Abstract
The RD2 region of the internalization-associated gene prtF1, which encodes the fibronectin-binding repeat domain type 2 of protein F1, plays a crucial role in the entry of group A streptococci (GAS) into epithelial cells. A molecular study of the variability of the RD2 region was carried out with 77 independent Italian GAS, 66 erythromycin resistant (ER) and 11 erythromycin susceptible (ES), which had previously been investigated for the association between erythromycin resistance and ability to enter human respiratory cells. The amplicons obtained from PCR analysis of the RD2 region were consistent with a number of RD2 repeats ranging from one to five, more frequently four (n = 30), three (n = 27), and one (n = 18). A new method to type cell-invasive GAS (RD2 typing) was developed by combining PCR analysis of the RD2 region and restriction analysis of PCR products with endonucleases HaeIII, DdeI, and HinfI. Overall, 10 RD2 types (a to j) were distinguished (all detected among the 66 ER isolates, four detected among the 11 ES isolates). Comparison and correlation of RD2 typing data with the genotype and phenotype of macrolide resistance and with data from PCR M typing and SmaI macrorestriction analysis allowed us to identify 41 different clones (31 among the 66 ER isolates and 10 among the 11 ES isolates). Three major clones accounted for 40% of the isolates (47% of ER strains). Some ES isolates appeared to be related to ER isolates with identical combinations of RD2 type and emm type. While simultaneous use of different typing methods is essential for a thorough investigation of GAS epidemiology, RD2 typing may be especially helpful in typing cell-invasive GAS.
In the past few years, fresh evidence has suggested that group A streptococci (GAS), traditionally viewed as highly adhesive extracellular pathogens, can in fact be efficiently internalized by and survive within human cells of respiratory-tract origin, albeit with marked differences from one strain to another (3, 16, 19). GAS entry into epithelial cells is mediated by a subclass of adhesins referred to as invasins; among these, a crucial role is played by F1, a high-affinity fibronectin-binding protein (13, 14, 23) encoded by the prtF1 gene, and its allelic variant SfbI (20, 32), encoded by sfbI. F1/SfbI is expressed in approximately 70% of clinical GAS isolates (3).
A prominent feature shared by the high-affinity fibronectin-binding proteins of GAS and the fibronectin-binding proteins of other gram-positive cocci is a structure containing tandem repeats 32 to 50 amino acids long found adjacent to the conserved C-terminal cell attachment domain (8, 25). In particular, the GAS protein F1 contains two fibronectin-binding domains, of which the one located toward the C terminus of the molecule—repeat domain type 2 (RD2)—has been reported to contain five repeats (four complete repeats of 37 amino acids and a fifth incomplete repeat of 32 amino acids toward the C terminus) (28). Accordingly, the RD2 region of prtF1 has been reported to consist of five repeats, four measuring 111 bp and the fifth (at the 3′ end) measuring 96 bp (21, 28). In fact, the number of repeats is variable, ranging at least from one to six (17, 21, 22), but this feature is unrelated to the ability to bind fibronectin (21).
Other surface components of GAS that participate in interactions with eukaryotic cells include the M protein, a major surface antigen and virulence factor of GAS. To date, more than 100 serotypes have been identified based on serological reactivity with the variable N termini of M proteins or, more recently, on analysis of the 5′ sequences of the genes encoding M proteins (emm genes) (1, 4). Different serotypes may recognize different receptors on the surface of eukaryotic cells, and some, like M1 and M6, may function as invasins (3, 4). The presence of prtF1 and ability to bind fibronectin correlate with the M type of various GAS strains (21).
The ability of throat GAS to enter pharyngeal cells in vivo may enable them to avoid host defenses as well as those antibiotics that, like β-lactams, are confined to extracellular fluids. While this may explain the failure of penicillin to cure a number of streptococcal pharyngites (9), it might also favor convalescent and persistent throat carriage of GAS (29). Indeed, the prtF1 gene seems to be prevalent among GAS isolated from asymptomatic carriers (22). Moreover, intracellular GAS might constitute a reservoir of persisting bacteria in vivo with the potential to cause reinfections (24). Thus, special concern has been raised by the recent finding of an unexpected, significant association between erythromycin resistance and ability to enter human respiratory cells among GAS isolated in Italy (6). Strains in which these two features are combined may escape β-lactams because of intracellular location and macrolides—which, unlike β-lactams, enter eukaryotic cells and are active in intracellular compartments—because of resistance, resulting in difficulty of eradication. This may have facilitated the diffusion of erythromycin-resistant (ER) GAS in Italy. Here, GAS resistance to macrolides is widespread—an overall rate of >42% has been reported in a recent nationwide survey (34)—and extensive studies have confirmed the genotypic and phenotypic heterogeneity of ER GAS (11). The methylase gene erm(B) can be associated with constitutive (cMLS phenotype) as well as inducible (iMLS-A phenotype) resistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics. Another methylase gene, erm(A), originally designated erm(TR) (30), is associated with inducible expression of high- or low-level erythromycin resistance (iMLS-B and iMLS-C phenotypes, respectively, differing in the presence in the former of a novel erythromycin efflux system) (10). Finally, the conventional efflux gene, mef(A), is associated with low-level resistance only to 14- and 15-membered macrolides (M phenotype), but is occasionally found in addition to methylase genes in isolates of other phenotypes.
This study was designed to investigate the genetic diversity of the Italian GAS that were found to possess the prtF1 gene in the course of a previous study of the association between erythromycin resistance and human cell invasiveness (6). The present work, which focused on the variability of the RD2 region of prtF1, led to the development of a new method to type cell-invasive GAS based on the combination of PCR analysis of RD2 and restriction enzyme cleavage analysis of PCR products. The results were correlated both with previously investigated features (cell invasion efficiency and genotype and phenotype of macrolide resistance) and with results of two typing methods that we tested herein, PCR M typing with emm-specific oligonucleotide primers and SmaI macrorestriction fragment pattern analysis by pulsed-field gel electrophoresis (PFGE).
MATERIALS AND METHODS
Bacterial strains and early characterization.
Seventy-seven GAS, including 66 ER (erythromycin MIC, ≥1 μg/ml) and 11 erythromycin-susceptible (ES; MIC, ≤0.5 μg/ml) strains, were studied. They were selected from the 126 GAS strains isolated throughout Italy in 1997 to 1998 from children with pharyngitis and investigated for the association between erythromycin resistance and ability to enter human respiratory cells (6). In particular, these 77 GAS (Table 1) are the ones which in the previous study were found to carry the internalization-related gene prtF1. All had already been characterized for erythromycin susceptibility and resistance—including MIC, detection of resistance genes erm(A), erm(B), and mef(A), and attribution to the cMLS, iMLS-A, iMLS-B, iMLS-C, or M phenotype—and scored for respiratory cell invasion efficiency as highly invasive (n = 64) or weakly invasive (n = 13) (6).
TABLE 1.
Distribution of RD2 repeat numbers among the 77 prtF1-positive GAS strains subdivided according to their genotypes and phenotypes of erythromycin resistance
| Genotype/phenotype of erythromycin resistance | No. of strains
|
|||||
|---|---|---|---|---|---|---|
| Total | With indicated number of RD2 repeats:
|
|||||
| 1 | 2 | 3 | 4 | 5 | ||
| erm(B)/cMLS | 8 | 1 | 1 | 3 | 3 | |
| erm(B) mef(A)/cMLS | 2 | 2 | ||||
| erm(B)/iMLS-A | 12 | 12 | ||||
| erm(B) mef(A)/iMLS-A | 1 | 1 | ||||
| erm(A)/iMLS-B | 13 | 13 | ||||
| erm(A)/iMLS-C | 8 | 8 | ||||
| mef(A)/M | 22 | 15 | 1 | 5 | 1 | |
| Totals | ||||||
| ER strains | 66 | 16 | 1 | 25 | 23 | 1 |
| ES strains | 11 | 2 | 2 | 7 | ||
Isolates were maintained in glycerol at −70°C and subcultured twice on blood agar before testing. Todd-Hewitt broth and agar (Oxoid Ltd., Basingstoke, United Kingdom) were used for routine culture.
RD2 repeats.
The RD2 region of prtF1 was detected by PCR with the pair of primers (each measuring 24 nucleotides) reported by Neeman et al. (22). These two primers, derived from those originally established by Natanson et al. (21) and reported to be complementary to the flanking region of RD2 (22), in fact partially overlap the end of RD2 by eight (5′ primer) or nine (3′ primer) nucleotides (28). PCR conditions were described previously (22). The number of RD2 repeats of prtF1 was determined on the basis of amplicon size, taking into account that one repeat was 111 or 96 bp long (21, 28). Marker XIV (Roche Molecular Biochemicals, Mannheim, Germany) was used as DNA size markers.
Restriction enzyme cleavage analysis of PCR products.
The PCR products obtained from the amplification of the RD2 region of prtF1 were subjected to restriction analysis with endonucleases DdeI, HaeIII, and HinfI (New England Biolabs, Beverly, Mass.). A 10-μl aliquot of the PCR product was digested with the endonuclease according to the manufacturer's recommendations. Restriction fragments were separated by agarose (1%) gel electrophoresis and visualized under UV light by staining with ethidium bromide. In some experiments, restriction fragments were separated by acrylamide (12.5%) gel electrophoresis in a Hoefer miniVE System (Amersham Biosciences Europe, Freiburg, Germany) at a constant voltage of 70 V for 3 h and visualized by silver staining. Gels were photographed with Polaroid B/W 667 film and an MP-4 Land Camera. GeneRuler 100-bp DNA Ladder Plus (M-Medical Genenco, Cornaredo, Italy) or 50-bp Ladder (Amersham) was used as molecular size markers.
M typing.
The PCR M typing method described by Vitali et al. (35) was used. This method, recently employed to assess the emm gene distribution among ER and ES Italian GAS (36), is directed at amplifying the N-terminal region of the emm gene with a set of 21 emm-specific oligonucleotide primers specific for emm1, emm2, emm3, emm4, emm5, emm6, emm8, emm11, emm12, emm18, emm22, emm24, emm28, emm29, emm48, emm75, emm77, emm78, emm87, emm89, and emm94.
Macrorestriction analysis and PFGE.
Preparation of genomic DNA and digestion with SmaI endonuclease (New England Biolabs), analysis of PFGE patterns, and method of reporting were as described elsewhere (26). The type was designated with a capital letter following the scheme previously adopted for Italian ER GAS (26). PFGE types recognized in single isolates were reported as one-strain types. Isolates were designated as PFGE untypeable when their DNA was not digested by SmaI.
Statistics.
Fisher's exact test was applied with the fisher.test procedure of the S-Plus statistical package (31).
RESULTS
PCR analysis of the RD2 region of the prtF1 gene.
PCR analysis of the RD2 region yielded products ranging from approximately 125 to 570 bp (Fig. 1A), consistent with between one and five RD2 repeats. Strains with four or three repeats were more frequent (30 and 27, respectively); one repeat was found in 18 strains, and two and five repeats were found in one strain each. Correlations between genotypes and phenotypes of erythromycin resistance and numbers of RD2 repeats in the 77 test strains are detailed in Table 1. Fisher's exact test evidenced a highly significant association (P < 0.001) between the number of RD2 repeats and the genotype of erythromycin resistance.
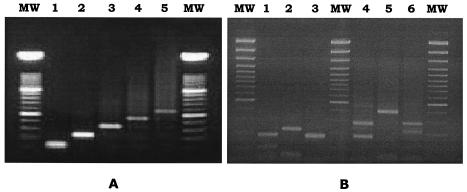
FIG. 1.
Size variation of PCR products of prtF1 among the 77 GAS isolates (A) and HaeIII restriction profiles obtained from the PCR products of the 46 isolates whose amplicons were digested by HaeIII (B). (A) Lane 1, amplicon size of 127 bp, suggesting the presence of a single RD2 repeat (n = 18); lane 2, amplicon size of 238 bp, suggesting the presence of two RD2 repeats (n = 1); lane 3, amplicon size of 349 bp, suggesting the presence of three RD2 repeats (n = 27); lane 4, amplicon size of 460 bp, suggesting the presence of four RD2 repeats (n = 30); lane 5, amplicon size of 571 bp, suggesting the presence of five RD2 repeats (n = 1). (B) Lanes 1 to 3, restriction profiles obtained from strains with three RD2 repeats (lane 1, three bands of 59, 111, and 179 bp; lane 2, three bands of 59 and 68 bp [comigrating] and 222 bp; and lane 3, two comigrating bands of 170 and 179 bp); lanes 4 and 5, restriction profiles obtained from strains with four RD2 repeats (lane 4, two bands of 170 and 290 bp, and lane 5, two bands of 59 and 401 bp); lane 6, restriction profile obtained from the single strain with five RD2 repeats (three bands of 59, 222, and 290 bp). Lane MW, DNA size markers.
Restriction analysis of PCR products.
HaeIII restriction was successfully performed on the PCR products of 46 of the 77 test strains, the amplicons of 31 strains remaining undigested. The 46 digested strains yielded six different restriction profiles (Fig. 1B). Three profiles were from strains with three RD2 repeats (amplicon size, 349 bp): one was characterized by three bands of 59, 111, and 179 bp (Fig. 1B, lane 1); another by three bands, including two comigrating bands of 59 and 68 bp and a band of 222 bp (Fig. 1B, lane 2); and the third by two comigrating bands of 170 and 179 bp (Fig. 1B, lane 3). Bands comigrating in the agarose gel were resolved and visualized by acrylamide gel electrophoresis and silver staining (data not shown). Two other restriction profiles were from strains with four RD2 repeats (amplicon size, 460 bp): one exhibited two bands of 170 and 290 bp (Fig. 1B, lane 4) and the other two bands of 59 and 401 bp (Fig. 1B, lane 5). The sixth restriction profile was from the single strain with five RD2 repeats (amplicon size, 571 bp) and showed three bands of 59, 222, and 290 bp (Fig. 1B, lane 6).
DdeI restriction was successfully performed on the PCR products of 59 of the 77 test strains; the amplicons of 18 strains (those with one RD2 repeat) remained undigested. The 59 digested strains yielded four different restriction profiles exactly matching the strains with two, three, four, and five repeats, respectively.
HinfI restriction of the PCR products was successful in all 77 test strains and yielded five different restriction profiles matching exactly the strains with different numbers (one to five) of repeats.
RD2 typing.
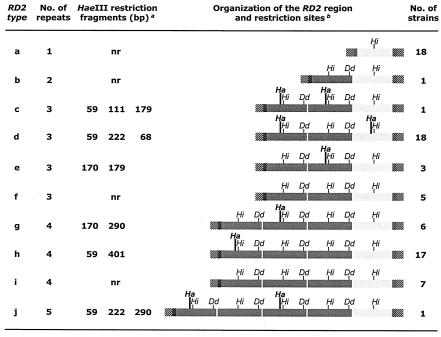
RD2 typing was obtained by combining PCR analysis of the RD2 region of the prtF1 gene and restriction analysis of PCR products with DdeI, HaeIII, and HinfI. Ten RD2 types (a to j) were distinguished among the 77 GAS strains (Fig. 2). Type a included all the isolates with one RD2 repeat, whose PCR products were consistently resistant to HaeIII and DdeI restriction. Type b was represented by the sole isolate with two RD2 repeats, whose PCR product was resistant to HaeIII restriction. Types c, d, and e corresponded to the three different HaeIII restriction profiles detected in the strains with three RD2 repeats. Type f included the remaining isolates with three RD2 repeats, whose PCR products were resistant to HaeIII restriction. Types g and h corresponded to the two HaeIII restriction profiles detected in the strains with four RD2 repeats. Type i consisted of the remaining isolates with four RD2 repeats, whose PCR products were resistant to HaeIII restriction. Finally, type j was represented by the single strain with five RD2 repeats.
FIG. 2.
RD2 types and organization of the RD2 region in the 77 prtF1-positive GAS based on PCR and restriction enzyme cleavage analysis of PCR products. The organization of the RD2 region of GAS strains was outlined on the basis of the results of RD2 typing and the published sequence of a five-repeat prtF1 gene (28). Superscript letters: a, The reported sizes of PCR products include the primers, which are known to partially overlap the ends of RD2 by eight (5′ primer) or nine (3′ primer) nucleotides (28). nr, not restricted by HaeIII; b, Light shading, 96-bp repeat; dark shading, 111-bp repeat; hatching, primer. Vertical bars indicate restriction sites (Ha, HaeIII; Dd, DdeI; and Hi, HinfI) within individual repeats.
Organization of the RD2 region.
The organization of the RD2 region of the 77 GAS strains was outlined based on the results of RD2 typing plus the published sequence of a five-repeat prtF1 gene (28) which appeared to correspond to the RD2 type j of our collection.
As shown in Fig. 2, an incomplete, 96-bp repeat was present, alone (type a) or preceded by one to four 111-bp repeats (types b to j), in all strains at the 3′ end of the RD2 region. The 111-bp repeat exhibited two variants: one with no HaeIII restriction site and one with a HaeIII restriction site at 43 bp. The HaeIII restriction profile of RD2 type d could be explained by the presence of a second variant of the 96-bp repeat, similarly featuring a HaeIII site at 43 bp. One HinfI restriction site was present in all (both 96-bp and 111-bp) repeats at 51 bp, whereas one DdeI restriction site was present in the 111-bp repeats only at 106 bp.
emm types and PFGE types.
Sixteen emm types were found among the 77 prtF1-positive GAS, nine of which (emm1, emm2, emm8, emm22, emm28, emm48, emm61, emm77, and emm89) were present only in ER strains, three (emm11, emm78, and emm87) only in ES strains, and four (emm4, emm6, emm12, and emm75) in both ER and ES strains. The prevalent types were emm77 (n = 18), emm4 (n = 15), and emm89 (n = 12).
Twenty-seven PFGE types, 19 of which were detected in single isolates, were identified among the 77 prtF1-positive GAS. Seven isolates [all of the mef(A)/M genotype/phenotype of erythromycin resistance] were untypeable, i.e., their DNA was not restricted by SmaI. None of the 11 ES GAS shared a PFGE type with any ER isolate: nine isolates had one-strain PFGE types, and two were the only representatives of a unique type (PFGE type R).
Clones.
The distribution of typing data (RD2 type, emm type, and PFGE type) among the 77 prtF1-positive GAS strains subdivided on the basis of their genotypes and phenotypes of erythromycin resistance is detailed in Table 2. Fisher's exact test yielded a highly significant association (P < 0.001) between RD2 type and genotype of erythromycin resistance.
TABLE 2.
Distribution of typing characteristics (RD2 type, emm type, and PFGE type) among the 77 prtF1-positive GAS strains subdivided on the basis of their genotypes and phenotypes of erythromycin resistancea
| Genotype/phenotype of erythromycin resistance (no. of strains) | No. of strainsb | RD2 type | emm type | PFGE typec |
|---|---|---|---|---|
| erm(B)/cMLS (8) | 2 | f | 22 | E |
| 2 | i | 22 | E | |
| 1 | a | 4 | B | |
| 1 | b | 28 | ostd | |
| 1 | c | 61 | oste | |
| 1 | i | 22 | ost | |
| erm(B) mef(A)/cMLS (2) | 1 | g | 12 | ost |
| 1 | i | 48 | ost | |
| erm(B)/iMLS-A (12) | 8 | h | 89 | A |
| 1 | h | 4 | A | |
| 1 | h | 89 | B | |
| 2 (1) | h | 89 | J | |
| erm(B) mef(A)/iMLS-A (1) | 1 | h | 1 | A |
| erm(A)/iMLS-B (13) | 13 (1) | d | 77 | D |
| erm(A)/iMLS-C (8) | 2 | d | 77 | D |
| 2 | d | 77 | O | |
| 1 | d | 77 | ost | |
| 1 | f | 22 | ost | |
| 1 | f | 22 | ost | |
| 1 | f | 22 | ost | |
| mef(A)/M (22) | 10 (2) | a | 4 | B |
| 2 | a | 4 | ut | |
| 1 | a | 12 | B | |
| 1 | a | 12 | ut | |
| 1 | a | 89 | B | |
| 1 | e | 75 | ut | |
| 2 (1) | g | 12 | ut | |
| 1 | i | 8 | G | |
| 1 | i | 48 | G | |
| 1 (1) | i | 2 | ostf | |
| 1 (1) | j | 6 | ut | |
| ES strains (11) | 2 (2) | h | 6 | R |
| 1 | a | 4 | ost | |
| 1 (1) | a | 11 | ost | |
| 1 | e | 75 | ost | |
| 1 (1) | e | 75 | ost | |
| 1 | g | 12 | ost | |
| 1 (1) | g | 12 | ost | |
| 1 | g | 87 | ost | |
| 1 | h | 78 | ost | |
| 1 (1) | h | 78 | ost |
Lines correspond to clones (n = 45).
Numbers in parentheses indicate the number of strains with weak cell invasion efficiency.
ost, one-strain type; ut, untypeable (DNA not restricted by SmaI).
Identical to the PFGE type designated F in a previous study (26).
Identical to the PFGE type designated I in a previous study (26).
Identical to the PFGE type designated C in a previous study (26).
GAS isolates with a unique combination of a given erythromycin resistance genotype and phenotype, a given RD2 type, a given emm type, and a given PFGE type were considered to represent a clone. By this criterion, 41 different clones were recognized (31 among the 66 ER isolates and 10 among the 11 ES isolates), each corresponding to one line in Table 2. While 30 (39%) of the 77 GAS were single isolates representing single clones, three major clones—(i) erm(A)/iMLS-B, RD2 type d, emm77, PFGE type D (n = 13); (ii) mef(A)/M, RD2 type a, emm4, PFGE type B (n = 10); and (iii) erm(B)/iMLS-A, RD2 type h, emm89, PFGE type A (n = 8)—accounted for 40% of the 77 GAS (47% of the 66 ER strains). Of the nine ES clones with one-strain PFGE types, five displayed combinations of RD2 type and emm type also recorded in ER clones.
DISCUSSION
In gram-positive bacteria, C-terminally anchored surface proteins are a well-defined category characterized by regions with tandem sequence repeats and a conserved anchor: the vast majority of these proteins are multifunctional and contain domains that bind molecules found in body secretions, such as immunoglobulins, albumin, fibronectin, and fibrinogen (8). The F1/SfbI invasin of GAS belongs to this category: although it works primarily as an adhesin—mediating, through its fibronectin-binding repeat region, bacterial attachment to host cells and subsequent upper respiratory tract colonization (3, 4, 13)—its binding to fibronectin ends up efficiently triggering the invasion process (33). F1/SfbI also binds to the Fc fragment of human immunoglobulins, interfering with Fc receptor-mediated phagocytosis and antibody-dependent cell cytotoxicity by macrophages (18). At present, F1/SfbI is regarded not only as an emerging virulence determinant of GAS, but also as a promising candidate antigen for developing anti-GAS vaccines (12, 27). Our present findings about the variability of the RD2 region of the prtF1 gene and the relationships between RD2 and emm typing might contribute to the investigation of the new vaccine.
The present study shows that thorough analysis and molecular characterization of the RD2 repeats of prtF1 may represent a valuable tool to gain insights into the association between cell invasiveness and erythromycin resistance of GAS, i.e., the subject of the investigation (6) that yielded the 77 prtF1-positive GAS tested here. This is especially true of inducibly erythromycin-resistant isolates, of which all those carrying the erm(B) methylase (iMLS-A phenotype; n = 13) had four RD2 repeats and belonged to RD2 type h, whereas all those carrying the erm(A) methylase (iMLS-B and iMLS-C phenotypes; n = 21) had three repeats. The picture was more varied among constitutively resistant and M-phenotype strains and among ES isolates. It is worth noting that the RD2 region of prtF1 has been reported to be composed of a number of repeats ranging between two and six (21, 22), whereas in this study we show that a single repeat (the one measuring 96 bp) is also a common finding, especially among M-phenotype GAS.
Remarkably, HaeIII restriction analysis of the PCR products obtained from the amplification of the RD2 region made it possible to discriminate between two principal RD2 repeat variants, (i) one with and (ii) one without a HaeIII site at 43 bp. Fully in agreement with the HaeIII mapping of the RD2 region of our single strain bearing five repeats (RD2 type j) (Fig. 2), the published sequence of RD2 displays the HaeIII site (GGCC) at 43 bp in the first and the third of the five repeats, whereas an alternative sequence (GGTC) is present in the other three repeats (28).
Most of the emm and PFGE types detected among the prtF1-positive throat clinical GAS examined in this study have previously been found among Italian throat isolates investigated for emm (5, 36) or PFGE (26) typing, respectively. In a recent German study of throat GAS, prtF1 was detected only in strains with emm12 and emm6 (2), whereas in a similar Japanese study the prevalent emm types associated with prtF1 were emm12 and emm4 (17). Overall, our findings show that the previously demonstrated ability of GAS to invade respiratory cells (6) is shared by prtF1-positive strains with a variety of emm types.
Our findings suggest that each different approach to GAS typing may provide a specific piece of information, indicating that simultaneous use of different methods is essential to thoroughly investigate GAS epidemiology. In this study, extensive comparative analysis of RD2 typing with PCR M typing, PFGE typing, and previously investigated features (cell invasion efficiency, macrolide resistance genotype, and phenotype) (6) concurred to outline the polyclonal nature of cell-invasive GAS circulating in Italy. As many as 41 different clones, of which 30 were represented by single isolates, were recognized among the 77 prtF1-positive GAS tested. On the other hand, in line with previous findings on ER GAS (26), a small number of clones was predominant, three major clones accounting for 40% of all the GAS tested (and for 47% of the ER strains). The 11 ES GAS were largely unrelated: only two strains of RD2 type h represented a single clone with a new PFGE type. However, some ES isolates appeared to be related to ER isolates that shared identical combinations of RD2 type and emm type.
This polyclonal spread of GAS with different combinations of resistance and virulence genes in Italy suggests that horizontal acquisition of macrolide resistance determinants by cell-invasive strains may have played a role in the association between erythromycin resistance and ability to enter human respiratory cells reported among Italian GAS (6). This conclusion is consistent with recent studies emphasizing the genetic diversity among GAS and suggesting that horizontal transfer and recombination of virulence genes have played a major role in generating this diversity (15). As speculated by Feil and Spratt for other pathogens (7), this may have resulted in a streptococcal population composed of two parts: a background population consisting of a large number of unrelated, relatively rare clones (the majority), and clusters of closely related, ecologically successful clones (the predominant ones).
Acknowledgments
This work was partly supported by the Italian Ministry of Education, University and Research (grant MUVAR01302) and the European Union (project ARTRADI, grant QLK2-CT-2002-00843).
REFERENCES
- 1.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt C. M., F. Allerberger, B. Spellerberg, R. Holland, R. Lutticken, and G. Haase. 2001. Characterization of consecutive Streptococcus pyogenes isolates from patients with pharyngitis and bacterological treatment failure: special reference to prtf1 and sic/drs. J. Infect. Dis. 183:670-674. [DOI] [PubMed] [Google Scholar]
- 3.Cleary, P. P., and D. Cue. 2000. High frequency invasion of mammalian cells by β-hemolytic streptococci, p. 137-166. In T. A. Oelschlaeger and J. Hacker (ed.), Bacterial invasion into eukaryotic cells. Subcellular biochemistry, vol. 33. Kluwer Academic/Plenum Publishers, New York, N.Y. [DOI] [PubMed]
- 4.Courtney, H. S., D. L. Hasty, and J. B. Dale. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann. Med. 34:77-87. [DOI] [PubMed] [Google Scholar]
- 5.Dicuonzo, G., G. Gherardi, G. Lorino, S. Angeletti, M. De Cesaris, E. Fiscarelli, D. E. Bessen, and B. Beall. 2001. Group A streptococcal genotypes from pediatric throat isolates in Rome, Italy. J. Clin. Microbiol. 39:1687-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Facinelli, B., C. Spinaci, G. Magi, E. Giovanetti, and P. E. Varaldo. 2001. Association between erythromycin resistance and ability to enter human respiratory cells in group A streptococci. Lancet 358:30-33. [DOI] [PubMed] [Google Scholar]
- 7.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 8.Fischetti, V. A. 2000. Surface proteins on gram-positive bacteria, p. 11-24. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 9.Gillespie, S. H. 1998. Failure of penicillin in Streptococcus pyogenes pharyngeal infection. Lancet 352:1954-1956. [DOI] [PubMed] [Google Scholar]
- 10.Giovanetti, E., A. Brenciani, R. Burioni, and P. E. Varaldo. 2002. A novel efflux system in inducibly erythromycin-resistant strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 46:3750-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman, C. A., S. R. Talay, G. Molinari, E. Medina, and G. S. Chhatwal. 1999. Protective immune response against Streptococcus pyogenes in mice after nasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 179:901-906. [DOI] [PubMed] [Google Scholar]
- 13.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jadoun, J., V. Ozeri, E. Burstein, E. Skutelsky, E. Hanski, and S. Sela. 1998. Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J. Infect. Dis. 178:147-158. [DOI] [PubMed] [Google Scholar]
- 15.Kehoe, M. A., V. Kapur, A. M. Whatmore, and J. M. Musser. 1996. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 4:436-443. [DOI] [PubMed] [Google Scholar]
- 16.LaPenta, D., D. C. Rubens, E. Chi, and P. P. Cleary. 1994. Group A streptococci efficiently invade human respiratory epithelial cells. Proc. Natl. Acad. Sci. USA 91:12115-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, X., H. Kikuta, N. Ishiguro, M. Yoshioka, T. Ebihara, T. Murai, I. Kobayashi, and K. Kobayashi. 2002. Association of the prtF1 gene (encoding fibronectin-binding protein F1) and the sic gene (encoding the streptococcal inhibitor of complement) with emm types of group A streptococci isolated from Japanese children with pharingitis. J. Clin. Microbiol. 40:3835-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina, E., S. R. Talay, G. S. Chhatwal, and C. A. Guzman. 1998. Fibronectin-binding protein I of Streptococcus pyogenes is a promising adjuvant for antigens delivered by mucosal route. Eur. J. Immunol. 28:1069-1077. [DOI] [PubMed] [Google Scholar]
- 19.Molinari, G., and G. S. Chhatwal. 1999. Streptococcal invasion. Curr. Opin. Microbiol. 2:56-61. [DOI] [PubMed] [Google Scholar]
- 20.Molinari, G., S. R. Talay, P. Valentin-Weigand, M. Rohde, and G. S. Chhatwal. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 65:1357-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natanson, S., S. Sela, A. E. Moses, J. M. Musser, M. G. Caparon, and E. Hanski. 1995. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J. Infect. Dis. 171:871-878. [DOI] [PubMed] [Google Scholar]
- 22.Neeman, R., N. Keller, A. Barzilai, Z. Korenman, and S. Sela. 1998. Prevalence of internalization-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet 352:1974-1977. [DOI] [PubMed] [Google Scholar]
- 23.Okada, N., I. Tatsuno, E. Hanski, M. Caparon, and C. Sasakawa. 1998. Streptococcus pyogenes protein F promotes invasion of HeLa cells. Microbiology 144:3079-3086. [DOI] [PubMed] [Google Scholar]
- 24.Österlund, A., R. Popa, T. Nikkilä, A. Scheynius, and L. Engstrand. 1997. Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope 107:640-647. [DOI] [PubMed] [Google Scholar]
- 25.Ozeri, V., I. Rosenshine, D. F. Mosher, R. Fässler, and E. Hanski. 1998. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol. 30:625-637. [DOI] [PubMed] [Google Scholar]
- 26.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:55-61. [DOI] [PubMed] [Google Scholar]
- 27.Schulze, K., E. Medina, G. S. Chhatwal, and C. A. Guzman. 2003. Stimulation of long-lasting protection against Streptococcus pyogenes after intranasal vaccination with non adjuvanted fibronectin-binding domain of the SfBI protein. Vaccine 21:1958-1964. [DOI] [PubMed] [Google Scholar]
- 28.Sela, S., A. Aviv, A. Tovi, I. Burstein, M. G. Caparon, and E. Hanski. 1993. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10:1049-1055. [DOI] [PubMed] [Google Scholar]
- 29.Sela, S., R. Neeman, N. Keller, and A. Barzilai. 2000. Relationship between asymptomatic carriage of Streptococcus pyogenes and the ability of the strains to adhere and be internalised by cultured epithelial cells. J. Med. Microbiol. 49:499-502. [DOI] [PubMed] [Google Scholar]
- 30.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S-Plus 6 for Windows. 2001. Guide to statistics, vol. 1. Insightful Corporation, Seattle, Wash.
- 32.Talay, S. R., P. Valentin-Weigand, K. N. Timmis, and G. S. Chhatwal. 1994. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol. Microbiol. 13:531-539. [DOI] [PubMed] [Google Scholar]
- 33.Talay, S. R., A. Zock, M. Rohde, G. Molinari, M. Oggioni, G. Pozzi, C. A. Guzman, and G. S. Chhatwal. 2000. Cooperative binding of human fibronectin to Sfbl protein triggers streptococcal invasion into respiratory epithelial cells. Cell. Microbiol. 2:521-535. [DOI] [PubMed] [Google Scholar]
- 34.Varaldo, P. E., E. A. Debbia, G. Nicoletti, D. Pavesio, S. Ripa, G. C. Schito, G. Tempera, and the Artemis-Italy Study Group. 1999. Nationwide survey in Italy of treatment of Streptococcus pyogenes pharyngitis in children: influence of macrolide resistance on clinical and microbiological outcomes. Clin. Infect. Dis. 29:869-873. [DOI] [PubMed] [Google Scholar]
- 35.Vitali, L. A., C. Zampaloni, M. Prenna, and S. Ripa. 2002. PCR M typing: a new method for rapid typing of group A streptococci. J. Clin. Microbiol. 40:679-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zampaloni, C., P. Cappelletti, M. Prenna, L. A. Vitali, and S. Ripa. 2003. emm gene distribution among erythromycin-resistant and -susceptible Italian isolates of Streptococcus pyogenes. J. Clin. Microbiol. 41:1307-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]