Abstract
Type IV pili are thin filaments that extend from the poles of a diverse group of bacteria, enabling them to move at speeds of a few tenths of a micrometer per second. They are required for twitching motility, e.g., in Pseudomonas aeruginosa and Neisseria gonorrhoeae, and for social gliding motility in Myxococcus xanthus. Here we report direct observation of extension and retraction of type IV pili in P. aeruginosa. Cells without flagellar filaments were labeled with an amino-specific Cy3 fluorescent dye and were visualized on a quartz slide by total internal reflection microscopy. When pili were attached to a cell and their distal ends were free, they extended or retracted at rates of about 0.5 μm s−1 (29°C). They also flexed by Brownian motion, exhibiting a persistence length of about 5 μm. Frequently, the distal tip of a filament adsorbed to the substratum and the filament was pulled taut. From the absence of lateral deflections of such filaments, we estimate tensions of at least 10 pN. Occasionally, cell bodies came free and were pulled forward by pilus retraction. Thus, type IV pili are linear actuators that extend, attach at their distal tips, exert substantial force, and retract.
Keywords: Pseudomonas aeruginosa, twitching motility, fluorescence
Motile bacteria swim, swarm, or glide. Gliding refers to movement on a solid surface, such as glass or hard agar, without flagella, usually in the direction of the long axis of a cell (1). One form of gliding, known either as twitching motility or as social gliding, depends on 6-nm diameter filaments called type IV pili that extend from the pole of a cell (2–6). Twitching cells can exhibit chemotaxis (7) and/or phototaxis (8). Twitching motility is required for formation of biofilms (9), and social gliding is important for formation of fruiting bodies (10).
We studied twitching motility in Pseudomonas aeruginosa, a common Gram-negative bacterium that acts as an opportunistic human pathogen. It causes bacteremia in burn victims, infections of the urinary tract in catheterized patients, chronic lung infections in patients with cystic fibrosis, and acute ulcerative keratitis in users of extended-wear soft-contact lenses (11, 12). P. aeruginosa has a single flagellum and multiple type IV pili; thus, it can move by swimming or twitching motility. Pili-mediated adhesion is a known virulence factor (13) and the molecular genetics of pilus assembly has been studied extensively (14). It is thought that twitching motility allows cells to spread on body surfaces during infection (6).
Type IV pili have been seen with an electron microscope (15, 16) but have not been visualized in vivo. Recently, the motion of bacterial flagella was analyzed by labeling intact cells with amino-specific fluorescent dyes (17). By labeling cells of P. aeruginosa with an amino-specific Cy3, we have observed directly pilus extension, pilus retraction, and retraction-mediated cell movement. Our work confirms and extends that of Mertz et al. (18), who used an optical trap to measure the force of retraction of pili of Neisseria gonorrhoeae linked by anti-pilin Abs to latex beads, and of Sun et al. (19), who analyzed the motion of cells tethered by pili in Myxococcus xanthus.
Materials and Methods
Bacterial Strains.
All P. aeruginosa strains were derived from the clinical isolate PAK (S. Lory, Harvard Medical School, Boston). Cells of P. aeruginosa swim by means of a single polar flagellum. Twitching motility is flagella-independent; therefore, synthesis of this filament was suppressed by insertion of a gentamycin-resistance cassette into the flagellin (fliC) locus. We generated strain PAKfliC∷gmR by electroporation of plasmid pMS590 (S. Lory). In a similar fashion, the fliC locus was disrupted in PAKΔpilA (20) to generate PAKfliC∷gmRΔpilA.
Cy3 Labeling.
A culture was grown in 20 ml of LB (1% Bacto–tryptone/0.5% yeast extract/0.5% NaCl) at 37°C with rotary shaking at 250 rpm. Cells (2 ml) were harvested in exponential phase (OD600 = 2.0) and washed 3 times in labeling buffer (50 mM KPO4, pH 8.0/5 mM MgCl2/25 μM EDTA) by gentle centrifugation. After the third wash, the pellet was resuspended in 0.5 ml of labeling buffer containing 1 vial of Cy3 monofunctional succinimidyl ester (enough dye to label 1 mg of protein; Amersham Pharmacia), an amino-specific fluorescent dye. After 1 h at room temperature, samples were washed 3 times and resuspended in a minimal growth medium—mineral salts (21) supplemented with a nitrogen and carbon source (0.2% KNO3/0.5% sodium succinate; ref. 22). A 1-μl sample was placed on a quartz slide and covered with an 18-mm square glass coverslip sealed at its edges with Valap (1:1:1 petroleum jelly/lanolin/paraffin), yielding a fluid depth of about 3 μm.
Fluorescence Microscopy.
The condenser of a Nikon Optiphot microscope was replaced with a 2.5-cm 60° silica prism. The quartz slide was mated to the top of this prism with glycerol. Light from a 532-nm diode laser (BWT-20; 23 mW; B & W Tek, Newark, DE) was reflected by a mirror into the side of the prism at an angle just yielding total internal reflection at the quartz–water interface. The light was polarized circularly (with a quarter-wave plate), attenuated by half to reduce bleaching (with a neutral-density filter), and focused (with an 88-mm focal-length lens) to a small spot. Cells were observed with a Nikon PlanApo 60/1.2 water-immersion objective, with a long-pass emission filter (Chroma HQ545 LP) inserted between the objective and the camera. The prism and objective turret were temperature-controlled and the temperature at the sample was checked with a small thermocouple. Images were recorded on Hi-8 tape with a CCD camera run with automatic gain control in 10-frame integration mode (WV-BP550, Panasonic).
Image Analysis.
Tapes were digitized at 1-s intervals with a G3 Power Macintosh equipped with a Scion (Frederick, MD) LG-3 frame grabber, using Scion image (version 1.62c). Distance measurements were made with the line tool of this program calibrated against an objective micrometer. For each extension or retraction event, only the initial and final lengths were used to calculate rates. If the motion was discontinuous, e.g., if a pilus retracted, attached at its tip, came free, and then continued to retract, the rate was computed as the average over successive intervals. Filament straightness was judged by eye and straightedge with enlarged prints. The mean velocity of a cell was determined from measurements of displacements of its body made over at least 10 consecutive intervals.
Results
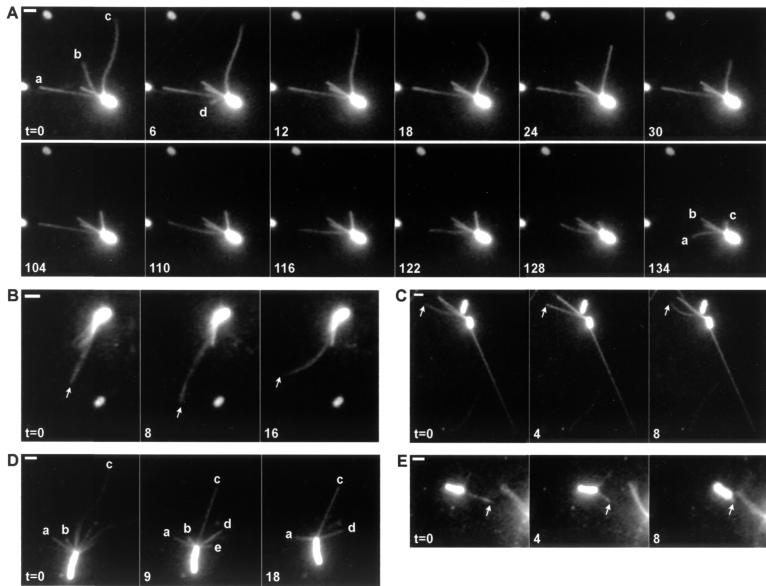
Twitching motility in P. aeruginosa occurs efficiently at either a plastic–agar or glass–agar interface, where single cells can move at speeds up to 0.2 μm s−1 (4). Using a nonflagellate strain, we monitored pilus function at a quartz–water interface, which simplified imaging. When cells of this strain were labeled with Cy3, they showed filaments emerging from the poles, usually at one end only. No filaments were observed on a pilA mutant, demonstrating that the filaments were type IV pili. Filaments attached to a cell body could extend and retract, and if free at their distal tips, exhibit thermal fluctuations in shape (Fig. 1 A–C). When a filament attached to the quartz surface at its distal tip, it was pulled straight. After release of the tip, it could continue to retract. Alternatively, the cell body was pulled forward (Fig. 1 D and E).
Figure 1.
Visualization of Cy3-labeled pili on nonflagellate cells of P. aeruginosa. (A and B) Filament extension and retraction. (C) Filaments under tension. (D and E) Cell movement. Elapsed time (t) in s. (Bars = 2 μm.)
Pili on individual cells changed their lengths independently. For example, in Fig. 1A for t = 0 to 30 s, filament a remained extended and taut, whereas filament c retracted. Filament d extended during the first 6 s and then retracted, disappearing by t = 12 s. Filament c attached briefly at its distal tip at about t = 24 s. Between t = 30 and 104 s (not shown), filaments a and b remained the same length, whereas filament c continued to shorten. At t = 104 s, filament c attached at its tip. At t = 110 s, filament a let go and began to retract. Between t = 122 and 128 s, filament c let go and retracted.
Alternations between extension and retraction could be quite abrupt. For example, one filament in Fig. 1B (tip marked by arrow) extended during the first 8 s and retracted during the next 8 s. Also, when pili emerged from the cell body they looked as bright as pili that already had extended. This is shown by filament d in Fig. 1A. These results imply that fluorescent subunits are recycled, consistent with current models of pilus assembly that use a membrane pool of pilin subunits (23–25). However, the cell bodies were so highly fluorescent that we were not able to tell where this pool might be located.
Pili that were broken off of cells exhibited thermal fluctuations in shape but never changed their length, as shown in Fig. 2. Computation of the cosine correlation function for this filament gave a persistence length of about 5 μm, similar to that of actin (26).
Figure 2.
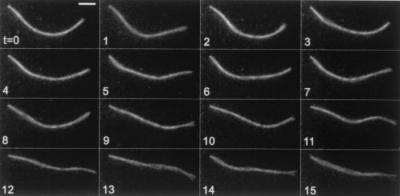
A broken pilus exhibiting thermal fluctuations in shape over a time span of 15 s. This filament was attached to the substratum at its left end. Elapsed time (t) in s. (Bar = 2 μm.)
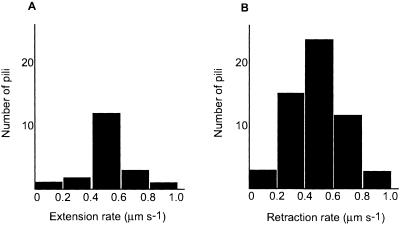
We determined the extension and retraction rates for individual pili found on 29 different cells, obtaining the results shown in Fig. 3. The mean extension rate was 0.50 ± 0.18 μm s−1 (mean ± SD; n = 17) and the mean retraction rate was 0.48 ± 0.18 μm s−1 (n = 57). The two distributions had similar shapes.
Figure 3.
Histograms of rates of pilus extension and retraction. (A) Extension of 17 pili. (B) Retraction of 57 pili.
Cells moved by extending pili, letting them attach at their distal ends, and retracting them. Extension did not seem to cause any cell movement; pili probably are too flexible to push a cell forward. Retraction, however, generated motion that was discontinuous. Motion of the cell body occurred in excursions of about 1–6 μm. The velocity for 11 such events was 0.31 ± 0.21 μm s−1 (mean ± SD), which is similar to the speed observed for twitching motility (3, 6). Examples are shown in Fig. 1 D and E. In Fig. 1D, filaments a and b both shortened while remaining attached at their tips. Filaments c and d were initially free but then attached and pulled the cell forward. Filaments b and e retracted fully, disappearing from view. In Fig. 1E, a single filament (arrow) pulled the cell forward. Because the cell began by moving to the right rather than downwards to the right, a second filament must have been involved early on. We do not know whether the differences in retraction rates of free and attached pili are significant; however, we would expect retraction rates to be smaller under load. The load depends, in turn, on how the cell body interacts with the substratum. Because most cells failed to move at all, the interaction with quartz must be relatively strong.
Evidently, the distal tip of a pilus serves as a nonspecific adhesin. Broken pili often attached to quartz at one end, as shown by the filament in Fig. 2. Pili that protruded from cells also attached to quartz, but only at their distal ends (Fig. 1 A and C–E). Lee et al. (27) have demonstrated that the C-terminal disulfide loop region of the pilin subunit of P. aeruginosa is exposed at the distal tip of the filament and can bind certain glycosphingolipids.
We estimated the retraction force by examining the lateral displacements of filaments at their midpoints, assuming that they are flexible, like threads. This displacement, caused by thermal motion, is smaller the higher the tension. If a thread of length, L, is put under tension, F, and then is pulled sideways in the middle a small distance, δy, the restoring force is (4F/L)δy. The thread behaves like a spring with spring constant (4F/L). The energy stored in this spring for total displacement y is (2F/L)y2. By equipartition, the mean value of this energy is equal to kT/2, where k is Boltzmann's constant and T is the absolute temperature (28). Therefore, F = kTL/(4〈y2〉), where 〈y2〉 is the mean-square displacement. None of the attached filaments seemed to flex. Assuming that we could have seen displacements of one-fourth of the width of the image of a filament (y = 0.05 μm), we obtain for the filament of greatest length (L = 25 μm; Fig. 1C) F > 10 pN. This estimate is lower than forces observed for some retraction events in N. gonorrhoeae of 80 pN or more (18). However, those experiments were done by measuring the retraction force between an immobilized cell and a 1-μm bead coated with anti-pilin mAb held by an optical trap. Therefore, more than one pilus might have been involved. Unambiguous results could be obtained by combining our techniques, i.e., by pulling beads attached to single fluorescent pili.
Discussion
Pilus retraction was first proposed by Marvin and Hohn (29) to account for the disappearance of F pili after infection with bacteriophage and for the association of mating cells during conjugation. According to their model, adsorption of phage to the side or tip of a pilus, or binding of an F− cell to the tip, triggers retraction. Subsequent studies in Escherichia coli (30, 31), P. aeruginosa (32–34), and Caulobacter crescentus (35, 36) strengthened the hypothesis that pilus retraction plays a role in the translocation of pili-specific phage to the cell surface. The role of pilus retraction in cell motility was first proposed by Bradley (3). Our experiments leave little doubt that twitching motility in P. aeruginosa is driven by type IV pilus retraction. We expect our method of visualizing bacterial pili to be widely applicable. Using fluorescent labels, one now should be able to visualize movement of both phages and pili during infection, or of cells and pili during conjugation. It should also be possible to look at extension and retraction of pili in M. xanthus to test the hypothesis that cells glide forward and backward by activating pili at opposite ends of the cell (19). Similar experiments could be done with P. aeruginosa. Finally, we can learn whether groups of cells glide or twitch by extending pili from one to another (10).
Movie files are available at http://www.rowland.org.
Acknowledgments
We thank M. Wolfgang and S. Lory for strains and advice; W. S. Ryu for computations of persistence lengths and for comments on the manuscript; and A. L. Greenwood for comments on the manuscript also. This work was supported by the Rowland Institute for Science.
References
- 1.Henrichsen J. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strom M S, Lory S. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 3.Bradley D E. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 4.Semmler A B, Whitchurch C B, Mattick J S. Microbiology. 1999;145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 5.Wall D, Kaiser D. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 6.Henricksen J. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 7.Kearns D B, Robinson J, Shimkets L J. J Bacteriol. 2001;183:763–767. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaya D, Watanabe N, Ogawa T, Grossman A R. Proc Natl Acad Sci USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Toole G A, Kolter R. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 10.Spormann A M. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyczak J B, Cannon C L, Pier G B. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 12.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, et al. Nature (London) 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 13.Hahn H P. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 14.Mattick J S, Whitchurch C B, Alm R A. Gene. 1996;179:147–155. doi: 10.1016/s0378-1119(96)00441-6. [DOI] [PubMed] [Google Scholar]
- 15.Bradley D E. Genet Res. 1972;19:39–51. [Google Scholar]
- 16.Weiss R L. J Gen Microbiol. 1971;67:135–143. doi: 10.1099/00221287-67-2-135. [DOI] [PubMed] [Google Scholar]
- 17.Turner L, Ryu W S, Berg H C. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertz A J, So M, Sheetz M P. Nature (London) 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Zusman D R, Shi W. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 20.Kagami Y, Ratliff M, Surber M, Martinez A, Nunn D N. Mol Microbiol. 1998;27:221–233. doi: 10.1046/j.1365-2958.1998.00679.x. [DOI] [PubMed] [Google Scholar]
- 21.Moulton R C, Montie T C. J Bacteriol. 1979;137:274–280. doi: 10.1128/jb.137.1.274-280.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craven R, Montie T C. J Bacteriol. 1985;164:544–549. doi: 10.1128/jb.164.2.544-549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 24.Wolfgang M, van Putten J P, Hayes S F, Dorward D, Koomey M. EMBO J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser D. Curr Biol. 2000;10:R777–R780. doi: 10.1016/s0960-9822(00)00764-8. [DOI] [PubMed] [Google Scholar]
- 26.Ott A, Magnasco M, Simon A, Libchaber A. Phys Rev E. 1993;48:R1642–R1645. doi: 10.1103/physreve.48.r1642. [DOI] [PubMed] [Google Scholar]
- 27.Lee K K, Sheth H B, Wong W Y, Sherburne R, Paranchych W, Hodges R S, Lingwood C A, Krivan H, Irvin R T. Mol Microbiol. 1994;11:705–713. doi: 10.1111/j.1365-2958.1994.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 28.Reif F. Statistical Physics. New York: McGraw–Hill; 1965. p. 250. [Google Scholar]
- 29.Marvin D A, Hohn B. Bacteriol Rev. 1969;33:172–209. doi: 10.1128/br.33.2.172-209.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson A. J Virol. 1972;10:835–843. doi: 10.1128/jvi.10.4.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novotny C P, Fives-Taylor P. J Bacteriol. 1974;117:1306–1311. doi: 10.1128/jb.117.3.1306-1311.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradley D E. Biochem Biophys Res Commun. 1972;47:142–149. doi: 10.1016/s0006-291x(72)80021-4. [DOI] [PubMed] [Google Scholar]
- 33.Bradley D E. J Gen Microbiol. 1972;72:303–319. doi: 10.1099/00221287-72-2-303. [DOI] [PubMed] [Google Scholar]
- 34.Bradley D E. Virology. 1974;58:149–163. doi: 10.1016/0042-6822(74)90150-0. [DOI] [PubMed] [Google Scholar]
- 35.Sommer J M, Newton A. J Bacteriol. 1988;170:409–415. doi: 10.1128/jb.170.1.409-415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skerker J M, Shapiro L. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]