Identical or similar PCR genotypes with one or two band differences in the family members obtained with the outward primers were regarded as being P. gingivalis transmission. Figure 5 shows a comparison of the genotypes between the probands and their family members. The parent-child transmission of P. gingivalis was confirmed in 11 of 14 (78.6%) families, whereas the interspousal transmission of this bacterium occurred in 5 of 16 (31.25%) married couples (Table 3).
FIG. 5.
Comparison of the P. gingivalis genotypes between the probands and their family members. PCR was performed with the primers PI1RC and PI2RC after the DNA was extracted from the subgingival samples. The genotypes of the nine parent-child pairs (A) and the nine spouse pairs (B) are shown. The left-hand lanes of the parent-child pairs (A) are from the parent. Lanes 1 to 6 in panel A and lanes 8 and 9 in panel B were regarded as reflecting an identical genotype between the study pairs.
DISCUSSION
The method for the strain-specific differentiation of P. gingivalis is useful for a transmission study. A rapid and reliable assay with the IS1126-specific primers for PCR was developed here to identify and differentiate the P. gingivalis strains in the plaque samples without a prior cultivation. PCR with the inward primers specific for IS1126 could be used to specifically identify P. gingivalis and to detect the presence of this bacterium in plaque samples. In addition, strain differentiation was possible, with the outward primers, which amplified the DNA fragments between the adjacent IS1126 genes in the P. gingivalis genome. The utility of this method was confirmed by comparing the genotypes of the isolates with those of the plaque samples from which the isolates were cultured.
Several methods have been used for molecular fingerprinting and typing of P. gingivalis strains. These methods include restriction endonuclease analysis (10, 27, 31), restriction fragment length polymorphism (RFLP) (8), ribotyping (27, 31), AP-PCR (1, 27, 31), multilocus enzyme electrophoresis (19), heteroduplex analysis (26), and fimA genotyping (23). The genotypic characterization of the P. gingivalis strains has revealed extensive heterogeneity. Most of these methods require a prior cultivation of the bacteria for analysis, and AP-PCR must be optimized by screening various primer sets.
IS1126 was reported in multicopies throughout the P. gingivalis genome. In the RFLP typing of the laboratory strains of P. gingivalis with IS1126 as a probe, the chromosomal DNA digests with PstI produced 25 to 35 hybridizing bands (8). In this study, IS1126 RFLP showed the strain-specific band patterns. The IS1126 RFLP of P. gingivalis has been suggested as a useful tool for strain typing, as well as an epidemiological study (8). IS fingerprint analysis was also performed for strain differentiation in the other bacteria, including Mycobacterium avium and Vibrio cholerae (6, 13). Among the periodontal pathogens, A. actinomycetemcomitans has been reported to harbor the IS element, and 27 clinical isolates of this bacterium were divided into seven groups by RFLP by using the IS200-like sequence (ISAa1) (14).
Since IS is a mobile element that translocates between or in the genome, its stability is an important factor for application to strain differentiation. Dong et al. (8) assessed the relative stability of IS1126 by comparing the RFLP patterns of the copies of the same strain from different laboratories and observed one or two band differences. Meanwhile, a one-band gain and a one-band loss was observed in W50BEI, which is a spontaneous nonpigmented, avirulent mutant, compared to the P. gingivalis strain W50 (8). The mutant had been isolated from the P. gingivalis strain W50 after 49 to 73 generations of growth in a chemostat (22). These results suggest the relative stability of IS1126, but a small change can occur through the transposition or recombination between the IS copies by some factors, particularly the age of a stock culture and the number of passages. In our study, the genotyping of P. gingivalis by PCR by using the IS1126-specific primers was carried out on 4 laboratory strains and 37 clinical isolates cultured from the 12 periodontitis patients. All of the bacteria showed individual characteristic band patterns but, in most cases, the isolates that originated from the same patient showed identical patterns. However, a one-band difference was observed in some isolates from the same patient. The PCR patterns of the isolates and the plaque samples from which the isolates were cultured were also compared. Almost identical patterns were detected. Furthermore, the band pattern between the plaque samples taken from the different sample sites of the same patient was the same. These results show that periodontitis patients appear to harbor the P. gingivalis strains of a single genotype, when they are analyzed by using the IS1126-specific primers, which detected the size and the number of the adjacent IS1126 by PCR. This result made the comparison of genotypes between the plaque samples from family members and thus interpretation of the analysis easier. It has been reported that most infected people harbor only a single genotype of P. gingivalis with detection rates ranging from 61 to 90% (2, 18, 27).
The transmission of the periodontal pathogens has been studied mostly in A. actinomycetemcomitans and P. gingivalis. The intrafamilial transmission of these bacteria may in part explain the familial tendency of periodontitis and may have important prophylactic and treatment implications. The vertical transmission (parent-child transmission) rate of A. actinomycetemcomitans varied from 0 to 60%, and the horizontal transmission rate between siblings or between spouses ranged from 14 to 60% (2, 11, 25, 29). In P. gingivalis, interspousal transmission was detected in 20 to 75% of cases by AP-PCR or a heteroduplex analysis of the ribosomal ISR. However, transmission from parent to child appeared to be uncommon (2, 31). Nevertheless, the finding that concordance in the colonization of P. gingivalis was frequently observed for children and their mothers by detecting the presence of P. gingivalis by PCR assay may implicate the vertical transmission of this bacterium (30, 33). In this context, information on the stability of the colonization with P. gingivalis in children could help in selecting the subjects' age for a vertical transmission study. Lamell et al. (16) reported a positive relationship between increasing age and persistent colonization with P. gingivalis by heteroduplex analysis of ribosomal ISR. According to those results, P. gingivalis appeared to colonize only transiently in children and was more stable in the late teenage years. In our study, the age of the “children” involved in parent-child transmission ranged from 19 to 37 years. Although the sample size was small, these results showed that the majority (11 of 14) of parent-child pairs had an identical P. gingivalis genotype. The high vertical transmission rate of P. gingivalis in our study relied upon the selection of the older children. The transmission rate may be also influenced by the detection methods, the size of the family, their daily habits, and the living conditions, as well as the geographic location and the ethnicity of the study population. Interspousal transmission rate in our study is within the rage of the rate reported in other studies (2, 30).
In summary, IS1126-based PCR of P. gingivalis resulted in the production of two to seven amplicons, which gave unique electrophoretic patterns in each strain. Repeated assays with the same samples revealed reproducible data. This method showed a one-step fingerprinting process from the plaque samples after DNA extraction. A prior culture, cloning, or hybridization was unnecessary. Since most samples harbored a single unique genotype of P. gingivalis, this method is useful particularly for a transmission study for which a highly discriminating method is required. The application of this method to a transmission study with a large sample number will provide more insight into the transmission mode of P. gingivalis.
Acknowledgments
This study was supported by grant 01-PJ5-PG3-20507-0049 from the Ministry of Health and Welfare of Korea.
REFERENCES
- 1.Asikainen, S., C. Chen, and J. Slots. 1996. Likelihood of transmitting Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in families with periodontitis. Oral Microbiol. Immunol. 11:387-394. [DOI] [PubMed] [Google Scholar]
- 2.Asikainen, S. and C, Chen. 1999. Oral ecology and person-to-person transmission of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontol. 2000 20:65-81. [DOI] [PubMed] [Google Scholar]
- 3.Beck, J. D. 1994. Methods of assessing risk for periodontitis and developing multifactorial models. J. Periodontol. 65:468-478. [DOI] [PubMed] [Google Scholar]
- 4.Beck, J., R. Garcia, G. Heiss, P. S. Vokonas, and S. Offenbacher. 1996. Periodontal disease and cardiovascular disease. J. Periodontol. 67:1123-1137. [DOI] [PubMed] [Google Scholar]
- 5.Beck, J. D., G. Slade, and S. Offenbacher. 2000. Oral disease, cardiovascular disease and systemic inflammation. Periodontol. 2000 23:110-120. [DOI] [PubMed] [Google Scholar]
- 6.Bik, E. M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, B.-K., S. H. Park, Y. J. Yoo, S. H. Choi, J. K. Chai, K. S. Cho, and C. K. Kim. 2000. Detection of major putative periodontopathogens in Korean advanced adult periodontitis patients using a nucleic acid-based approach. J. Periodontol. 71:1387-1394. [DOI] [PubMed] [Google Scholar]
- 8.Dong, H., T. Chen, F. E. Dewhirst, R. D. Fleischmann, C. M. Fraser, and M. J. Duncan. 1999. Genomic loci of the Porphyromonas gingivalis insertion element IS1126. Infect. Immun. 67:3416-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genco, C. A., C. W. Cutler, D. Kapczynski, K. Maloney, and R. R. Arnold. 1991. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genco, R. J., and B. G. Loos. 1991. The use of genomic DNA fingerprinting in studies of the epidemiology of bacteria in periododntitis. J. Clin. Periodontol. 18:396-405. [DOI] [PubMed] [Google Scholar]
- 11.Greenstein, G., and I. Lamster. 1997. Bacterial transmission in periodontal diseases: a critical review. J. Periodontol. 68:421-431. [DOI] [PubMed] [Google Scholar]
- 12.Griffen, A. L., M. R. Becker, S. R. Lyons, M. L. Moeschberger, and E. J. Leys. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 36:3239-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrero, C., C. Bernasconi, D. Burki, D. T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashida, H., H. Hotokezaka, N. Ohara, T. Koseki, T. Nishihara, O. Takagi, and T. Yamada. 1998. Differentiation of clinical isolates of Actinobacillus actinomycetemcomitans using an insertion sequence, ISAa1. Oral Microbiol. Immunol. 13:120-123. [DOI] [PubMed] [Google Scholar]
- 15.Laine, M. L., B. J. Appelmelk, and A. J. van Winkelhoff. 1997. Prevalence and distribution of six capsular serotypes of Porphyromonas gingivalis in periodontitis patients. J. Dent. Res. 76:1840-1844. [DOI] [PubMed] [Google Scholar]
- 16.Lamell, C. W., A. L. Griffen, D. L. McClellan, and E. J. Leys. 2000. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J. Clin. Microbiol. 38:1196-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J. W., B.-K. Choi, Y. J. Yoo, S. H. Choi, K. S. Cho, J. K. Choi, and C. K. Kim. 2003. Distribution of periodontal pathogens in Korean aggressive periodontitis. J. Periodontol. 74:1329-1335. [DOI] [PubMed] [Google Scholar]
- 18.Leys, E. J., J. H. Smith, S. R. Lyons, and A. L. Griffen. 1999. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J. Clin. Microbiol. 37:3906-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loos, B. G., D. W. Dyer, T. S. Whittam, and R. K. Selander. 1993. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect. Immun. 61:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maley, J., and S. J. Roberts. 1994. Characterisation of IS1126 from Porphyromonas gingivalis W83: a new member of the IS4 family of insertion sequence elements. FEMS Microbiol. Lett. 123:219-224. [DOI] [PubMed] [Google Scholar]
- 22.McKee, A. S., A. S. McDermid, R. Wait, A. Baskerville, and P. D. Marsh. 1988. Isolation of colonial variants of Bacteroides gingivalis W50 with a reduced virulence. J. Med. Microbiol. 27:59-64. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa, I., A. Amano, R. K. Kimura, T. Nakamura, S. Kawabata, and S. Hamada. 2000. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J. Clin. Microbiol. 38:1909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Offenbacher, S., H. L. Jared, P. G. O'Reilly, S. R. Wells, G. E. Salvi, H. P. Lawrence, S. S. Socransky, and J. D. Beck. 1998. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann. Periodontol. 3:233-250. [DOI] [PubMed] [Google Scholar]
- 25.Preus, H. R., J. J. Zambon, R. G. Dunford, and R. J. Genco. 1994. The distribution and transmission of Actnobacillus actinomycetemcomitans in families with established adult periodontitis. J. Periodontol. 65:2-7. [DOI] [PubMed] [Google Scholar]
- 26.Rumpf, R. W., A. L. Griffen, and E. J. Leys. 2000. Phylogeny of Porphyromonas gingivalis by ribosomal intergenic spacer region analysis. J. Clin. Microbiol. 38:1807-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teanpaisan, R., and C. W. Douglas. 1999. Molecular fingerprinting of Porphyromonas gingivalis by PCR of repetitive extragenic palindromic (REP) sequences and comparison with other fingerprinting methods. J. Med. Microbiol. 48:741-749. [DOI] [PubMed] [Google Scholar]
- 28.Thorisdottir, A. S., L. L. Carias, and S. H. Marshall. 1994. IS6770, an enterococcal insertion-like sequence useful for determining the clonal relationship of clinical enterococcal isolates. J. Infect. Dis. 170:1539-1548. [DOI] [PubMed] [Google Scholar]
- 29.Tinoco, E. M., M. Sivakumar, and H. R. Preus. 1998. The distribution and transmission of Actinobacillus actinomycetemcomitans in families with localized juvenile periodontitis. J. Clin. Periodontol. 25:99-105. [DOI] [PubMed] [Google Scholar]
- 30.Tuite-McDonnell, M., A. L. Griffen, M. L. Moeschberger, R. E. Dalton, P. A. Fuerst, and E. J. Leys. 1997. Concordance of Porphyromonas gingivalis colonization in families. J. Clin. Microbiol. 35:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Steenbergen, T. J., C. Menard, C. J. Tijhof, C. Mouton, and J. De Graaff. 1993. Comparison of three molecular typing methods in studies of transmission of Porphyromonas gingivalis. Med. Microbiol. 39:416-421. [DOI] [PubMed] [Google Scholar]
- 32.Van Winkelhoff, A. J., M. L. Laine, M. F. Timmerman, G. A. Van der Weijden, F. Abbas, E. G. Winkel, E. M. Arief, and U. Van der Velden. 1999. Prevalence and serotyping of Porphyromonas gingivalis in an Indonesian population. J. Clin. Periodontol. 26:301-305. [DOI] [PubMed] [Google Scholar]
- 33.Yang, E. Y., A. C. Tanner, P. Milgrom, S. A. Mokeem, C. A. Riedy, A. T. Spadafora, R. C. Page, and J. Bruss. 2002. Periodontal pathogen detection in gingiva/tooth and tongue flora samples from 18- to 48-month-old children and periodontal status of their mothers. Oral Microbiol. Immunol. 17:55-59. [DOI] [PubMed] [Google Scholar]