Abstract
Multiplex PCR was established for differential diagnosis of taeniasis and cysticercosis, including their causative agents. For identification of the parasites, multiplex PCR with cytochrome c oxidase subunit 1 gene yielded evident differential products unique for Taenia saginata and Taenia asiatica and for American/African and Asian genotypes of Taenia solium with molecular sizes of 827, 269, 720, and 984 bp, respectively. In the PCR-based detection of tapeworm carriers using fecal samples, the diagnostic markers were detected from 7 of 14 and 4 of 9 T. solium carriers from Guatemala and Indonesia, respectively. Test sensitivity may have been reduced by the length of time (up to 12 years) that samples were stored and/or small sample volumes (ca. 30 to 50 mg). However, the diagnostic markers were detected by nested PCR in five worm carriers from Guatemalan cases that were found to be negative by multiplex PCR. It was noteworthy that a 720 bp-diagnostic marker was detected from a T. solium carrier who was egg-free, implying that it is possible to detect worm carriers and treat before mature gravid proglottids are discharged. In contrast to T. solium carriers, 827-bp markers were detected by multiplex PCR in all T. saginata carriers. The application of the multiplex PCR would be useful not only for surveillance of taeniasis and cysticercosis control but also for the molecular epidemiological survey of these cestode infections.
Taenia solium, Taenia saginata, and Taenia asiatica are known as causative agents of taeniasis in humans. T. solium also causes cysticercosis in humans. In particular, neurocysticercosis caused by larval T. solium cysticerci developed in the central nervous system is the most serious disease characterized by diverse neurologic symptoms, most commonly epileptic seizure (5, 20). In contrast to cysticercosis, taeniasis is relatively innocuous, with the adult stages of these cestodes infecting the small intestine of humans and causing a few specific symptoms, such as abdominal pain and nausea (15). However, gravid proglottids filled with eggs expelled from tapeworm carriers serve as a new source of infection for intermediate hosts, particularly in developing countries where sanitary conditions are poor. Therefore, early detection and adequate treatment of taeniasis is important for the prevention of cysticercosis infections. Furthermore, reliable epidemiological information for use in the effective control of taeniasis or cysticercosis, including accurate tools for parasite identification, is needed. To date, proglottids and scolices expelled from tapeworm carriers or cysticerci collected from intermediate hosts have been identified morphologically. In Asian regions, however, T. saginata and T. asiatica are frequently confused due to their morphological similarities. Moreover, a recent study demonstrates that two distinct genotypes of T. solium exist, i.e., Asian and American/African genotypes (14).
DNA differential diagnosis is considered very useful for the accurate identification of human taeniid cestode samples. In order to overcome the limitations of identification of taeniid cestodes based on the morphology, various molecular approaches have been developed, including the use of DNA probes (4, 6, 7, 9, 16, 17), PCR, or PCR coupled to restriction fragment length polymorphism (3, 8, 13, 18, 23). Such molecular approaches have been recently reviewed (11). However, most of these studies have focused on the differentiation of T. solium from T. saginata and, to date, no copro-PCR test for human taeniasis has been developed. Most recently, a new method based on the thymine bases of mitochondrial genes has been developed for comprehensive differential diagnosis of T. saginata, T. asiatica, and two genotypes of T. solium (22, 23). That method, although it provides precise diagnostic results, is somewhat complicated. In the present study, a more simple and reliable multiplex PCR has been established for differential diagnosis of causative agents of taeniasis and cysticercosis. In addition, the method was also assessed for specific detection of Taenia spp. DNA in fecal samples from tapeworm carriers.
MATERIALS AND METHODS
Parasite materials.
For molecular identification of taeniid cestode parasites, a total of 57 taeniid parasite materials, including proglottids, cysticerci, and eggs, were examined (Table 1). Proglottids were obtained from tapeworm carriers. Of 26 cysticerci examined, 20 were collected from naturally infected intermediate hosts, and the remaining 6 were from nonobese diabetic/Shi-severe combined immunodeficiency (NOD/Shi-scid) mice as animal models for cysticercosis (10). A taeniid egg sample from Yunnan province, China was also analyzed as same as other materials. The viable eggs were prepared from gravid proglottids collected from different tapeworm carriers, and then oncospheres hatched in vitro were injected into the peritoneal cavity of NOD/Shi-scid mice. After 5 to 6 months, cysticerci developed in such mice were recovered and identified individually by multiplex PCR. All parasite materials were kept in absolute ethanol at −30°C for DNA preparation after collection.
TABLE 1.
Taeniid cestode samples examined in the present study
| Species (n) | Developmental stage (no. of samples examined) | Locality | Accession no.c for cox1 | Lane no.d | Comment |
|---|---|---|---|---|---|
| T. saginata (13) | Cysticercus (1) | Brazil (Mato Grosso do Sul) | AB107246 | 1 | |
| Proglotiid (1) | Brazil | AB107237 | 2 | ||
| Proglottid (1) | Ecuador | AB107238 | 3 | ||
| Proglottid (1) | Ethiopia | AB107241 | 4 | Expelled after treatment in Japan | |
| Cysticercus (1) | Belgium | AB107242 | 5 | Developed in NOD/Shi-scid mice | |
| Proglottid (1) | Indonesia (Bali) | AB107240 | 6 | ||
| Proglottid (3) | Thailand (Bangkok) | AB107244 and AB107245 | 7 | ||
| Proglottid (2) | Nepal (Katumandu) | AB107243 | 8 | ||
| Cysticercus (1) | China (Xinjiang) | AB066495 | 9 | ||
| Proglottid (1) | China (Yunnan) | AB107247 | 10 | ||
| T. saginata and T. asiatica eggsa | China (Yunnan) | AB107247 and AB107235 | 11 | ||
| T. asiatica (13) | Cysticercus (1) | Taiwan | AB066494 | 12 | Developed in NOD/Shi-scid mice |
| Proglottid (1) | Taiwan | AB107234 | |||
| Cysticercus (2) | South Korea | AB107236 | 13 | Developed in NOD/Shi-scid mice | |
| Cysticercus (1)b | China (Yunnan) | AB107235 | 14 | Developed in NOD/Shi-scid mice | |
| Proglottid (7) | China (Yunnan) | AB107234 | 15 | ||
| Cysticercus (1)b | Indonesia (Bali) | AB107236 | 16 | Developed in NOD/Shi-scid mice | |
| T. solium (30) | Cysticercus (1) | Brazil | AB066492 | 17 | |
| Cysticercus (1) | Bolivia (Santa Cruz) | AB066491 | 18 | ||
| Proglottid (1) | Peru | AB066490 | 19 | ||
| Cysticercus (2) | Ecuador | AB066491 | 20 | ||
| Cysticercus (1) | Mexico | AB066490 | 21 | ||
| Proglottid (1) | Mexico | AB066490 | 22 | ||
| Cysticercus (2) | Mozambique | AB066493 | 23 | ||
| Cysticercus (1) | Tanzania | AB066493 | 24 | ||
| Cysticercus (1) | Cameroon | AB066490 | 25 | ||
| Cysticercus (1) | Thailand | AB066487 | 26 | ||
| Cysticercus (1)b | Indonesia (Irian Jaya) | AB066488 | 27 | Developed in NOD/Shi-scid mice | |
| Proglottid (1) | Indonesia (Irian Jaya) | AB066488 | 28 | ||
| Cysticercus (4) | India (Panjab) | AB066489 | 29 | ||
| Cysticercus (2) | India (Vellore) | AB066489 | |||
| Proglottid (7) | China (Yunnan) | AB066486 | 30 | ||
| Proglottid (1) | China (Yunnan) | AB066486 | 31 | ||
| Cysticercus (2) | China (Jiling) | AB066485 | 32 |
The eggs were injected into immunodeficient mice for DNA analysis of developed cysticerci.
The materials were lysed in 0.02 N NaOH at 90°C for 10 min.
The numbers were deposited in DDBJ/EMBL/GenBank databases.
These numbers correspond to the lanes in Fig. 1.
Fecal samples.
T. solium-infected fecal samples were obtained from the following sources: 14 from Guatemala were collected in 1991 and 1994 and 9 from Papua (formerly Irian Jaya), Indonesia, were collected in 2000 (Table 2). In the cases from Guatemala, all samples were from worm carriers diagnosed by copro-antigen detection where T. solium was identified morphologically after treatment and stored at −20 to −30°C prior to testing (2). In one sample from Bali, Indonesia, a T. solium carrier was suspected. One sample was from a carrier who expelled only immature (nongravid) tapeworms, including four scolices and strobillae from at least seven worms (code F13 in Table 2). The fecal samples from Papua, Indonesia, where T. saginata has not been detected, had been proven to be positive for T. solium by using copro-antigens based on diagnosis by using a commercial kit (Ag-ELISA) that utilized anti-T. solium antibodies (Virotech, Rüsselshein, Germany). In addition, stool samples from four volunteers infected with T. saginata in 2002 were also examined. The fecal samples from seven noninfected volunteers from the United Kingdom collected in 2002 were used as negative controls. These fecal samples were also stored at −20 to −30°C until used. Fecal samples from Guatemala and the United Kingdom were tested as a blind test.
TABLE 2.
Summary of multiplex PCR diagnoses for detection of tapeworm carriers
| Code | Yra | Status | Origin of carrier | Copro-antigenc | Multiplex PCR finding |
|---|---|---|---|---|---|
| F1 | 1994 | T. solium | Guatemala | +b | −e |
| F2 | 1994 | T. solium | Guatemala | + | T. soliumd |
| F4 | 1994 | T. solium | Guatemala | + | T. solium |
| F6 | 1994 | T. solium | Guatemala | + | T. soliumd |
| F7 | 1994 | T. solium | Guatemala | + | T. soliumd |
| F8 | 1994 | T. solium | Guatemala | + | T. soliumd |
| F9 | 1991 | T. solium | Guatemala | + | − |
| F10 | 1994 | T. solium | Guatemala | + | T. solium |
| F12 | 1994 | T. solium | Guatemala | + | T. solium |
| F13 | 1994 | T. solium | Guatemala | + | T. soliumd |
| G1 | 1994 | T. solium | Guatemala | + | T. solium |
| G6 | 1994 | T. solium | Guatemala | + | T. solium |
| G7 | 1994 | T. solium | Guatemala | + | T. solium |
| G8 | 1994 | T. solium | Guatemala | + | T. solium |
| K03 | 2000 | T. solium | Indonesia (Papua) | + | T. solium |
| K38 | 2000 | T. solium | Indonesia (Papua) | + | T. solium |
| K44 | 2000 | T. solium | Indonesia (Papua) | + | − |
| K50 | 2000 | T. solium | Indonesia (Papua) | + | − |
| K54 | 2000 | T. solium | Indonesia (Papua) | + | T. solium |
| K56 | 2000 | T. solium | Indonesia (Papua) | + | − |
| K87 | 2000 | T. solium | Indonesia (Papua) | + | T. solium |
| K90 | 2000 | T. solium | Indonesia (Papua) | + | − |
| K10 | 2000 | T. solium | Indonesia (Papua) | + | − |
| F3 | 1995 | T. solium | Indonesia (Bali) | NDc | T. saginata |
| N1 | 2002 | T. saginata | United Kingdom | + | T. saginata |
| N2 | 2002 | T. saginata | Japan | − | T. saginata |
| N3 | 2002 | T. saginata | Brazil | + | T. saginata |
| N4 | 2002 | T. saginata | China | − | T. saginata |
| F5 | 2002 | Noninfectious | United Kingdom | ND | − |
| F11 | 2002 | Noninfectious | United Kingdom | ND | − |
| G3 | 2002 | Noninfectious | United Kingdom | ND | − |
| G4 | 2002 | Noninfectious | United Kingdom | ND | − |
| G9 | 2002 | Noninfectious | United Kingdom | ND | − |
| G10 | 2002 | Noninfectious | United Kingdom | ND | − |
| G12 | 2002 | Noninfectious | United Kingdom | ND | − |
Year in which the stool samples were collected.
Data from Guatemala are based on the report by Allan et al. (2).
ND, not done. +, positive; −, negative.
Detected by nested PCR with genotype-specific primers.
−, negative.
DNA extraction.
Mitochondrial DNA (mtDNA) from parasite materials was prepared by using DNeasy tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction. However, three cysticerci obtained from NOD/Shi-scid mice were lysed in 50 μl of 0.02 N sodium hydroxide at 90°C for 10 min, and the resulting supernatants were used directly as template DNA (Table 1). Copro-DNAs from fecal samples of tapeworm carriers were extracted by using the QIAamp DNA Stool Minikit, which requires at least 0.2 g of feces (Qiagen). Only small amounts (ca. 30 to 50 mg) in five of nine fecal samples from Papua, Indonesia, were available, and these were probably insufficient to extract copro-DNA. In addition, to evaluate the detection limit of taeniid DNA in fecal samples, a given number of T. saginata viable eggs (1, 2, 4, 10, 20, 200, and 2,000) were experimentally mixed with 0.2 g of stool samples from a noninfected volunteer under the microscope, and DNA derived from the eggs was extracted by using the same kit.
Multiplex PCR.
Cytochrome c oxidase subunit 1 gene (cox1) was used as a target gene. Based on the nucleotide sequences of cox1 from human taeniid cestodes, the following forward primers were designed to be amplified different sizes of products; 5′-TTGATTCCTTCGATGGCTTTTCTTTTG-3′, specific for T. saginata (Tsag, positions 322 to 348 from AB066495); 5′-ACGGTTGGATTAGATGTTAAGACTA-3′, specific for T. asiatica (Tasia, positions 880 to 904 from AB066494); 5′-GGTAGATTTTTTAATGTTTTCTTTA-3′, specific for the T. solium American/African genotype (Tsol/Amer, positions 429 to 453 from AB066485 to AB006489); and 5′-TTGTTATAAATTTTTGATTACTAAC-3′, specific for the T. solium Asian genotype (Tsol/Asia, positions 165 to 189 from AB066490 to AB066492). Species- or genotype-specific nucleotides were introduced at the 3′ end of each forward primer. Reverse primer (Rev, 5′-GACATAACATAATGAAAATG-3′, positions 1148 to 1129) were common to all species. A PCR cocktail contained mixed primers and 0.5 U of the Ex TaqDNA polymerase Hot Start (TaKaRa, Tokyo, Japan) in 25 μl of a reaction mixture. Standard multiplex PCR protocols consisted of 35 cycles of denaturation (30 s at 94°C), annealing (30 s at 60°C), and extension (90 s at 72°C), plus one cycle of 5 min at 72°C. Subsequently, PCR-amplified products were electrophoresed on 0.9 to 1.0% agarose gels. In the multiplex PCR with copro-DNA samples for detection of worm carriers, a volume of PCR cocktail was 50 μl to minimize the effect of inhibitory substances contained in the copro-DNA samples. Annealing was performed at 56°C. In multiplex PCR-negative samples from Guatemala, nested PCR with species- and genotype-specific primers was followed by conventional PCR with Cox1/F and Cox 1/R primers (14).
DNA sequencing and sequence analysis.
T. solium examined in the present study had been identified previously based on the nucleotide sequences of cox1 and cytochrome b gene (14, 22). In the present study, in order to confirm whether diagnostic results obtained by multiplex PCR were reliable or not, the complete nucleotide sequences of cox1 from T. saginata (12 of 13 samples) and T. asiatica (12 of 13 samples) were determined. For this purpose, cox1 (∼1.8-kb fragment) was amplified with Cox I/F and Cox I/R primers, and samples for DNA sequencing were prepared by using an ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster, Calif.) if necessary. In addition, the products amplified by multiplex and nested PCR with copro-DNA samples were also directly sequenced. In some cases, the PCR fragments were subcloned into pT7Blue T-vector (Novagen, Darmstadt, Germany) if necessary. DNA sequencing was performed on an ABI Prism 310 genetic analyzer, and the nucleotide sequence data were analyzed by using DNASTAR (version 3.75).
Nucleotide sequences.
The nucleotide sequences determined in the present study have been deposited in DDBJ/EMBL/GenBank databases under accession numbers AB107237 to AB107247 (T. saginata) and AB107234 to AB107236 (T. asiatica).
RESULTS
Differentiation of the causative agents of taeniasis and cysticercosis by multiplex PCR.
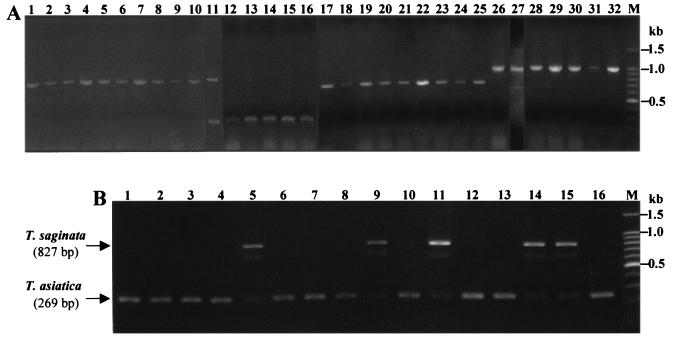
The amplification of diagnostic cox1 fragments, particularly for the Asian genotype of T. solium, was dependent on the nucleotide sequences of the primers used. Although three forward primers (positions 165 to 189, 666 to 690, and 701 to 723) for T. solium Asian genotype were tested, the use of one primer (positions 165 to 189) provided the best amplification of target gene fragments (data not shown). The successful amplification of diagnostic products was also dependent on the ratio of the forward and reverse primers and the optimal ratio was 1:2:4 for Tsag and Tsol/Amer (0.2 pmol), Tasia and Tsol/Asia (0.4 pmol), and a reverse primer (0.8 pmol), respectively. Under this condition, multiplex PCR with mtDNA prepared from parasite materials yielded the most evident results. As shown in Fig. 1A, the diagnostic products with molecular sizes of 827, 269, 720, and 984 bp were amplified in T. saginata (lanes 1 to 10) and T. asiatica (lanes 12 to 16) and in the American/African (lanes 17 to 25) and Asian (lanes 26 to 32) genotypes of T. solium, respectively. Two diagnostic bands (827 and 269 bp) were detected when a taeniid egg sample from Yunnan province, China, was tested (Fig. 1A, lane 11). This suggested that T. saginata and T. asiatica were mixed in the original proglottid samples. In order to verify this, therefore, the taeniid oncospheres hatched in vitro were injected into NOD/Shi-scid mice and allowed to develop in such mice. As shown in Fig. 1B, multiplex PCR with individual mtDNA prepared from a total 28 cysticerci recovered from two mice has been proven that T. saginata and T. asiatica were included in the original sample. Of 28 cysticerci, 12 and 16 were determined to be T. saginata (Fig. 1B, lanes 5, 9, 11, 14, and 15) and T. asiatica (Fig. 1B, lanes 1 to 4, 6 to 8, 10, 12, 13, and 16), respectively. The representative data are shown (Fig. 1B). These diagnostic results obtained by multiplex PCR were also supported by those based on the nucleotide sequence analysis of cox1 from individual 28 cysticerci.
FIG. 1.
(A) Multiplex PCR with mtDNA prepared from causative agents of taeniasis or cysticercosis. Lanes 1 to 10, T. saginata; lane 11, a mixture of T. saginata and T. asiatica; lanes 12 to 16, T. asiatica; lanes 17 to 25, American/African genotype of T. solium; lanes 26 to 32, Asian genotype of T. solium. The lane numbers correspond to the lanes shown in Table 1. (B) Confirmation by multiplex PCR of cysticerci developed in immunodeficiency mice. The cysticerci originated from the eggs in lane 11 of panel A. Lanes 1 to 4, 6 to 8, 10, 12, 13, and 16, T. asiatica; lanes 5, 9, 11, 14, and 15, T. saginata. M, DNA size markers (100-bp ladder [Promega]).
Differential diagnosis of tapeworm carriers by multiplex PCR with copro-DNA.
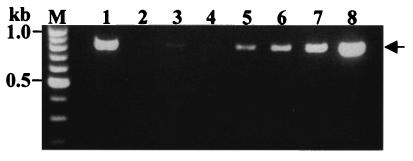
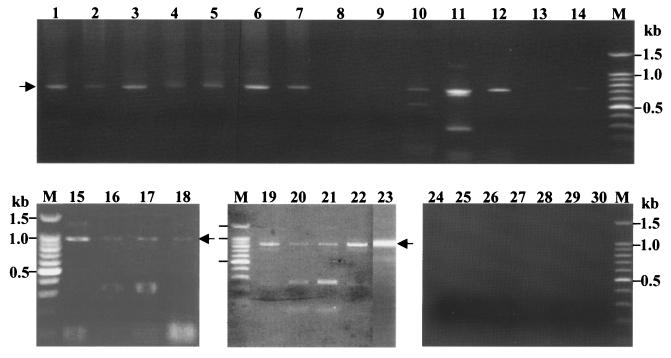
In order to evaluate the detection limit of taeniid DNA in fecal samples of tapeworm carriers, multiplex PCR was performed with DNA samples prepared from a given number of T. saginata eggs. As shown in Fig. 2, 827-bp products unique for T. saginata were amplified in a dose-dependent fashion. The target band was detectable if at least five eggs were contained in 1 g of feces (lane 2); however, multiplex PCR provided more reliable diagnostic results if more than 50 eggs were present in 1 g of fecal sample (lane 5). Figure 3 shows the results obtained by multiplex PCR with copro-DNA extracted from fecal samples of tapeworm carriers. These results are also summarized in Table 2. The diagnostic products were detected from 7 of 14 T. solium American genotype carriers from Guatemala (lanes 1 to 7) by multiplex PCR. However, diagnostic products were detected from five of the remaining seven samples found to be negative by multiplex PCR when nested PCR with Tsol/Amer and Rev primers was performed (lanes 9 to 12 and lane 14, also very faint in the lane 9). Any products from these negative seven cases were not amplified when nested PCR with either Tsag or Tsol/Asia primer sets was performed (data not shown). It was noted that a faint cox1 fragment was detected from a Guatemalan worm carrier who expelled only immature (non gravid) tapeworms (Fig. 3, lane 14, and Table 2, code F13). In T. solium Asian genotype carriers from Papua, Indonesia, 984-bp products were detected in four of nine fecal samples (lanes 15 to 18), but five of nine stool samples were not detected (data not shown). In contrast to the T. solium carriers, the 827-bp diagnostic markers were detected in all T. saginata carriers (lanes 19 to 23). One sample from Bali, Indonesia, was considered to be from a T. solium carrier; however, it was diagnosed as T. saginata by multiplex PCR (lane 23). In cases of negative controls from the United Kingdom, the product was not amplified by either multiplex PCR (lanes 24 to 30) or nested PCR using any of the primer sets (data not shown).
FIG. 2.
Detection limit of target gene by multiplex PCR with DNA samples prepared from taeniid eggs. Target cox1 fragments were amplified by a dose-dependent fashion (indicated by an arrow). Lane 1, positive control with mtDNA from a T. saginata proglottid, lanes 2 to 8, 5, 10, 20, 50, 102, 103, and 104 eggs/g of feces, respectively.
FIG. 3.
Differential diagnosis of tapeworm carriers by multiplex and nested PCR with copro-DNA. Diagnostic markers with molecular sizes of 720, 984, and 827 bp were detected from tapeworm carriers with the T. solium American/African genotype from Guatemala (lanes 1 to 7), and nested PCR results with Tsol/Amer and Rev primers products are shown in lanes 8 to 14. The worm carriers with the Asian genotype of T. solium from Indonesia and T. saginata are shown in lanes 15 to 18 and lanes 19 to 23, respectively. Lanes 24 to 30 are from negative controls. M, DNA size markers.
DISCUSSION
Differentiation of T. solium and T. saginata based on the DNA analysis has been possible for several years (3, 4, 6-9, 13, 17-18, 22). Identification of Asian Taenia spp. and Asian versus American/African genotypes of T. solium, as well as application for stool samples has not previously been reported. Multiplex PCR for the differential diagnosis of T. saginata and T. solium was described by González et al. (8). The method coupled with restriction fragment length polymorphism has been further applied for differentiation of geographical isolates of T. saginata and T. solium (8), and it appears useful for the diagnosis of taeniasis and cysticercosis in Europe, America, and Africa, where T. saginata and T. solium (American/African genotype) are distributed. In Asian regions, on the other hand, since T. saginata, T. asiatica, and T. solium (Asian genotype) are distributed sympatrically, methods capable of differentiating these taeniid cestodes are necessary. Indeed, taeniid proglottids collected from different worm carriers in Yunnan province, China, where T. solium is also endemic, were originally thought to be only T. saginata on the basis of proglottid morphology; however, it has subsequently been proven to be a mixture of T. saginata and T. asiatica by multiplex PCR. The diagnostic results obtained by multiplex PCR were same as those based on the nucleotide sequences of cox1 from taeniid cestode samples examined, indicating that the multiplex PCR can be used with a high degree of accuracy.
In DNA differential diagnosis of human taeniid cestodes, it is interesting to determine whether distinct genotypes or geographical variations exist in T. saginata and T. asiatica. The DNA sequencing of cox1 revealed that T. saginata showed slight polymorphism (0.2 to 0.6%) among the 10 different geographical isolates tested (Table 1, AB06695 and AB107237 to AB107247); however, distinct phylogenetic genotypes such as those seen in T. solium were not detected. The cox1 genes fromT. asiatica samples collected from four different localities were almost identical (with a similarity of 99.8%, Table 1, AB066494 and AB107234 to AB107236). It has previously been reported that geographically different strains or isolates have been detected in T. saginata (7, 21), however such variations observed in T. saginata and T. asiatica did not interfere DNA differential diagnosis by multiplex PCR used here.
Target gene fragments were detectable if at least 5 eggs of T. saginata were present in 1 g of fecal sample. It has been reported that even one T. solium egg could be detected by PCR, however 50 or more eggs were needed for more reliable diagnostic (4). In the current study, neither eggs of T. solium nor T. asiatica were available, however a similar sensitivity might be expected in cases of T. solium or T. asiatica eggs. Interestingly, as indicated in lane 14 in Fig. 3, taeniid DNA was detected from egg-free fecal samples (same as code F13 in Table 2). Given the turnover of taeniid parasite material into the feces, there is very good chance that taeniid DNA is present in feces where taeniid eggs are not. A similar situation has been observed in a volunteer T. saginata carrier from the United Kingdom, where it was possible to detect DNA in feces by PCR prior to patency (unpublished data). This suggests that DNA from non-egg sources is present in feces from tapeworm carriers. No diagnostic marker was amplified from five of nine carriers with the T. solium Asian genotype from Indonesia; however, this was considered due to small amounts of fecal samples. In contrast, 720-bp diagnostic markers were detected from 7 of 14 carriers with the T. solium American genotype from Guatemala. Two samples (Fig. 3, lanes 8 and 13, and Table 2, codes F1 and F9) from Guatemala were both determined to be negative by multiplex PCR and by nested PCR. In contrast, T. solium carriers, all four T. saginata samples from human volunteer infections, which were freshly collected and with which there were no problems with sample volume, were positive. Indeed, the sensitivity of the multiplex PCR technique seems to be dependent on the conditions of sample storage and the volumes of the fecal samples. In the present study, fecal samples from Guatemala stored at −20 to −30°C for 12 years and small volumes of stool samples from Indonesia had to be used. In recent research on taeniasis in Indonesia, fecal samples stored in 80 to 99.5% ethanol after collection were used and provided more reliable diagnostic results (T. Wandra, unpublished data). Thus, PCR diagnosis using copro-DNA for the detection of taeniasis can be tested under different circumstances if the stool samples are preserved in ethanol.
As summarized in Table 2, it appears that the sensitivity of multiplex/nested PCR may be somewhat lower in comparison with that of copro-antigen detection test; however, PCR diagnosis has an advantage in its ability to differentiate human taeniid cestode species. The copro-antigen assay is genus specific, not species specific, detecting both T. solium and T. saginata worm carriers (1). In PCR-based diagnosis, the ability to differentially diagnose tapeworm carriers, particularly T. solium carriers, has important implications: greater clinical relevance (given the greater clinical risks associated with T. solium cysticercosis due to the autoinfection) and epidemiological relevance (given the epidemiological importance of the ability to differentiate taeniasis to the species level). The main risk factor for acquiring cysticercosis in humans and pigs is the presence of T. solium tapeworm carriers within the household (19) but not T. saginata and T. asiatica. In the present study, the diagnostic markers were detected from one case of T. solium carriers who was egg-free, implying that it is possible to detect worm carriers prior to patency. In areas where taeniasis is endemic, therefore, multiplex PCR diagnosis will be useful for taeniasis control that aims to detect tapeworm carriers and treat them. The use of praziquantel at a single low dose is strongly recommended for the treatment of taeniasis due to either T. saginata or T. solium (12). PCR-based diagnosis is also applicable, even in areas where this organism is not endemic, for diagnosing tapeworm carriers among immigrants or tourists who have returned from such regions in order to avoid locally acquired taeniasis or cysticercosis.
Acknowledgments
We thank T. Ikejima, P. Dekumyoy, S. S. Margono, S. P. Sinha Babu, A. Oommen, G. Singh, P. C. Fan, K. Eom, V. C. W. Tsang, A. Plancarte, W. Benitez-Ortiz, C. M. Nunes, M. Vilhena, S. Geerts, A. A. Kassuku, J. Garcia Noval, M. Velasquez Tohom, S. S. Afonso, A. Zoli, and S. Miura for kindly providing parasite materials or stool samples.
This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan and by the Japan Society for Promotion of Science to A.I. (grant 14256001).
REFERENCES
- 1.Allan, J. C., G. Avila, J. Garcia-Noval, A. Flisser, and P. S. Craig. 1990. Immunodiagnosis of taeniasis by coproantigen detection. Parasitology 101:473-477. [DOI] [PubMed] [Google Scholar]
- 2.Allan, J. C., M. Velasquez-Tohom, J. Garcia-Noval, A. R. Torres, P. Yurrita, C. Fletes, F. de Mata, H. de Soto de Alfaso, and P. S. Craig. 1996. Epidemiology of intestinal taeniasis in four rural Guatemalan communities. Ann. Trop. Med. Parasitol. 90:157-165. [DOI] [PubMed] [Google Scholar]
- 3.Bowles, J., and D. P. McManus. 1994. Genetic characterization of the Asian Taenia, a newly described taeniid cestode of humans. Am. J. Trop. Med. Hyg. 50:33-44. [PubMed] [Google Scholar]
- 4.Chapman, A., V. Vallejo, K. G. Mossie, D. Ortiz, N. Agabian, and A. Flisser. 1995. Isolation and characterization of species-specific DNA probes from Taenia solium and Taenia saginata and their use in an egg detection assay. J. Clin. Microbiol. 33:1283-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flisser, A. 1998. Neurocysticercosis in Mexico. Parasitol. Today 4:131-137. [DOI] [PubMed] [Google Scholar]
- 6.Flisser, A., A. Reid, Z. E. Garcia, and D. P. McManus. 1988. Specific detection of Taenia saginata eggs by DNA hybridization. Lancet ii:1429-1430. [DOI] [PubMed] [Google Scholar]
- 7.González, L. M., E. Montero, L. J. S. Harrison, R. M. E. Parkhouse, and T. Gárate. 2000. Differential diagnosis of Taenia saginata and Taenia solium infection by PCR. J. Clin. Microbiol. 38:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González, L. M., E. Montero, S. Puente, R. López-Velez, M. Hernández, E. Sciutto, L. J. S. Harrison, R. M. E. Parkhouse, and T. Gárate. 2002. PCR tools for the differential diagnosis of Taenia saginata and Taenia solium taeniasis/cysticercosis from different geographical locations. Diagn. Microbiol. Infect. Dis. 42:243-249. [DOI] [PubMed] [Google Scholar]
- 9.Harrison, L. J. S., J. Delgado, and R. M. E. Parkhouse. 1990. Differential diagnosis of Taenia saginata and Taenia solium with DNA probes. Parasitology 100:459-461. [DOI] [PubMed] [Google Scholar]
- 10.Ito, A., K. Nakaya, Y. Sako, M. Nakao, and M. Ito. 2001. NOD-scid mouse as an experimental animal model for cysticercosis. Southeast Asian J. Trop. Med. Public Health 32(Suppl. 2):83-89. [PubMed] [Google Scholar]
- 11.Ito, A., and P. S. Craig. 2003. Immunodiagnostic and molecular approaches for the detection of taeniid cestode infections. Trends Parasitol. 19:377-381. [DOI] [PubMed] [Google Scholar]
- 12.Ito, A., C. Urbani, JM. Qiu, D. A. Vuitton, DG. Qiu, D. D. Heath, P. S. Craig, F. Zheng, and P. M. Schantz. 2003. Control of echinococcosis and cysticercosis: a public health challenge to international cooperation in China. Acta Trop. 86:3-17. [DOI] [PubMed] [Google Scholar]
- 13.Mayta, H., A. Talley, R. H. Gilman, J. Jimenez, M. Verastegui, M., Ruiz, H. H. Garcia, and A. E. González. 2000. Differentiating Taenia solium and Taenia saginata infections by simple hematoxylin-eosin staining and PCR-restriction enzyme analysis. J. Clin. Microbiol. 38:133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakao, M., M. Okamoto, Y. Sako, H. Yamasaki, K. Nakaya, and A. Ito. 2002. A phylogenetic hypothesis for the distribution of two genotypes of the pig tapeworm Taenia solium worldwide. Parasitology 124:657-662. [DOI] [PubMed] [Google Scholar]
- 15.Pawlowski, Z. S., and M. G. Schultz. 1972. Taeniasis and cysticercosis (Taenia saginata). Adv. Parasitol. 10:269-343. [DOI] [PubMed] [Google Scholar]
- 16.Rishi, A. K., and D. P. McManus. 1987. DNA probes which unambiguously distinguish Taenia solium from T. saginata. Lancet i:1275-1276. [DOI] [PubMed] [Google Scholar]
- 17.Rishi, A. K., and D. P. McManus. 1988. Molecular cloning of Taenia solium genomic DNA and characterization of taeniid cestodes by DNA analysis. Parasitology 97:161-176. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Hidalgo, R., D. Geysen, W. Benitez-Ortiz, S. Geerts, and J. Brandt. 2002. Comparison of conventional techniques to differentiate between Taenia solium and Taenia saginata and an improved polymerase chain reaction-restriction fragment length polymorphism assay using a mitochondrial 12S rDNA fragment. J. Parasitol. 88:1007-1011. [DOI] [PubMed] [Google Scholar]
- 19.Sarti, E., P. M. Schantz, A. Plancarte, M. Wilson, O. Gutierrez, A. S. Lopez, J. Roberts, and A. Flisser. 1992. Prevalence and risk factors for Taenia solium taeniasis and cysticercosis in humans and pigs in a village in Morelos, Mexico. Am. J. Trop. Med. Hyg. 46:677-685. [DOI] [PubMed] [Google Scholar]
- 20.Simanjuntak, G. M., S. S. Margono, M. Okamoto, and A. Ito. 1997. Taeniasis/cysticercosis in Indonesia as an emerging disease. Parasitol. Today 13:321-323. [Google Scholar]
- 21.Wouters, G., J. Brandt, and S. Geerts. 1987. Observations on possible strain differences in Taenia saginata, p. 76-80. In S. Geerts, V. Kumar, and J. Brandt (ed.), Helminth zoonoses. Martinus Nijhoff Publishers, Dordrecht, The Netherlands.
- 22.Yamasaki, H., M. Nakao, Y. Sako, K. Nakaya, M. O. Sato, W. Mamuti, M. Okamoto, and A. Ito. 2002. DNA differential diagnosis of human cestodes by base excision sequence scanning thymine-base reader analysis with mitochondrial genes. J. Clin. Microbiol. 40:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamasaki, H., M. Nakao, Y. Sako, K. Nakaya, M. O. Sato, W. Mamuti, N. Xiao, M. Okamoto, and A. Ito. 2002. DNA Differential diagnosis of human taeniid cestodes using restriction fragment length polymorphism and base excision sequence scanning T-base system, p. 189-193. In Proceedings of the 10th International Congress of Parasitology. Monduzzi Editore, Boronia, Italy.