Abstract
The VERSANT hepatitis B virus (HBV) 3.0 Assay (branched DNA [bDNA]) (referred to herein as VERSANT 3.0) was evaluated at four external sites for analytical sensitivity, specificity, reproducibility, linearity of quantification, and limits of detection. In addition, each of the test evaluation sites provided HBV DNA-positive clinical samples that were previously analyzed by one of three commercially available HBV DNA quantitative tests: Digene Hybrid Capture II HBV DNA Test (Digene); VERSANT HBV DNA 1.0 Assay (bDNA) (VERSANT 1.0); and COBAS AMPLICOR HBV Monitor Test (COBAS AMPLICOR). These samples were reexamined using VERSANT 3.0. The results from these studies showed that VERSANT 3.0 has high specificity (99.3%), excellent reproducibility (between-run coefficient of variation [CV] = 1.6 to 9.4%; within-run CV = 6.5 to 20.7%), and a broad linear range of quantification (2.0 × 103 to 1.0 × 108 HBV DNA copies/ml) that facilitate the monitoring of HBV DNA levels at diagnosis and throughout the course of treatment. In general, correlation was very good between results obtained from clinical samples analyzed by VERSANT 3.0 and the comparative HBV DNA quantitative assays (VERSANT 1.0, R2 = 0.900; Digene, R2 = 0.985; COBAS AMPLICOR, R2 = 0.771). The greatest differences in comparative quantitation occurred at HBV DNA levels approaching the limits of the dynamic ranges for the comparative assays. The performance characteristics of the new VERSANT 3.0 assay demonstrated that it provides a reliable and robust method for routinely monitoring serum HBV DNA levels in assessing disease activity and determining response to antiviral treatment.
Infection with hepatitis B virus (HBV) remains a difficult worldwide challenge to public health. The World Health Organization estimates that more than one-third of the world's population has been infected with HBV (25). Epidemiological trends suggest there are currently 400 million HBV chronic carriers worldwide, with over 1 million deaths annually due to HBV-associated liver disease (13, 23). HBV is the leading cause of cirrhosis and hepatocellular carcinoma globally (25; Centers for Disease Control and Prevention hepatitis fact sheet [www.cdc.gov/hepatitis]). The development and utilization of molecular diagnostic assays for the detection and quantification of HBV genomes have provided insight into the natural history of HBV and the pathogenesis of HBV infection as well as facilitated the monitoring of viral response to treatment (15, 17). In addition, a quantitative evaluation of HBV DNA concentrations can provide valuable information on the levels of viral replication and may be useful as a prognostic indicator of liver disease (4, 21). A number of commercial assays are currently available for the quantification of HBV DNA in patient serum or EDTA-plasma, including hybridization-, signal-, and target-amplification-based technologies (5, 11, 16-18, 22). Selection of the optimal assay is dependent on the intrinsic performance characteristics of the methodology as well as the necessity to make appropriate clinical decisions in the context of HBV-associated disease (4, 15, 21).
The VERSANT HBV 3.0 Assay (referred to herein as VERSANT 3.0) is a third-generation branched-DNA (bDNA) assay for the direct quantification of HBV DNA in human serum and plasma. After HBV genomic DNA is released from the virions, the viral DNA is captured by a set of specific, synthetic oligonucleotide capture probes fixed in a microtiter well. A set of target probes (or label extender probes) then hybridizes to both the captured viral DNA and unique preamplifier probes. The capture probes and the target probes bind to conserved DNA regions throughout the entire HBV genome. The amplifier probes subsequently hybridize to the preamplifier probes, forming a bDNA complex. Multiple copies of an alkaline phosphatase-labeled probe are then hybridized to this immobilized complex. Detection is achieved by incubating the alkaline phosphatase-bound complex with a chemiluminescent substrate. The intensity of light emission is directly related to the amount of HBV DNA present in each sample, and results are recorded as relative light units by the luminometer. A standard curve is defined by light emission from quantitative standards containing known concentrations of recombinant DNA. Concentrations of HBV DNA in specimens are determined from this standard curve. This third-generation sandwich nucleic acid hybridization procedure differs from earlier bDNA assays by using the unique preamplifier probes to increase the number of labeled probes that can bind to the target, thereby augmenting the signal amplification and the assay sensitivity. The label extender probes are designed such that two probes must be bound to adjacent regions of the target for efficient hybridization to the preamplifier molecule to occur. This new bDNA technology has been described extensively in previous peer-reviewed publications (1, 2, 8, 9, 12, 14, 18, 19, 22).
This study describes the findings of a multicenter evaluation of VERSANT 3.0 (Bayer HealthCare LLC, Diagnostics Division, Tarrytown, N.Y.). Analytical performance parameters including specificity, reproducibility, linearity, limits of detection, and quantification were examined and compared to those of three other commercially available assays.
MATERIALS AND METHODS
Testing facilities and samples.
Evaluation of performance characteristics of the VERSANT 3.0 and analysis of clinical specimens was performed by independent clinical investigators at four field testing sites: The Academic Medical Center (Amsterdam, The Netherlands), Mayo Clinic (Rochester, Minn.), Singapore General Hospital (Singapore, Singapore), and the University of British Columbia (Vancouver, British Columbia, Canada). Each testing site was provided with a nine-member quantitative control (QC) panel consisting of whole HBV particles (live virus, not genotyped) and recombinant HBV DNA (HBV subtype adw2) diluted in human HBV-seronegative plasma for experiments evaluating analytical performance characteristics of the VERSANT 3.0 assay. Target concentrations (HBV DNA copies per milliliter) based on stock materials with known concentrations and assigned values based upon analysis of each QC panel member with a single validated kit lot (G002) are reported in Table 1.
TABLE 1.
Target concentrations and value assignments of the QC panel
| Panel member | Target concn (HBV DNA copies/mL) | Value assignment (HBV DNA copies/mL) |
|---|---|---|
| QC1 (rDNAa) | 110,000,000 | 106,332,819 |
| QC2 (rDNA) | 10,000,000 | 9,666,620 |
| QC3 (virus) | 2,000,000 | 1,954,243 |
| QC4 (rDNA) | 1,000,000 | 966,662 |
| QC5 (virus) | 100,000 | 97,712 |
| QC6 (virus) | 10,000 | 9,771 |
| QC7 (virus) | 4,000 | 3,908 |
| QC8 (virus) | 2,000 | 1,954 |
| QC9 (virus) | 800 | 782 |
rDNA, rRNA gene.
Each site was also provided with 150 unique serum samples from healthy donors. These samples were purchased from ProMedDx (Norton, Mass.) and were seronegative for HBV (negative for HBsAg and anti-HBc antibody), HCV, human immunodeficiency virus, and human T-cell leukemia virus. Each evaluation site tested a different subset of these serum samples (total n = 600).
All testing sites provided additional HBV DNA-positive samples with previous nucleic acid test results obtained using the Digene Hybrid Capture II HBV DNA Test (Digene), VERSANT HBV DNA 1.0 Assay (bDNA) (VERSANT 1.0), or COBAS AMPLICOR HBV MONITOR Test (COBAS AMPLICOR). These samples were reanalyzed using the VERSANT 3.0 assay, and results were compared to the original results. The institutional review boards of each testing site approved the use of specimens utilized in this study.
VERSANT 3.0.
VERSANT 3.0 has a reportable range of results from 2.0 × 103 to 1.0 × 108 HBV DNA copies/ml. The assay uses a small sample volume (50 μl undiluted) and is currently available for research use only. The ease of use and throughput of the VERSANT 3.0 are greatly facilitated by use of a semiautomated instrument (Bayer System 340). This instrument system provides a platform wherein the multiple incubations (hybridizations), washes, final luminescence readout, as well as data reduction are all automated. The instrument can process two 96-microwell plates per run (i.e., 84 results per microwell plate or 168 results per single run).
Analytical performance.
The analytical performance characteristics of VERSANT 3.0 were evaluated by each testing site analyzing one full microwell plate of the nine-member QC panel samples (shown in Table 1) on five different days. A full microwell plate run consisted of testing each QC panel member prepared in four to seven replicates as well as 30 seronegative samples. For each run, uniform plate maps were used by all testing sites to ensure that the QC panel members and seronegative samples were placed in different microwell locations for each assay run. Eleven blank wells were left as placeholders on each plate. Therefore, at the completion of five runs, there were 20 to 35 results for each QC panel member and 150 results from the unique seronegative samples from each testing site.
Assessment of specificity, reproducibility, linearity, limit of detection, and sensitivity.
Specificity of the VERSANT 3.0 was determined using the results of the seronegative specimens analyzed at each testing site (n = 150 unique samples per site). Specificity was defined as the proportion of seronegative samples yielding negative results according to an assay detection cutoff (DC) of 2,000 HBV DNA copies/ml. The point estimate and the exact 95% lower confidence limits for the overall specificity were calculated.
The nine-member QC panel (Table 1) was used to assess the reproducibility, linearity, and limits of detection for VERSANT 3.0. The QC panel (QC1 to QC9, with titers ranging from 800 to 110,000,000 copies/ml) extended over the reportable range of the assay. The reproducibility variance components were estimated using restricted maximum-likelihood estimation, and the total variance (which accounts for between-run and within-run effects at all testing sites) was estimated as the sum of the individual variance components. The variance components and the total variance were converted into coefficients of variation (CV).
Linearity of quantification by VERSANT 3.0 was evaluated as the log recovery for each QC panel member. The log recovery is defined as the difference between mean quantified log value and the assigned log value for each panel member.
The quantitative range of VERSANT 3.0 was defined by the lowest and highest concentrations of HBV DNA evaluated with acceptable linearity, accuracy, and precision. The analytical sensitivity of VERSANT 3.0 is expressed as the lower limit of detection (LOD). The LOD is defined as the lowest concentration of HBV that yields a quantifiable result in 95% of the time. Therefore, a signal at the LOD can be reliably distinguished from background signal in HBV DNA-negative specimens 95% of the time. To estimate this concentration, an interpolation between the 5th percentiles of the quantitative distribution of two QC panel members whose sensitivity brackets 95% was used. Data from all four testing centers were combined to provide a total volume of data sufficient for statistical estimation of the performance specifications of VERSANT 3.0.
Comparative analysis.
Correlation between quantitative HBV concentrations obtained from VERSANT 3.0 and those previously determined by other HBV quantitative assays was assessed. Each testing site provided HBV DNA-positive samples with previous nucleic acid test results (111 to 147 specimens per site stored at −70 to −80°C ranging from less than 3 to 24 months). These samples chosen by each testing site had HBV DNA concentrations that overlapped the dynamic range of VERSANT 3.0 (2.0 × 103 to 1.0 × 108 HBV DNA copies/ml). These serum or plasma samples were reanalyzed using VERSANT 3.0, and the results were compared to the initial results obtained from one of the following assays: standard and ultrasensitive Digene (n = 247; Mayo Clinic and University of British Columbia), VERSANT 1.0 (n = 185; Singapore General Hospital and Academic Medical Center), or COBAS AMPLICOR (n = 79; Academic Medical Center). Comparisons between VERSANT 3.0 and the initial assays were achieved by plotting the paired observed values expressed in log10 units for each sample. Furthermore, consistency of the differences between the log10 quantitative values obtained from VERSANT 3.0 and the other assays was assessed according to the analysis proposed by Bland and Altman (3) (Bland-Altman plot). This analysis consisted of constructing a scatter plot depicting the difference in log10 values for each matched pair of quantitative results versus the average of the log10 quantitative values for each pair. The resulting scatter plot would show two horizontal reference lines at levels equal to the upper and lower limits of the 95% two-sided confidence limits for individual differences, and 95% of the differences are expected to be between these two confidence limits. A horizontal reference line at the average difference would be included. If the relationship between the quantitative values were consistent across the range of concentrations for these matched pairs of results, the scatter plot would exhibit no systematic trends in the distribution of differences as a function of the averages. A regression line fitting the differences versus the averages is added to the scatter plot as well as the regression equation. This analysis would determine if the average log10 difference of results was consistent across concentrations spanning the dynamic range of results in common to the two assays being compared.
RESULTS
Analytical specificity.
Collectively, a total of 600 unique, HBV-seronegative specimens were tested by the four testing sites. Three of the testing sites found that 149 of 150 (99.3%) specimens analyzed using the VERSANT 3.0 assay had results below the DC. The fourth site determined that 148 of 150 (98.7%) seronegative samples generated results below the DC. The overall estimate for specificity was 99.2%, with a one-sided 95% lower confidence limit of 98.3%.
The five negative samples with detectable HBV DNA had initial titers of 2,167, 2,199, 2,668, 3,121, and 35,900 copies/ml. These five samples were returned to Bayer, and these samples plus the matching stored samples were tested in triplicate. All test results showed HBV DNA titers of <2,000 copies/ml.
Analytical precision (reproducibility).
Table 2 contains the data on assay reproducibility including site, between-run, and within-run variance components, as well as the total CV for each QC panel member. Over the dynamic range of VERSANT 3.0 (2 × 103 to 1 × 108 HBV DNA copies/ml), the total CV due to all sources of variation ranged from 10.6 to 26.5% at 2 × 103 HBV DNA copies/ml. For the single QC panel member (QC9, 782 copies/ml) with an HBV DNA concentration of <2,000 copies/ml and outside of the quantitative range of the assay, the overall variation was 32.7%.
TABLE 2.
Reproducibility of quantitation of the QC panel by VERSANT 3.0
| Dilution panel member | n | % CV
|
|||
|---|---|---|---|---|---|
| Between site | Between run | Within run | Total | ||
| QC1 | 80 | 9.8 | 9.4 | 8.4 | 16.0 |
| QC2 | 80 | 13.3 | 8.7 | 8.1 | 17.9 |
| QC3 | 80 | 10.0 | 7.2 | 7.4 | 14.4 |
| QC4 | 79 | 6.4 | 5.4 | 6.5 | 10.6 |
| QC5 | 80 | 8.6 | 7.7 | 7.9 | 14.1 |
| QC6 | 80 | 15.7 | 8.1 | 17.4 | 25.0 |
| QC7 | 120 | 16.1 | 1.6 | 17.0 | 23.6 |
| QC8 | 120 | 13.3 | 9.1 | 20.7 | 26.5 |
| QC9 | 140 | 10.6 | 10.2 | 28.9 | 32.7 |
Analytical accuracy (linearity).
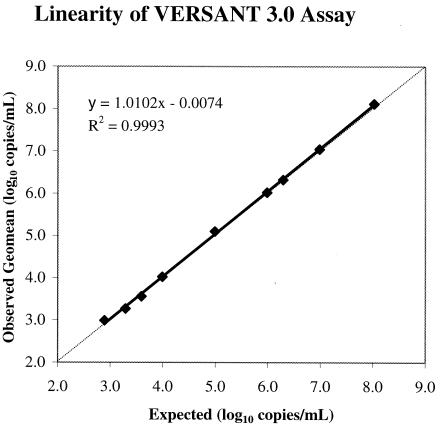
Figure 1 shows the results of the linearity assessment for VERSANT 3.0. The expected log10 values versus observed mean log10 quantitative values for each QC panel member were plotted. Linear regression fit and the middle of the data points are indicated. Absolute deviations from the assigned values were less than 0.12 log10 for QC panel members QC1 to QC8 (within the quantitative range of VERSANT 3.0). The linear regression plot had a slope of 1.01 with a correlation coefficient (R2) of 0.99. Thus, good linearity of VERSANT 3.0 was observed throughout the quantitative range of 3.3 to 8.0 log10 copies/ml for the QC panel members at all four independent testing sites.
FIG. 1.
Observed log10 mean quantitation versus log10 concentration for VERSANT 3.0.
Quantitative range.
The quantitative range of VERSANT 3.0 is defined by the lowest and highest concentrations of HBV DNA detectable with acceptable accuracy and precision. Table 3 shows the total CV, percent log10 recovery, and estimated proportion of quantitative values falling within 0.5 log10 of the value assigned for each QC panel member. The estimated proportions of the quantitative values for the QC panel members falling within 0.5 log10 from the assigned values are all 100%. These results indicate acceptable linearity and reproducibility throughout the quantitative range of the QC panel. Based upon the total precision and accuracy in the range spanning from QC9 (800 copies/ml) to QC1 (110,000,000 copies/ml), VERSANT 3.0 has an HBV DNA quantitative range of approximately 5 log10 (2.0 × 103 to 1.0 × 108) copies/ml.
TABLE 3.
Reproducibility and accuracy of VERSANT 3.0
| Panel member | Value assignment (HBV DNA copies/ml) | Observed geomean (HBV DNA copies/ml) | n | Total % CV | Log10 % recovery | Estimated proportion of log10 Quant within 0.5 log from value assignment (%) |
|---|---|---|---|---|---|---|
| QC1 | 106,332,819 | 131,394,840 | 80 | 16.0 | 0.09 | 100 |
| QC2 | 9,666,620 | 11,057,682 | 80 | 17.9 | 0.06 | 100 |
| QC3 | 1,954,243 | 2,084,651 | 80 | 14.4 | 0.03 | 100 |
| QC4 | 966,662 | 1,053,900 | 79 | 10.6 | 0.04 | 100 |
| QC5 | 97,712 | 125,825 | 80 | 14.1 | 0.11 | 100 |
| QC6 | 9,771 | 10,511 | 80 | 25.0 | 0.03 | 100 |
| QC7 | 3,908 | 3,641 | 120 | 23.6 | −0.03 | 100 |
| QC8 | 1,954 | 1,856 | 120 | 26.5 | −0.02 | 100 |
| QC9 | 782 | 978 | 140 | 32.7 | 0.10 | 100 |
Analytical sensitivity.
For each QC panel member, the number of replicate samples with HBV DNA quantitative values above the detection cutoff (2,000 copies/ml) was determined and expressed as a percentage. QC panel members with HBV DNA concentrations above 3,900 copies/ml (QC1 to QC7) had >99% of samples quantified above the DC of 2,000 copies/ml. Panel members QC8 (1,954 copies/ml) and QC9 (782 copies/ml) were observed to have 45.8 and 2.9% of replicates, respectively, quantified above the cutoff value. The LOD, defined as the lowest concentration of HBV DNA at which the percentage of samples detected above the cutoff value is ≥95%, was estimated by an interpolation between the 5th percentiles of the quantitative distribution of two panel members whose sensitivity brackets 95%. The two panel members were QC7 (99.2% detected) and QC8 (45.8% detected), and the LOD estimate was 3,321 copies/ml with an approximate 95% upper confidence limit of 4,233 copies/ml.
Comparative analysis.
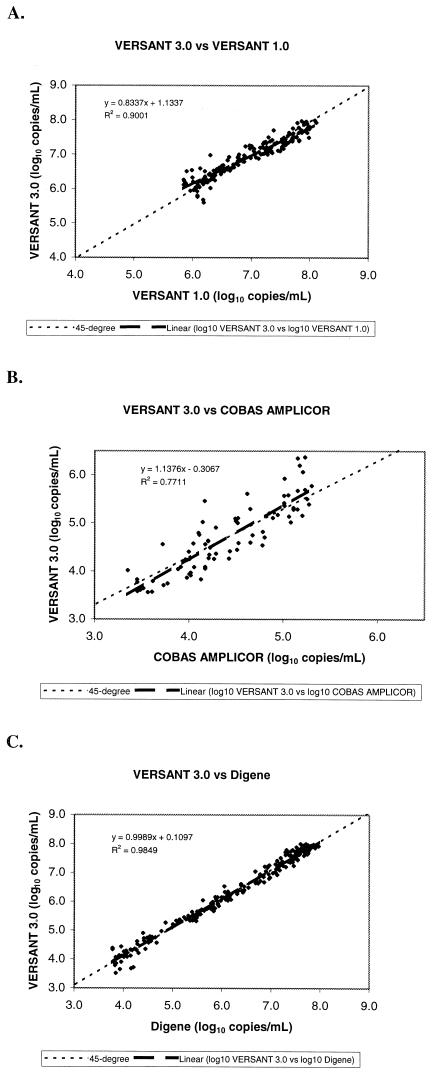
Figure 2 shows the three scatter plots of the log10 quantitative values from VERSANT 3.0 versus the log10 quantitative values obtained using each of the comparative assays. For each scatter plot, a linear regression was fitted to the data to determine the slope, and a 45° line through the middle of the data was used to evaluate distribution of the quantitative measures around the line.
FIG. 2.
Correlation of log10 quantitative HBV DNA titers between VERSANT 3.0 and VERSANT 1.0 (A), COBAS AMPLICOR (B), and Digene (C).
When samples previously analyzed by VERSANT 1.0 were tested and compared with VERSANT 3.0, the linear regression fitted to the data resulted in a slope of 0.834 and a 95% confidence interval (95% CI) of 0.793 to 0.874 (Fig. 2A). The slope was slightly significantly different from a value of 1, indicating a somewhat less than linear relationship between the two assays. Data points were narrowly distributed around the 45° line through the middle of the scatter plot, suggesting that the quantitative results from the two assays were in close agreement, with minimal bias between the two assays.
When VERSANT 3.0 was used to test HBV DNA-positive samples previously analyzed by COBAS AMPLICOR, the scatter plot of the results generated a linear regression slope of 1.138 with a 95% CI of 0.997 to 1.278 (Fig. 2B). The slope was not significantly different from a value of 1, indicating a linear relationship between these two assays with overlap of the dynamic ranges. However, the data points were not closely distributed around the 45° line through the middle of the scatter plot, indicating bias between the two assays.
Comparison of results from clinical samples previously tested by standard and ultrasensitive Digene and reanalyzed by VERSANT 3.0 showed a linear regression slope of 0.999 with a 95% CI of 0.983 to 1.014 (Fig. 2C). This plot indicates a highly linear relationship between the two assays, with the 45° line scatter plot virtually identical to the regression line. Furthermore, the data points were narrowly distributed around the 45° line, suggesting that the quantitative results from the two assays were in very close agreement with very little bias between the two assays.
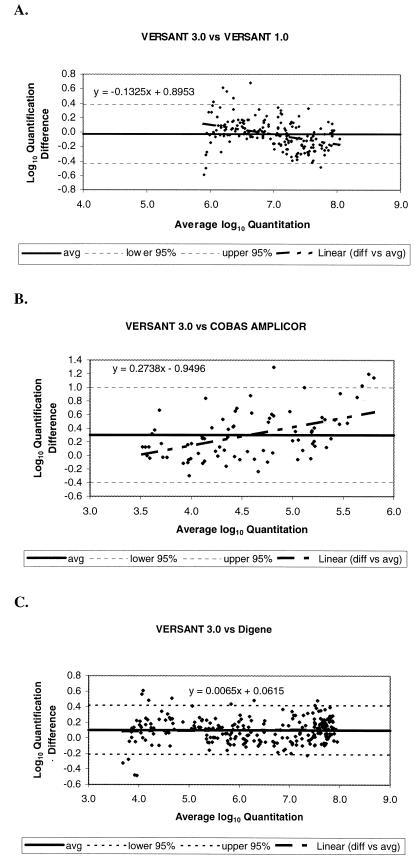
Consistency of the differences in log10 quantitative values between the assays across their overlapping dynamic ranges is assessed by the Bland-Altman plots shown in Fig. 3. The average log10 difference between the data pairs from the two bDNA assays was −0.029 log10 (Fig. 3A). The 95% CI for the individual log10 differences was −0.431 to 0.374. The regression between the log10 differences and log10 averages had a slope of 0.133 that was significantly different from 0. This analysis indicates that the average difference between the two assays is expected to decrease slightly as the concentration of virus increases. Across the range of available data in this study (5.0 to 8.9 log10 copies/ml), the average log10 difference was predicted to range from 0.11 log10 to −0.17 log10.
FIG. 3.
Differences in log10 quantitative HBV DNA titers between VERSANT 3.0 and VERSANT 1.0 (A), COBAS AMPLICOR (B), and Digene (C) in relation to average log10 quantitation.
In a similar analysis for samples tested by COBAS AMPLICOR and VERSANT 3.0, the average log10 difference between the two assays across their overlapping dynamic range was 0.301, with a 95% CI of −0.171 to 0.371 for the individual log10 differences (Fig. 3B). The regression between the log10 differences and log10 averages had a slope of 0.2738 that was statistically significantly different from 0. This trend suggests that the average difference between the two assays is expected to increase with increasing HBV DNA concentrations. The average log10 difference was predicted to be from 0.014 to 0.639 log10 across the range of data in this experiment (3.5 to 5.8 log10 copies/ml).
The average log10 difference between samples quantified by VERSANT 3.0 and Digene across an overlapping dynamic range was 0.103 log10 (Fig. 3C). The 95% CI for the individual log10 differences was −0.212 to 0.417. The regression between the log10 differences and log10 averages had a slope of 0.007 and was not statistically significantly different from 0. These results indicate that the difference between these two assays is consistent across their overlapping dynamic range. This final result differs from the nonlinear trends observed in the comparison of results between VERSANT 3.0 and VERSANT 1.0 or COBAS AMPLICOR.
DISCUSSION
The measurement of viral nucleic acid in serum and plasma has been found to be a valuable tool for the clinical management of viral infections. The availability of molecular diagnostic assays designed to quantify levels of HBV DNA has enhanced the understanding of the clinical manifestations of HBV infection and has facilitated the monitoring of viral response to therapeutic interventions (4, 22). It is generally believed that suppression of HBV DNA levels in plasma to low concentrations correlates with nonprogressive disease (4, 15). This therapeutic threshold is not well defined (104 to 106 HBV DNA copies/ml) and may be dependent upon HBV genotype and the presence of core promoter and precore mutations. Earlier viral load assays were able to quantify HBV DNA concentrations down to 105 or 106 copies/ml (21).
VERSANT 3.0 is a new bDNA assay with improved analytical sensitivity and specificity due to the use of preamplifier probes and label extender probes. The hybridization of capture and label extender probes to multiple conserved regions of the target genome provides efficient detection and quantification of the target with minimal effect from viral genomic diversity. Developmental studies revealed that HBV DNA concentrations were all within 0.5 log10 of expected values throughout a 4-log10 quantitative range for HBV genotypes A to F (unpublished data). With an estimated specificity of 99.2% and a lower 95% confidence limit of 98.3%, VERSANT 3.0 is slightly more specific than the COBAS AMPLICOR assay (97%; 95% CI, 94 to 100%) and has a specificity similar to that of the standard and ultrasensitive Digene (99.2%; 95% CI, 97.7 to100%) (22).
VERSANT 3.0 was found to be linear (slope = 1.01, R2 = 0.999) and precise (total CV ≤ 26.5%) within the assay quantitative range of 2 × 103 to 108 copies/ml. Furthermore, the LOD for this assay was determined to be 3.32 × 103 copies/ml, which is significantly improved over those of VERSANT 1.0 (range of 7.0 × 105 to 5.0 × 109 copies/ml) and Digene (standard range of 1.4 × 105 to 1.7 × 109 copies/ml; ultrasensitive range of 4.7 × 103 to 5.6 × 107 copies/ml). Of note is the difference between the LOD value and the lower limit (2 × 103 copies/ml) of the quantitative range of VERSANT 3.0. The former value was the result of combining the findings from 20 assay runs at all four testing sites using only one reagent lot, while the latter is the DC titer determined by many assay runs, different operators, and several lots of reagents during the development of this assay by the manufacturer. Although VERSANT 3.0 does not share the analytical sensitivity previously reported for the COBAS AMPLICOR assay (range of 2.0 × 102 to 2.0 × 105 copies/ml), it has a wider dynamic range that includes the current clinically relevant HBV DNA concentrations (104 to 108 copies/ml) (10, 16). The improved analytical sensitivity combined with the wide dynamic range of VERSANT 3.0 may facilitate the monitoring of HBV DNA levels in infected patients at the time of diagnosis and during treatment (including at levels below the current therapeutic threshold of 104 to 105 copies/ml) (6, 20).
Quantitative HBV DNA values generated from VERSANT 3.0 correlated very well with values previously determined by the standard and ultrasensitive Digene tests over the entire overlapping dynamic ranges of the two assays. The relationship between these two tests was highly linear, and there was little or no bias between the two methods. There was substantial bias between the VERSANT 3.0 and COBAS AMPLICOR assays, as results generated by the former were, on average, twice as high as the values quantified by the PCR assay. Furthermore, the log10 difference between these two assays was not consistent across the dynamic range, as indicated by the upward trend in the log10 difference as the HBV DNA concentration increased. This trend suggests that the average difference between HBV DNA concentrations measured using the two assays would be expected to increase as the quantitative values increase. Since VERSANT 3.0 was demonstrated to be highly linear (Fig. 1), one possible explanation for this upward trend in the relationship between VERSANT 3.0 and COBAS AMPLICOR could be the slight nonlinearity of the PCR assay as HBV DNA values approach the upper limit of its dynamic range.
Finally, the relationship between VERSANT 3.0 and VERSANT 1.0 was slightly nonlinear. However, there was only a small bias between the two assays, as the results generated by VERSANT 3.0 were, on average, 94% of corresponding quantitative values from VERSANT 1.0. The log10 difference between the two assays was not consistent across the dynamic range. The slight downward trend in the log10 difference as HBV DNA values increase could be attributed to the nonlinearity of VERSANT 1.0 as HBV DNA concentrations approach the lower limit of the dynamic range for this assay. Overall, the agreement between VERSANT 3.0 and the comparative HBV DNA quantitative assays was very good, with the greatest differences in quantitative values occurring as HBV DNA levels approached or exceeded the limits of the dynamic ranges for the comparative assays.
With the availability of new therapeutic strategies targeting HBV, the use of HBV DNA quantification by clinical laboratories to monitor and optimize treatment will increase (4, 7, 24). The results from this multicenter evaluation of the Bayer VERSANT 3.0 assay (bDNA) show that this test is highly specific and reproducible, with a wide dynamic range for accurate quantitation of HBV DNA in the serum or plasma of HBV-infected specimens. The extended linearity of this assay will facilitate monitoring of HBV viral loads prior to treatment as well as during therapy, when HBV DNA levels may fall below the therapeutic threshold of 104 to 105 copies/ml (6, 20). These characteristics make the VERSANT 3.0 assay (bDNA) an attractive test method for clinical laboratory applications.
Acknowledgments
We are grateful to the following individuals for their technical assistance and expertise in this study: Douglas Bleau, Yee Lee Tan, and Lorine Tanimoto.
REFERENCES
- 1.Beld, M., R. Sentjens, S. Rebers, C. Weegink, J. Weel, C. Sol, and R. Boom. 2002. Performance of the new Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS Amplicor HCV Monitor, version 2.0, assay. J. Clin. Microbiol. 40:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, T., C. Sarrazin, E. Herrmann, H. Hinrichsen, T. Gerlach, R. Zachoval, B. Wiedenmann, U. Hopf, and S. Zeuzem. 2003. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology 37:600-609. [DOI] [PubMed] [Google Scholar]
- 3.Bland, J. M., and D. G. Altman. 1995. Comparing two methods of clinical measurement: a personal history. Int. J. Epidemiol. 24(Suppl. 1):S7-S14. [DOI] [PubMed] [Google Scholar]
- 4.Conjeevaram, H. S., and A. S. Lok. 2003. Management of chronic hepatitis B. J. Hepatol. 38:S90-S103. [DOI] [PubMed] [Google Scholar]
- 5.De Lamballerie, X., P. Gallian, and P. H. De Micco. 1995. Evaluation of a chemiluminescent molecular hybridization assay for the detection and quantitation of hepatitis B virus-DNA in serum. New Microbiol. 18:207-213. [PubMed] [Google Scholar]
- 6.The EASL Jury. 2003. EASL International Consensus Conference on Hepatitis B. J. Hepatol. 39:S3-S25. [PubMed] [Google Scholar]
- 7.Galan, M. V., D. Boyce, and S. C. Gordon. 2001. Current pharmacotherapy for hepatitis B infection. Expert Opin. Pharmacother. 2:1289-1298. [DOI] [PubMed] [Google Scholar]
- 8.Gleaves, C. A., J. Welle, M. Campbell, T. Elbeik, V. Ng, P. E. Taylor, K. Kuramoto, S. Aceituno, E. Lewalski, B. Joppa, L. Sawyer, C. Schaper, D. McNairn, and T. Quinn. 2002. Multicenter evaluation of the Bayer VERSANT HIV-1 RNA 3.0 assay: analytical and clinical performance. J. Clin. Virol. 25:205-216. [DOI] [PubMed] [Google Scholar]
- 9.Hendricks, D. A., B. J. Stowe, B. S. Hoo, J. Kolberg, B. D. Irvine, P. D. Neuwald, M. S. Urdea, and R. P. Perrillo. 1995. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am. J. Clin. Pathol. 104:537-546. [DOI] [PubMed] [Google Scholar]
- 10.Kessler, H. H., K. Pierer, E. Dragon, H. Lackner, B. Santner, D. Stunzner, E. Stelzl, B. Waitzl, and E. Marth. 1998. Evaluation of a new assay for HBV DNA quantitation in patients with chronic hepatitis B. Clin. Diagn. Virol. 9:37-43. [DOI] [PubMed] [Google Scholar]
- 11.Krajden, M., J. Minor, L. Cork, and L. Comanor. 1998. Multi-measurement method comparison of three commercial hepatitis B virus DNA quantification assays. J. Viral Hepat. 5:415-422. [DOI] [PubMed] [Google Scholar]
- 12.Lai, C. L., M. Rosmawati, J. Lao, H. Van Vlierberghe, F. H. Anderson, N. Thomas, and D. Dehertogh. 2002. Entecavir is superior to lamivudine in reducing hepatitis B virus DNA in patients with chronic hepatitis B infection. Gastroenterology 123:1831-1838. [DOI] [PubMed] [Google Scholar]
- 13.Lok, A. S. 2000. Hepatitis B infection: pathogenesis and management. J. Hepatol. 32:89-97. [DOI] [PubMed] [Google Scholar]
- 14.Lok, A. S., M. G. Ghany, G. Watson, and B. Ayola. 1998. Predictive value of aminotransferase and hepatitis B virus DNA levels on response to interferon therapy for chronic hepatitis B. J. Viral Hepat. 5:171-178. [DOI] [PubMed] [Google Scholar]
- 15.Lok, A. S., E. J. Heathcote, and J. H. Hoofnagle. 2001. Management of hepatitis B: 2000—summary of a workshop. Gastroenterology 120:1828-1853. [DOI] [PubMed] [Google Scholar]
- 16.Lopez, V. A., E. J. Bourne, M. W. Lutz, and L. D. Condreay. 2002. Assessment of the COBAS Amplicor HBV Monitor Test for quantitation of serum hepatitis B virus DNA levels. J. Clin. Microbiol. 40:1972-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin, I. J., M. Poljak, K. Seme, V. Brinovec, M. Maticic, J. Meglic-Volkar, G. Lesnicar, and P. Gradisek. 2002. Comparative evaluation of three commercial assays for quantitative measurement of hepatitis B virus DNA in serum samples. Hepatogastroenterology 49:1390-1394. [PubMed] [Google Scholar]
- 18.Martinot-Peignoux, M., N. Boyer, M. Colombat, R. Akremi, B. N. Pham, S. Ollivier, C. Castelnau, D. Valla, C. Degott, and P. Marcellin. 2002. Serum hepatitis B virus DNA levels and liver histology in inactive HBsAg carriers. J. Hepatol. 36:543-546. [DOI] [PubMed] [Google Scholar]
- 19.Nolte, F. S. 1998. Branched DNA signal amplification for direct quantitation of nucleic acid sequences in clinical specimens. Adv. Clin. Chem. 33:201-235. [DOI] [PubMed] [Google Scholar]
- 20.Pawlotsky, J. M. 2003. Hepatitis B virus (HBV) DNA assays (methods and practical use) and viral kinetics. J. Hepatol. 39:S31-S35. [DOI] [PubMed] [Google Scholar]
- 21.Pawlotsky, J. M. 2002. Molecular diagnosis of viral hepatitis. Gastroenterology 122:1554-1568. [DOI] [PubMed] [Google Scholar]
- 22.Pawlotsky, J. M., A. Bastie, C. Hezode, I. Lonjon, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories: performance of three commercial assays. J. Virol. Methods 85:11-21. [DOI] [PubMed] [Google Scholar]
- 23.Pumpens, P., E. Grens, and M. Nassal. 2002. Molecular epidemiology and immunology of hepatitis B virus infection: an update. Intervirology 45:218-232. [DOI] [PubMed] [Google Scholar]
- 24.Wai, C. T., and A. S. Lok. 2002. Treatment of hepatitis B. J. Gastroenterol. 37:771-778. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization Department of Communicable Diseases Surveillance and Response. 2002. Hepatitis B. WHO/CDS/CSR/LYO/2002.2: Hepatitis B:8. World Health Organization, Geneva, Switzerland. [Online.] http://www.who.int/emc-documents/hepatitis/docs/whocdsrlyo20022/index.html.