Abstract
RNA viruses evolve rapidly. One source of this ability to rapidly change is the apparently high mutation frequency in RNA virus populations. A high mutation frequency is a central tenet of the quasispecies theory. A corollary of the quasispecies theory postulates that, given their high mutation frequency, animal RNA viruses may be susceptible to error catastrophe, where they undergo a sharp drop in viability after a modest increase in mutation frequency. We recently showed that the important broad-spectrum antiviral drug ribavirin (currently used to treat hepatitis C virus infections, among others) is an RNA virus mutagen, and we proposed that ribavirin's antiviral effect is by forcing RNA viruses into error catastrophe. However, a direct demonstration of error catastrophe has not been made for ribavirin or any RNA virus mutagen. Here we describe a direct demonstration of error catastrophe by using ribavirin as the mutagen and poliovirus as a model RNA virus. We demonstrate that ribavirin's antiviral activity is exerted directly through lethal mutagenesis of the viral genetic material. A 99.3% loss in viral genome infectivity is observed after a single round of virus infection in ribavirin concentrations sufficient to cause a 9.7-fold increase in mutagenesis. Compiling data on both the mutation levels and the specific infectivities of poliovirus genomes produced in the presence of ribavirin, we have constructed a graph of error catastrophe showing that normal poliovirus indeed exists at the edge of viability. These data suggest that RNA virus mutagens may represent a promising new class of antiviral drugs.
The rapid evolution of RNA viruses is apparently powered by the high mutation frequency in RNA virus populations (1–6). The high mutation frequency of animal RNA viruses has been primarily inferred from genetic markers, and such assays may not be accurate enough to establish the exact mutation frequencies. Here we report data defining the high mutation frequency of poliovirus, a model RNA virus, and then we proceed to examine the implications of that high mutation frequency regarding mutagenic antiviral drug interventions that might exploit the high mutation frequency to destroy the virus.
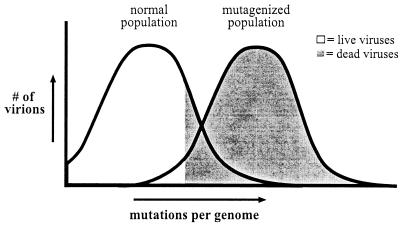
The quasispecies theory states that an RNA virus population does not consist of a single “wild-type” genotype but instead is an ensemble of related genotypes (7–11). This quasispecies is then capable of very rapid evolution in new environments because of the large number of potentially beneficial mutations already present within the population. Maintaining such a high mutation frequency, however, is dangerous for the virus. There is an intrinsic limit to the maximum variability of viral genetic information before it loses meaning (7, 12), and if an RNA virus quasispecies goes beyond that mutation limit, the population will no longer be viable. The phenomenon that occurs when the loss of genetic fidelity results in a lethal accumulation of errors has been termed “error catastrophe” (Fig. 1) (3, 7). Most cellular organisms have evolved a number of sophisticated processes to maintain their genetic information with high fidelity and stay far away from the threshold of error catastrophe. In contrast, it has been predicted that RNA viruses with high mutation frequencies exist close to the edge of error catastrophe and can be forced into error catastrophe by a moderate increase in mutation rate. Several indirect lines of evidence have been presented that support the existence of such an error catastrophe (13–17).
Figure 1.
Model of error catastrophe. The majority of viruses in a normal picornavirus population are viable (51). But a small increase in mutation frequency is predicted to push the virus population into error catastrophe (the mutagenized population, Right), where the number of errors per viral genome is sufficiently high to lethally mutate a majority of the virus population. White indicates live virus, gray indicates dead virus. Most animal RNA virus genomes are ≈10 kb long.
Our recent study demonstrated that the important broad-spectrum antiviral drug ribavirin [currently used clinically to treat hepatitis C virus (18, 19), respiratory syncytial virus (20), and lassa fever virus infections (21)] is an RNA virus mutagen (15). In that study, we observed a direct correlation between the mutagenic effect of ribavirin and the reduction of poliovirus titers, suggesting that ribavirin exerts its antiviral activity by lethally mutagenizing the virus population. However, a direct demonstration of error catastrophe/lethal mutagenesis has not been made for ribavirin or any RNA virus mutagen. That is to say, no study has provided direct proof that the antiviral effect of any drug is exerted via its mutagenic effects on the viral genetic material and not via secondary effects of the drug on cellular physiology or viability, or via inhibition of other aspects of the virus life cycle.
Here we demonstrate that ribavirin's antiviral activity is exerted through lethal mutagenesis, specifically destroying the infectivity of the poliovirus genomic RNA. We also describe direct evidence that the error catastrophe theory applies to poliovirus. Compiling data from experiments that use increasing levels of ribavirin mutagenesis, we are able to establish the relationship between mutation frequency and virus viability, showing a strikingly precipitous drop in virus viability at mutation levels only slightly higher than normal.
Materials and Methods
Cells and Viruses.
Human HeLa cells were propagated as previously described (15). All viral infections were performed by using a poliovirus stock grown from a plasmid clone-derived Mahoney strain poliovirus (pXpA). Inhibition of viral growth by ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide) was done in 3–12 × 106 HeLa cells in OptiMEM + 2% dialyzed FCS. Cells were pretreated with high-grade ribavirin for 15 h (kindly provided by Z. Hong, Schering–Plough) before infection, as previously described (15). Note that ribavirin obtained from Sigma is not sufficiently pure for in vivo experiments, as it causes significant toxicity at high concentrations (data not shown). Infections were done at a multiplicity of infection of 1–2.
RNA and virus were harvested at the time point of maximum RNA accumulation (6 h postinfection for 0 and 100 μM ribavirin and 10 h postinfection for 400 μM and 1,000 μM ribavirin). At time of harvest, cells and supernatant for each condition were divided into two equal samples, one half being the virus sample and the other half for RNA extraction. Virus samples were frozen for later analysis. PolyA+ RNA samples containing the viral genomes were collected from cytoplasmic lysates by using oligo dT25 DynaBeads (Dynal A.S., Oslo). RNA and virus stocks from a single series of harvests were used in the experiments reported in the presence of ribavirin, for internal consistency. Similar data were obtained by using additional RNA and virus stocks generated in the presence of ribavirin on additional days, although sequencing of those stocks was not performed (data not shown).
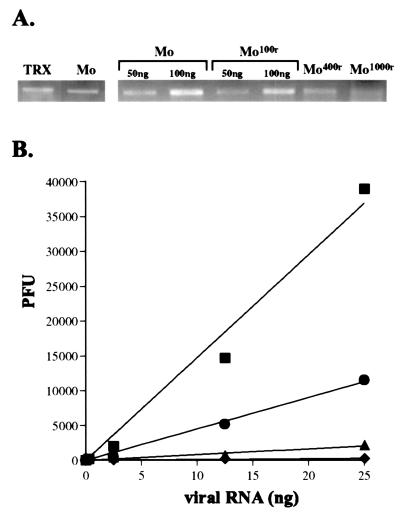
Quantification of reduction in total poliovirus RNA at each condition (Fig. 2A; Table 2) was normalized to the number of HeLa cells harvested in each condition [previously published experiments normalized to 18S rRNA (15)]. RNA was quantified three separate times, and the average is reported in Table 2. Total poliovirus titers (Table 2) were determined by plaque assay, as previously described (22). RNA infectivity (Figs. 3 and 4A) was determined by infectious center plaque assay, where the amount of RNA indicated at each data point (Figs. 2B and 4A) was electroporated into 1.2 × 106 HeLa cells. Electroporated cells were serially diluted, and each series of dilutions was added to a series of 10-cm plates of 50% confluent HeLa cells, allowed to adhere for 2 h, then overlaid with medium/agar and incubated at 37°C, 7% CO2 for 48 h before staining the plates and counting plaques, by using techniques previously described (23). A series of control experiments were done to confirm that plaque formation after cell electroporation was linear with cell dilution, RNA concentration, and RNA infectivity (data not shown).
Figure 2.
Direct antiviral effect of ribavirin on the viral genetic material. (A) Total poliovirus genomic RNA accumulation in infected cells, with and without ribavirin. On the Left, normal poliovirus genomic RNA (Mo) isolated from infected cells is normalized to a known quantity (100 ng) of in vitro-generated poliovirus genomes (TRX). On the Right, quantities of poliovirus genomic RNA isolated from infected cells in 100 μM ribavirin (Mo100r), 400 μM ribavirin (Mo400r), and 1,000 μM ribavirin (Mo1000r), are normalized to amounts of poliovirus genomic RNA from infected untreated cells (Mo). Comparisons are tabulated in Table 2. (B) Large reductions in specific infectivity of ribavirin mutagenized RNA virus genomes. Genomic poliovirus RNA from (■) untreated cells, (●) 100 μM ribavirin-treated cells, (▴) 400 μM ribavirin-treated cells, and (♦) 1,000 μM ribavirin-treated cells was tested for specific infectivity in a series of infectious center assays. Data are shown with a linear curve fit for each series. Assays were performed multiple times; data from a representative experiment is shown.
Table 2.
The antiviral effects of ribavirin can be directly attributed to lethal mutagenesis
| Normal | 100 μM ribavirin | 400 μM ribavirin | 1,000 μM ribavirin | |
|---|---|---|---|---|
| RNA-specific infectivity loss | — | 3.3 | 18 | 140 |
| Loss of total viral RNA | — | — | 6 | 16 |
| Total predicted titer reduction | 1 | 3.3 | 100 | 2,200 |
| Actual titer reduction* | 1 | 3.2 | 71 | 2,000 |
Untreated (“normal”) poliovirus titer in this experiment was 1.2 × 1010 PFU per plate of HeLa cells (6 × 106 cells). Data are the average of three experiments.
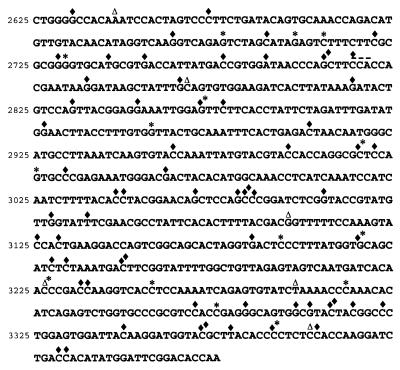
Figure 3.
Distribution of mutations found in the VP1 capsid gene without ribavirin (Δ), with 400 μM ribavirin mutagenesis (*), and 1,000 μM ribavirin mutagenesis (♦). Note that there are no particular mutational hotspots. Almost all mutations at Cs were C→U and mutations at Gs were G→A. Also note that in the presence of 400 μM ribavirin an unusual 3 nt deletion (indicated by —) was detected at a CCA sequence.
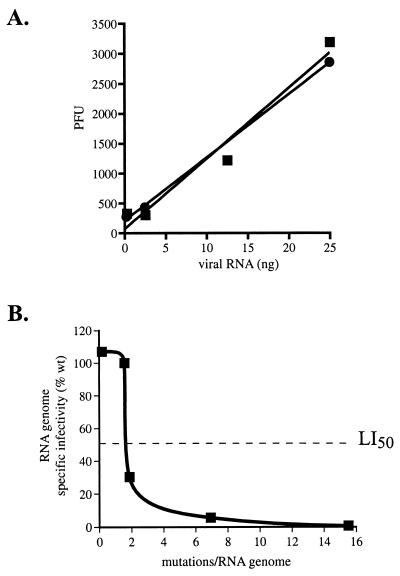
Figure 4.
(A) Specific infectivity of T7 transcribed poliovirus genomes compared with natural poliovirus genomes. This illustrates that between 0.21 and 2.1 mutations per 10,000 nt (Table 1), there is no significant detrimental effect on the viability (specific infectivity) of poliovirus genomes. In vitro-transcribed RNA (■); natural poliovirus RNA (●). Average data set is shown. (B) Relationship of mutation frequency to genomic RNA infectivity. Specific infectivity of normal poliovirus RNA was set to 100%. The graph shows that poliovirus populations exist near the edge of error catastrophe, as there is a rapid decline in RNA genome infectivity at levels of mutagenesis only slightly higher than normal. The LI50 (50% loss of specific infectivity) is defined as the mutation frequency at which 50% of the viral genomes are lethally mutated, indicated by the dashed line. Wild-type (wt or Mo) poliovirus genomes contain an average ≈1.5 mutations/genome, based on data from Table 1. Poliovirus genomes from cells treated with 100 μM ribavirin (Table 3) contain an average ≈1.9 mutations/genome. Poliovirus genomes from cells treated with 400 μM ribavirin (Table 3) contain an average ≈6.9 mutations/genome. Poliovirus genomes from cells treated with 1,000 μM ribavirin (Table 3) contain an average ≈15.5 mutations/genome.
Molecular Biology.
In vitro-transcribed poliovirus was generated by using T7 RNA polymerase, as previously described (23), on a pMoRA template [pXpA modified by a 5′ ribozyme and long 3′ polyA (24)]. Plasmid DNA was destroyed by DNaseI (Ambion, Austin, TX) treatment at 37 for 30′, and RNA was then precipitated with 5 M LiCl to eliminate any possible DNA contamination. Purity of all RNA samples (either in vitro transcribed or isolated from polio-infected cells) was confirmed by a negative PCR for poliovirus capsid DNA (data not shown). Oligo dT primed cDNA was synthesized from 0.4 μg RNA from the appropriate source (T7 transcripts, normal viral RNA, or ribavirin-treated RNA) by using Superscript II (Life Technologies, Grand Island, NY), and VP1-coding sequence was PCR amplified from 1/10th of the cDNA by using high fidelity PfuTurbo polymerase (Stratagene) in a 25-cycle reaction. The VP1 gene was cloned, and plasmid DNA was prepared from independent bacterial colonies. All DNA sequencing (Tables 1 and 3) was done from position 2625–3400 of the poliovirus genome by using BigDye terminator cycle sequencing (Applied Biosystems) and then analyzed with dnastar SeqManII (15). Sequence analysis of 23 (approximately half) of the normal poliovirus clones was published previously (15). Statistical analysis of significance (P values) of sequence data was done by Student's t test. Plus or minus ranges indicated for sequence data are estimated at double (0.41 errors/104 nt) the frequency of reverse transcription–PCR (RT-PCR)-introduced errors determined in Results (0.21 errors/104 nt).
Table 1.
Mutation frequency in a normal RNA virus population
| Transitions | Transversions | Total mutation frequency* | |
|---|---|---|---|
| RT-PCR background control | 0 | 0.21 | 0.21 |
| Normal poliovirus population | 1.88 | 0.24 | 2.12 |
Mutations per 10,000 nt sequenced.
Table 3.
Mutation frequency in ribavirin-treated RNA virus populations
| G→A | C→T | Total mutation frequency* | |
|---|---|---|---|
| Normal population | 0.5 | 1.2 | 2.1 |
| 100 μM ribavirin | — | 1.3 | 2.5 |
| 400 μM ribavirin | 4.4 | 5.0 | 9.3 |
| 1,000 μM ribavirin | 6.8 | 12.0 | 20.8 |
Mutations per 10,000 nt sequenced.
Results
Mutation Frequencies in Normal Poliovirus Populations and in Vitro-Generated Genomes.
One source of the impressive ability of RNA viruses to evolve is the apparently high mutation frequency in RNA virus populations. However, it has been argued that the available data do not accurately establish the mutation frequencies of animal RNA viruses, and the true mutation frequencies may be as low as 10−6 (25, 26). Most estimates of high RNA virus mutation frequencies have been based on genetic assays (1, 6, 27–34), which could give inaccurate estimates if the phenotypes are not completely tight. For example, the mutation frequency of poliovirus has been estimated at 2.1 × 10−4 by scoring for the presence of a single nucleotide mutation that confers a loss of dependence on guanidine (1). In that poliovirus strain, guanidine is necessary for poliovirus replication, but if that block is not absolute and the virus is capable of synthesizing ≈100 new genomes in the absence of guanidine (instead of the normal 100,000 copies), the genetic marker would be overestimated in the population by 100× and the true mutation frequency would be 2 × 10−6, not 2 × 10−4. Therefore, direct molecular evidence (sequencing data) is necessary to precisely determine the mutation frequency.
As the mutation frequency is a basic parameter of RNA virus genetics, it is important to clearly establish this parameter. Therefore, we set out to determine the mutation frequency in a poliovirus population by analyzing capsid gene sequences from a group of poliovirus clones. Poliovirus RNA was isolated from infected cells, and 55 independent poliovirus cDNA clones were obtained. Those poliovirus cDNAs were generated from viral RNA by RT-PCR. As it was important to determine the number of mutations introduced during the RT-PCR reactions, we first performed a background control experiment where we sequenced the capsid VP1 genes of 58 cDNA clones generated by RT-PCR reactions by using as templates in vitro generated poliovirus genomes transcribed from a plasmid. We sequenced a total of 48,000 nt from the background control clones and observed an RT-PCR mutation frequency of 0.21 per 10,000 nt (Table 1). Then we analyzed the capsid VP1 gene sequences derived from the 55 independent normal poliovirus clones. We sequenced a total of 42,000 nt of clones from a normal poliovirus population and observed a mutation frequency of 2.1 per 10,000 nt (Table 1), well above the background level (P < 0.006).
Direct Antiviral Effect of Mutagenesis.
Having established the high mutation frequency of a normal poliovirus population, we set out to test whether the virus exists near the threshold of error catastrophe. Several studies have shown a correlation between increased concentration of mutagen and loss of viral titer (13, 15–17) but not a direct demonstration that the antiviral effect was exerted via the mutagenesis of the viral RNA genetic material. The loss of titer could be because of inhibition of other virus processes (i.e., translation or RNA packaging) or because of secondary effects on cellular physiology/viability.
We recently demonstrated that ribavirin is an RNA virus mutagen (15). Here we have now carried out experiments designed to prove that lethal mutagenesis is the mechanism of action of ribavirin. We infected cells with poliovirus in the presence or absence of ribavirin. At the time point of maximum viral RNA replication, we measured viral titer by plaque assay and also isolated viral RNA. The amount of infectious virus produced was reduced 3.2-fold by 100 μM ribavirin, 71-fold by 400 μM ribavirin, and 2,000-fold by 1,000 μM ribavirin. Additionally, we quantified the amount of viral RNA isolated from infected cells in each condition by spectrophotometry and gel electrophoresis of purified viral RNA (Fig. 2A).
The specific infectivity of the poliovirus genomic RNA was then determined for each condition by transfection of isolated poliovirus genomic RNA into HeLa cells. Specific infectivity of genomic viral RNA is a direct measure of genome viability. Natural poliovirus genomic RNA had a specific infectivity of 1.5 × 106 plaque-forming units (PFU) per microgram genomes (Fig. 2B). Poliovirus RNA from cells treated with 100 μM ribavirin had a specific infectivity of 4.6 × 105 PFU/microgram genomes, a 3.3-fold reduction from wild-type levels. Poliovirus RNA from cells treated with 400 μM ribavirin had a specific infectivity of 8.4 × 104 PFU/microgram genomes, an 18-fold reduction from wild-type levels. Poliovirus RNA from cells treated with 1,000 μM ribavirin had a specific infectivity of 1.1 × 104 PFU/microgram genomes, a 140-fold reduction from wild-type levels, indicating that fewer than 1% of the viral genomes produced in the presence of 1,000 μM ribavirin were viable. These data demonstrate that treatment with ribavirin has a potent direct effect on the viral genetic material.
Strikingly, the full antiviral effect of ribavirin can be attributed to lethal mutagenesis of the viral genetic material. In the presence of 100 μM ribavirin, there was a 3.3-fold reduction in genome viability (Fig. 2B, Table 2), which can fully account for the 3.2-fold inhibition of infectious poliovirus titer (Table 2). In the presence of 400 μM ribavirin, there was an 18-fold reduction in genome viability (Fig. 2B, Table 2). Additionally, there was a 6-fold reduction in total genomic RNA (Fig. 2A, Table 2), which was likely due to the inactivation of many replicating viral genomes in the ribavirin-treated cells during the multiple rounds of replication and mutagenesis occurring in a single infectious cycle. The combined effects of the mutagen on loss in genome viability (18-fold) and reduction in genomic RNA production (6-fold) would result in an anticipated total reduction in infectious virus titer of ≈100-fold, which indeed accounts for the full loss of titer observed (71-fold; Table 2). In the presence of 1,000 μM ribavirin, there was a 140-fold reduction in genome viability (Fig. 2B; Table 2). Also there was a 16-fold reduction in total genomic RNA (Fig. 2A; Table 2), which again was likely due to the inactivation of many replicating viral genomes in the ribavirin-treated cells during the infectious cycle. The combined total of the loss in genome viability (140-fold) and the reduction in genomic RNA (16-fold) would result in an anticipated total reduction in infectious virus titer of ≈2,200-fold because of the direct effects of mutagenesis, which can account for the full loss of titer observed (2,000-fold; Table 2).
Poliovirus Replicates at the Edge of Error Catastrophe.
DNA copies of viral RNA from each mutagen condition were sequenced to determine the increase in mutation frequency caused by ribavirin. The mutation frequency was increased 1.2-fold (±0.2) in the presence of 100 μM ribavirin, 4.4-fold (±0.2) in the presence of 400 μM ribavirin, and 9.7-fold (±0.1) in the presence of 1,000 μM ribavirin (Table 3). A wide range of G→A and C→T mutations were observed in the region sequenced, indicating that the clones were obtained independently (Fig. 3). In addition, G→A mutations are predicted to be induced by incorporation of ribavirin triphosphate as a GTP nucleoside analog during positive strand RNA synthesis, whereas C→T mutations are predicted to be the result of ribavirin incorporation during negative strand synthesis (15).
Plotting the data obtained above as genome infectivity versus the average number of errors per poliovirus genome would provide a graph of error catastrophe. To construct this graph, we first determined the effect of a lower than normal mutation frequency on poliovirus specific infectivity. The specific infectivity of in vitro synthesized T7 genomic poliovirus transcripts, which have low (0.2 × 10−4/nt) mutation levels, is comparable to that of natural poliovirus RNA (Fig. 4A). With the inclusion of that data, we then constructed a graph of error catastrophe based on our experimental observations (Fig. 4B). The graph illustrates several interesting features of the relationship between mutation levels and virus RNA genome viability. First, there is no significant detrimental effect on the viability of poliovirus genomes at the normal mutation frequency (≈10% in this particular experiment). Therefore, the poliovirus genetic information is flexible enough to absorb up to an average of ≈1.5 mutations per genome without significant loss of function. Strikingly, a small increase in mutation frequency above the normal levels results in a large decline in viral infectivity (Fig. 4B). The LI50 (50% loss of infectivity), defined as the mutation frequency at which 50% of the viral genomes are lethally mutated, of poliovirus is ≈2.0 mutations per genome, less than 2-fold higher than the natural mutation frequency. Furthermore, 95% of the genomes are lethally mutated when there is a 4-fold increase in mutation frequency. Thus, we conclude that the virus has evolved to exist near the edge of error catastrophe.
Discussion
The error catastrophe theory has existed as a corollary of the RNA virus quasispecies theory for over 20 years, and several groups have previously published intriguing evidence that error catastrophe can occur in RNA virus populations (13–17). By using a potent RNA virus mutagen, ribavirin, here we have provided molecular evidence that poliovirus mutation frequencies are high (≈2.1 × 10−4), and we have described a direct molecular demonstration that error catastrophe occurs and can be used as an effective antiviral strategy.
Mutation Frequencies.
A variety of genetic markers have been used to estimate the mutation frequencies of different RNA viruses. Given that the mutation frequency is a basic parameter of RNA virus genetics, it was valuable to firmly establish the mutation frequency of a model RNA virus, poliovirus, at the molecular level. We therefore carried out extensive sequence analysis of natural poliovirus genomes and background control in vitro-generated genomes, and we have established that the mutation frequency is quite high, ≈2.1 × 10−4. Using a different virus (FMDV), Domingo's group has recently published similar sequencing data (17), with an estimated mutation frequency of 2.8–5.9 × 10−4. Although in that paper the frequency of reverse transcription and PCR errors was not determined, because they used experimental techniques similar to those used in this paper, the RT-PCR background control data presented here (Table 1) can probably be considered a reasonably accurate estimate of the RT-PCR induced mutations expected to be present in their FMDV study. Therefore the FMDV mutation frequencies published by Domingo's group should be an accurate representation of the true mutation frequency. In both cases, the ability to sustain a 2.1–5.9 × 10−4 mutation frequency in virus populations of greater than 109 PFU/ml clearly depends on viral genomes that have evolved to consist of highly flexible genetic information.
Parvin et al. published data in 1986 that are in apparent contradiction to the poliovirus mutation frequency data we present here, as they stated that they observed no mutations of 95,000 nt sequenced from 105 poliovirus VP1 gene clones (25). However, Parvin et al. stated in their methods section that, by using the traditional Sanger dideoxy DNA sequencing technique, they were uncertain about mutations at 320 sites in the poliovirus clones (25). Even if only 10% of those 320 “uncertain” sites were true mutations, the data of Parvin et al. would be very similar to ours, and therefore we suggest that any apparent discrepancy between the two studies can likely be dismissed by simply accounting for our use of the more accurate fluorescent dye sequencing techniques now available.
Our 2.1 × 10−4 mutation frequency estimation by sequence analysis corroborates much of the available genetic marker-based literature on animal RNA virus mutation frequencies (3, 8). Therefore, genetic markers can be considered reasonable tools for estimating RNA virus mutation frequencies. In our study, the majority of natural mutations observed were transition mutations (C→U or G→A, Table 1). Genetic marker data are consistent with this observation, as genetic markers scoring for specific transitions were observed at high frequencies in several independent studies [3 × 10−5 (15), 2 × 10−4 (1), 2 × 10−5 (32)], whereas a defined transversion event (A→C) was rarely detected in another study (26).
Error Catastrophe.
Ribavirin is an antiviral drug that is currently in widespread clinical use to treat hepatitis C, respiratory syncytial virus, and lassa fever virus infections (18–21). The mechanism of action of ribavirin has been unclear since its discovery almost 30 years ago (20, 35–39). We previously proposed that ribavirin's antiviral activity is exerted through its potent RNA virus mutagenesis, as a nucleoside analog that becomes incorporated into newly synthesized genomes by the viral RNA-dependent RNA polymerase (15). Here we provide molecular proof that ribavirin exerts its antiviral action directly on poliovirus's genetic material, lethally mutating the viral RNA genome (Fig. 2B). Most strikingly, ribavirin's mutagenic activity can fully account for its antiviral effect against poliovirus (Table 2).
Ribavirin's overall antiviral effect is the result of the combined effect of two components: a loss in specific infectivity of the viral genomes because of lethal mutagenesis, and a loss in total viral genomic RNA produced. We propose that the smaller of the two effects, the reduction in viral RNA accumulation, is also caused by the extensive drug-induced viral mutagenesis, resulting in very few of the viral RNAs produced in the presence of ribavirin remaining viable for replication. Within a single infectious cycle, the viral RNA genomes must go through multiple rounds of replication. Given that fewer than 1% of the total genomes produced in an infected cell are themselves infectious (in the presence of 1,000 μM ribavirin), the limited pool of genomes competent for replication within the infected cell will surely result in a reduction in total RNA genome accumulation, which is indeed seen in our experiments. However, it is possible that some of the loss in total genomic RNA is because of other effects of ribavirin in the cells, such as the 2-fold inhibition of translation (15) or ribavirin's cytostatic effects. Nevertheless, the majority of ribavirin's antiviral effect, and possibly all of the antiviral effect, is directly attributable to the lethal mutagenesis of the viral genomes.
Animal RNA viruses have been hypothesized to maintain themselves at the edge of error catastrophe (13). Fig. 4B maps the relationship between mutation frequency and virus genome viability. The graph illustrates that poliovirus appears to have evolved to exist near the edge of error catastrophe, as small increases in mutation frequency above the normal levels results in a striking decline in viral infectivity, with a greater than 95% loss in genome infectivity after a 4-fold increase in mutagenesis. Existing at the edge of error catastrophe is predicted to optimize the evolutionary fitness of the RNA virus quasispecies population by maximizing genetic variation without sacrificing viability (40). The data presented here demonstrate that high genetic variability, a biological property that is normally a major advantage for an RNA virus, can be turned into a weapon against the virus by increasing that mutation rate beyond tolerable levels and causing a genetic meltdown.
Unlike RNA viruses, DNA-based organisms generally have much lower mutation frequencies and do not exist near the error threshold. They appear to be able to absorb 300- to 5,000-fold higher increases in mutation frequencies before significant loss of viability is seen (41, 42), although DNA viruses may be an exception (3, 42).
Implications for Antiviral Strategies.
Ribavirin's activity as a nucleoside analog incorporated by the viral polymerase can explain the surprisingly broad spectrum action of ribavirin against members of almost all RNA virus families under laboratory cell culture conditions (39) (and clinical activity against three virus infections from diverse families), as the one common feature of RNA viruses is they possess an RNA-dependent RNA polymerase. A high mutation frequency and a susceptibility to error catastrophe also appear to be common traits of most RNA viruses (3, 8, 40). The experiments reported here used ribavirin concentrations in the range of 100–1,000 μM, comparable to concentrations observed in clinical settings (15, 36, 43). The effectiveness of ribavirin in vivo as an RNA virus mutagen may depend on the accumulation of ribavirin and ribavirin triphosphate in some tissues (such as liver, or respiratory tract epithelium when administered as an aerosol) but not others. Alternatively, differences in efficacy may be due to different rates of ribavirin incorporation by the RNA-dependent RNA polymerases of different viruses; or ribavirin may have different mechanisms of actions against different viruses (44, 45).
Ribavirin's lethal mutagenesis of poliovirus is probably enhanced by the well-characterized ability of ribavirin monophosphate to inhibit the cellular enzyme inosine monophosphate dehydrogenase (IMPDH) and thereby decrease cellular GTP pools (46–48). The decrease in cellular GTP pools likely increases the frequency of ribavirin incorporation as a mutagenic GTP analog.
Several RNA virus mutagens are known, but they generally exhibit substantial cellular toxicity (as nucleoside analogs presumably incorporated by cellular DNA and RNA polymerases) and are unacceptable for use in humans at the necessary doses (13, 14, 49, 50). Therefore, in the interest of developing new antiviral drugs that potentially exhibit activity against various RNA virus human pathogens, it may be possible to identify mutagenic nucleoside analogs that are highly specific for incorporation by viral RNA-dependent RNA polymerases. Such a drug development strategy should be plausible given the success in identifying and developing nucleoside analog anti-HIV therapies that are capable of specifically inhibiting that virus's RNA-dependent DNA polymerase.
Acknowledgments
We thank Judith Frydman and Don Ganem for helpful advice and comments. This work was supported by National Institutes of Health (NIH) Grant AI40085 (to R.A.) and NIH grants CA75118 and AI45818 (to C.E.C.). S.C. is a Howard Hughes Medical Institute doctoral fellow.
Abbreviations
- PFU
plaque-forming unit
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.de la Torre J C, Wimmer E, Holland J J. J Virol. 1990;64:664–671. doi: 10.1128/jvi.64.2.664-671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo E, Holland J J, Ahlquist P. RNA Genetics. Boca Raton, FL: CRC; 1988. [Google Scholar]
- 3.Domingo E, Holland J J. In: The Evolutionary Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 161–184. [Google Scholar]
- 4.Domingo E. Virology. 2000;270:251–253. doi: 10.1006/viro.2000.0320. [DOI] [PubMed] [Google Scholar]
- 5.Domingo E, Sabo D, Taniguchi T, Weissmann C. Cell. 1978;13:735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- 6.Eggers H J, Tamm I. Science. 1965;148:97–98. doi: 10.1126/science.148.3666.97. [DOI] [PubMed] [Google Scholar]
- 7.Eigen M, Biebricher C. In: RNA Genetics. Domingo E, Holland J J, Ahlquist P, editors. Boca Raton, FL: CRC; 1988. pp. 211–245. [Google Scholar]
- 8.Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 9.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 10.Holland J J, De La Torre J C, Steinhauer D A. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 11.Domingo E, Holland J J, Biebricher C, Eigen M. In: Molecular Basis of Virus Evolution. Gibbs A J, Calisher C H, Garcia-Arenal F, editors. Cambridge, U.K.: Cambridge Univ. Press; 1995. pp. 181–191. [Google Scholar]
- 12.Eigen M. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 13.Holland J J, Domingo E, de la Torre J C, Steinhauer D A. J Virol. 1990;64:3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C H, Gilbertson D L, Novella I S, Huerta R, Domingo E, Holland J J. J Virol. 1997;71:3636–3640. doi: 10.1128/jvi.71.5.3636-3640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotty S, Maag D, Arnold J J, Zhong W, Lau J Y, Hong Z, Andino R, Cameron C E. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 16.Loeb L A, Essigmann J M, Kazazi F, Zhang J, Rose K D, Mullins J I. Proc Natl Acad Sci USA. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sierra S, Davila M, Lowenstein P R, Domingo E. J Virol. 2000;74:8316–8323. doi: 10.1128/jvi.74.18.8316-8323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 19.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 20.Wyde P R. Antiviral Res. 1998;39:63–79. doi: 10.1016/s0166-3542(98)00029-1. [DOI] [PubMed] [Google Scholar]
- 21.McCormick J B, King I J, Webb P A, Scribner C L, Craven R B, Johnson K M, Elliott L H, Belmont-Williams R. N Engl J Med. 1986;314:20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- 22.Crotty S, Lohman B L, L, Tang S, Miller C J, Andino R. J Virol. 1999;73:9485–9495. doi: 10.1128/jvi.73.11.9485-9495.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gohara D W, Crotty S, Arnold J J, Yoder J D, Andino R, Cameron C E. J Biol Chem. 2000;275:25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 24.Herold J, Andino R. J Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parvin J D, Moscona A, Pan W T, Leider J M, Palese P. J Virol. 1986;59:377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sedivy J M, Capone J P, RajBhandary U L, Sharp P A. Cell. 1987;50:379–389. doi: 10.1016/0092-8674(87)90492-2. [DOI] [PubMed] [Google Scholar]
- 27.Blondel B, Crainic R, Fichot O, Dufraisse G, Candrea A, Diamond D, Girard M, Horaud F. J Virol. 1986;57:81–90. doi: 10.1128/jvi.57.1.81-90.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lubeck M D, Schulman J L, Palese P. Virology. 1980;102:458–462. doi: 10.1016/0042-6822(80)90114-2. [DOI] [PubMed] [Google Scholar]
- 29.Stec D S, Waddell A, Schmaljohn C S, Cole G A, Schmaljohn A L. J Virol. 1986;57:715–720. doi: 10.1128/jvi.57.3.715-720.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinhauer D A, de la Torre J C, Holland J J. J Virol. 1989;63:2063–2071. doi: 10.1128/jvi.63.5.2063-2071.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holland J J, de la Torre J C, Steinhauer D A, Clarke D, Duarte E, Domingo E. J Virol. 1989;63:5030–5036. doi: 10.1128/jvi.63.12.5030-5036.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de la Torre J C, Giachetti C, Semler B L, Holland J J. Proc Natl Acad Sci USA. 1992;89:2531–2535. doi: 10.1073/pnas.89.7.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minor P D, Schild G C, Bootman J, Evans D M, Ferguson M, Reeve P, Spitz M, Stanway G, Cann A J, Hauptmann R, et al. Nature (London) 1983;301:674–679. doi: 10.1038/301674a0. [DOI] [PubMed] [Google Scholar]
- 34.Sherry B, Mosser A G, Colonno R J, Rueckert R R. J Virol. 1986;57:246–257. doi: 10.1128/jvi.57.1.246-257.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.(2000) AFHS Drug Information (American Society of Hospital Pharmacists, Bethesda, MD).
- 36.Gilbert B E, Knight V. Antimicrob Agents Chemother. 1986;30:201–205. doi: 10.1128/aac.30.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith R A, Kirkpatrick W. Ribavirin, A Broad Spectrum Antiviral Agent. New York: Academic; 1980. [Google Scholar]
- 38.Smith R A, Knight V, Smith J A D. Clinical Applications of Ribavirin. Orlando, FL: Academic; 1984. [Google Scholar]
- 39.Sidwell R W, Huffman J H, Khare G P, Allen L B, Witkowski J T, Robins R K. Science. 1972;177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 40.Eigen M. Gene. 1993;135:37–47. doi: 10.1016/0378-1119(93)90047-7. [DOI] [PubMed] [Google Scholar]
- 41.Cupples C G, Miller J H. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drake J W, Holland J J. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller J P, Kigwana L J, Streeter D G, Robins R K, Simon L N, Roboz J. Ann NY Acad Sci. 1977;284:211–229. doi: 10.1111/j.1749-6632.1977.tb21953.x. [DOI] [PubMed] [Google Scholar]
- 44.Rankin J T, Jr, Eppes S B, Antczak J B, Joklik W K. Virology. 1989;168:147–158. doi: 10.1016/0042-6822(89)90413-3. [DOI] [PubMed] [Google Scholar]
- 45.Wray S K, Gilbert B E, Knight V. Antiviral Res. 1985;5:39–48. doi: 10.1016/0166-3542(85)90013-0. [DOI] [PubMed] [Google Scholar]
- 46.Smith R A, Sidwell R W, Robins R K. Annu Rev Pharmacol Toxicol. 1980;20:259–284. doi: 10.1146/annurev.pa.20.040180.001355. [DOI] [PubMed] [Google Scholar]
- 47.Streeter D G, Witkowski J T, Khare G P, Sidwell R W, Bauer R J, Robins R K, Simon L N. Proc Natl Acad Sci USA. 1973;70:1174–1178. doi: 10.1073/pnas.70.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wray S K, Gilbert B E, Noall M W, Knight V. Antiviral Res. 1985;5:29–37. doi: 10.1016/0166-3542(85)90012-9. [DOI] [PubMed] [Google Scholar]
- 49.Pringle C R. J Virol. 1970;5:559–567. doi: 10.1128/jvi.5.5.559-567.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burge B W, Pfefferkorn E R. Virology. 1966;30:204–213. doi: 10.1016/0042-6822(66)90096-1. [DOI] [PubMed] [Google Scholar]
- 51.Belsham G J, Bostock C J. J Gen Virol. 1988;69:265–274. doi: 10.1099/0022-1317-69-2-265. [DOI] [PubMed] [Google Scholar]