Abstract
Subgenus C human adenoviruses, which include serotypes 1, 2, 5, and 6, are often associated with respiratory illness, ocular infections, gastroenteritis, and systemic infection among immunocompromised patients. To address the problems associated with the conventional typing methods, we developed a fiber-based multiplex PCR assay for simple and specific identification of adenovirus type 1, 2, 5, and 6 field isolates. To design type-specific primers, adenovirus type 1 and 6 fiber genes were sequenced. The assay correctly identified prototype strains of adenovirus serotypes 1, 2, 5, 6, as well as 21 previously typed adenovirus field isolates. Mixing two different prototype DNAs produced two amplicons of different lengths, thus clearly distinguishing the prototypes. The results correlated 100% with serological tests and 95% with the previously described PCR-restriction fragment length polymorphism method. The detection of dual infection is an added benefit of the assay. No nonspecific amplification was detected with other adenovirus serotypes or with nonadenoviral DNA. Our fiber-based multiplex PCR assay will provide a convenient tool for type-specific identification of subgenus C adenovirus isolates in various clinical situations and in epidemiological investigations and is a better alternative than the hexon-based assay.
Fifty-one serotypes of human adenoviruses (Ads) have been divided into six subgenera, A through F, based on various biological and morphological criteria (9, 28). The members of subgenus C, Ad1, Ad2, Ad5, and Ad6, are the most frequently isolated human Ads, accounting for 59% of all Ad infections (27). Respiratory infections caused by these Ads may be indistinguishable from infections caused by other respiratory pathogens, such as influenza, parainfluenza, and respiratory syncytial viruses as well as certain bacteria (12). These Ads are the causative agents of pharyngeal conjunctival fever among children as well as of outbreaks of conjunctivitis related to swimming pools during the summer (4). Ad2 is the predominant serotype, next to Ad40 and Ad41, related to enteric infections among young children (11, 18, 24). In addition to diarrhea, subgenus C Ads are also linked to intussusception of the intestine caused by Ad-infected mesenteric lymph nodes (3, 7, 21). They are also the serotypes predominantly isolated among immunosuppressed patients and are responsible for fatal infections of transplant recipients (16, 19). Furthermore, subgenus C Ads seem to be associated with sudden infant death syndrome (2). Therefore, identification of the serotypes mentioned above is very important under different clinical conditions and in epidemiological studies.
Conventional methods for typing of Ads are usually based on neutralization (NT) and/or hemagglutination inhibition (HAI) assays, which are not only cumbersome but also time-consuming (14, 15, 17, 25). Restriction endonuclease analysis (REA) of DNA extracted from infected cells, although proven to be epidemiologically valuable, requires technical skill and time and therefore has limited clinical usefulness (10). In contrast, the PCR assay is a faster, sensitive method, which is quite useful in various clinical situations. To date, the fiber gene, encoding the fiber protein, which mediates cellular attachments and hemagglutination and which, together with the hexon, confers serotype specificity on Ads, has not been evaluated properly as a site for type-specific identification of Ads (20). In this study, the fiber gene is evaluated as a site for type-specific identification of the subgenus C Ads.
MATERIALS AND METHODS
Viruses.
Ad prototype (p) strains of serotypes Ad1 (adenoid 71), Ad2 (adenoid 6), Ad5 (adenoid 75), and Ad6 (tonsil 99) and members of other subgenera were obtained from the American Type Culture Collection (Rockville, Md.). Twenty-one geographically and temporally diverse field isolates (Ad1:5, Ad2:6, Ad5:4, Ad6:5, and mixed Ad1 and Ad2:1) that had previously been typed by NT were selected in order to evaluate the feasibility of this method.
Ad identification.
All strains were isolated in Hep2 or A549 cells and were typed by NT and/or HAI testing with Ad type-specific reference antisera.
DNA sequencing.
In order to design type-specific primers, the Ad1 and Ad6 fiber genes were sequenced from genomic DNAs extracted from infected cells as described previously (1). Briefly, confluent monolayers of Hep2 cells in 25-cm2 flasks were inoculated with virus stocks and incubated at 35°C. When 75 to 100% cytopathic effect was observed, cells were dislodged with a cell scraper and pelleted by low-speed centrifugation. The cells were washed twice with phosphate-buffered saline and then resuspended in 1 ml of lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 1% sodium dodecyl sulfate) for 15 min at room temperature. The suspension was incubated with proteinase K (Sigma Chemical, St. Louis, Mo.) at a concentration of 200 μg/ml at 37°C for 1 h. After the incubation, 5 M NaCl was added to a final concentration of 1 M, and the suspension was further incubated at 4°C overnight to precipitate cellular DNA. Then the suspension was centrifuged at 15,000 × g for 30 min. The supernatant was incubated with 30 μg of RNase A (Sigma Chemical) for 1 h and then extracted twice in phenol-chloroform. Then the supernatant was precipitated in 2 volumes of absolute ethanol. After drying, DNA was suspended in 50 μl of TE buffer (10 mM Tris-HCl [pH 7.4]-10 mM EDTA) and quantified spectrophotometrically. The fiber genes were sequenced by internal primers in both the sense and antisense directions. Selection of the primers was based on alignment of the sequences of the fiber genes of Ad2 and Ad5 available from GenBank (accession numbers J0107 and M18369, respectively) (5). The cycle sequencing reaction was carried out with the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit, and sequences were read in a Genetic Analyzer, model 310 (both from Applied Biosystems, Foster City, Calif.). DNASIS software (Hitachi Software Ltd., Tokyo, Japan) was used for sequence alignment and analysis. The amino acid sequences of these residues were deduced.
PCR assay. (i) DNA extraction.
Viral DNA from the field isolates was prepared by phenol-chloroform extraction and ethanol precipitation. In a microcentrifuge tube, 200 μl of culture fluid and the same volume of lysis buffer (10 mM Tris HCl [pH 7.6], 5 mM EDTA, 1% sodium dodecyl sulfate, and 200 μg of proteinase K/ml) were mixed and incubated for 1 h at 37°C. Then 20 μg of RNase A (Sigma Chemical) was added to the tube and incubated at 37°C for another hour. After incubation, the contents were extracted with same volume of phenol-chloroform mixture (100 μl of phenol and 100 μl of chloroform) for 10 min and then centrifuged at 8,000 × g for 10 min. This extraction process was repeated once. Finally, the supernatant containing genomic DNA was precipitated in 500 μl of absolute ethanol. After drying, the pellet was dissolved in distilled water.
(ii) Ad subgenus C type-specific primers.
Type-specific primer design was based on alignment of the fiber gene sequences of Ad1 and Ad6 (sequenced in our laboratory) and Ad2 and Ad5 (obtained from GenBank; accession numbers J0107 and M18369, respectively). The forward primer (AdCF) was common to all the serotypes of subgenus C, and the reverse primers (Ad1R, Ad2R, Ad5R, and Ad6R) were type specific (Table 1).
TABLE 1.
Oligonucleotide primers for multiplex PCR of subgenus C Ads
| Serotype | Primer | Polarity | Positiona | Sequence (5′-3′) | Amplicon length (bp) |
|---|---|---|---|---|---|
| AdCF | + | 657-677 | 5′-TGC TTG CGC THA AAA TGG GCA-3′ | ||
| Ad1 | Ad1R | − | 895-873 | 5′-CGA GTA TAA GAC GCC TAT TTA CA-3′ | 630 |
| Ad2 | Ad2R | − | 838-857 | 5′-CGC TAA GAG CGC CGC TAG TA-3′ | 204 |
| Ad5 | Ad5R | − | 1093-1111 | 5′-ATG CAA AGG AGC CCC GTA C-3′ | 455 |
| Ad6 | Ad6R | − | 1209-1188 | 5′-CTT GCA GTC TTT ATC TGA AGC A-3′ | 929 |
(iii) DNA amplification and detection.
PCR amplification was carried out in a 50-μl reaction mixture containing 1-μl aliquots of DNA, 5 μl of 10-fold-concentrated buffer, 0.5 μM each primer, 200 μM each deoxynucleoside triphosphate, and 1.25 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). Sterile distilled water was used as a negative control. For clinical samples, Ad2 DNA was used as a positive control. The assay was performed in a programmable heat block (model 9600-R; Perkin-Elmer). Thermal cycling consisted of a preliminary denaturation for 3 min at 94°C; 35 cycles of denaturation at 94°C for 1 min, annealing at 47°C for 1 min, and extension at 72°C for 2 min; and a final extension at 72°C for 7 min. Five microliters of reaction product was mixed with 1 μl of loading buffer (60% glycerol, 0.25% bromphenol blue, and 0.25% xylene cyanol) and then run on 1.5% horizontal agarose gel (containing 1 μg of ethidium bromide/ml) at 100 V for 50 min in a 50 mM Tris-borate-EDTA buffer (pH 8.0). The bands were visualized under a UV transilluminator and photographed with a charge-coupled device camera.
Specificity and limits of detection.
The specificity of the test was evaluated with pooled DNAs from subgenus A, B, D, E, and F Ads. DNAs from herpes simplex virus type I and type II, enterovirus, and Chlamydia species were also tested. The limits of detection of our multiplex PCR assay were determined by amplification of a known amount of purified Ad2 DNA to obtain a theoretical range of virus particle numbers, from 1010 to 101 per reaction mixture; 0.384 fg of Ad DNA corresponds to a single copy of linear double-stranded DNA that is approximately 35,000 bp long (30). After PCR amplification, 5 μl of the product was electrophoresed on a 1.5% agarose gel and stained with ethidium bromide.
Comparison of PCR assays.
In order to validate our fiber-based PCR method, we selected another assay, a combination of hexon-based PCR with restriction fragment polymorphism (PCR-RFLP), which has been described previously (26). Briefly, after nested PCR, 956-bp PCR products were digested with three restriction enzymes, HaeIII, HinfI, and EcoT14I, and restriction patterns were compared with the prototype patterns for typing.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the DDBJ/GenBank nucleotide sequence database with accession numbers AB108423 (Ad1) and AB108424 (Ad6).
RESULTS
Ad1 and Ad6 sequences.
Analysis of the Ad1 and Ad6 fiber proteins revealed that, as described previously (13), they possess three distinct regions: the amino-terminal tail, a central shaft, and a C-terminal knob. The open reading frames of Ad1 and Ad6 were 1,746 and 1,584 nucleotides (nt) long, with predicted products of 582 and 528 amino acids (aa), respectively. The Ad6 open reading frame was 162 nt (54 aa) shorter than that of Ad2 and 159 nt (53 aa) shorter than that of Ad5. The difference in fiber length was due to the relatively shorter shaft of the Ad6 fiber protein (918 nt, or 306 aa) in comparison to those of Ad1, Ad2, and Ad5 (1,061 nt, or 357 aa). However, the tails were the same length, with 132 nt (44 aa) (Table 2).
TABLE 2.
Lengths of fiber genes and proteins of subgenus C Ads
| Ad serotype | Length of fiber gene (protein)a:
|
|||
|---|---|---|---|---|
| Tail | Shaft | Knob | Overall | |
| Ad1 | 132 (44) | 1,071 (357) | 543 (181) | 1,746 (582) |
| Ad2 | 132 (44) | 1,071 (357) | 543 (181) | 1,746 (582) |
| Ad5 | 132 (44) | 1,071 (357) | 540 (180) | 1,743 (581) |
| Ad6 | 132 (44) | 918 (306) | 534 (178) | 1,584 (528) |
DNA lengths are given in nucleotides; protein lengths are given in amino acids.
Multiplex PCR assay.
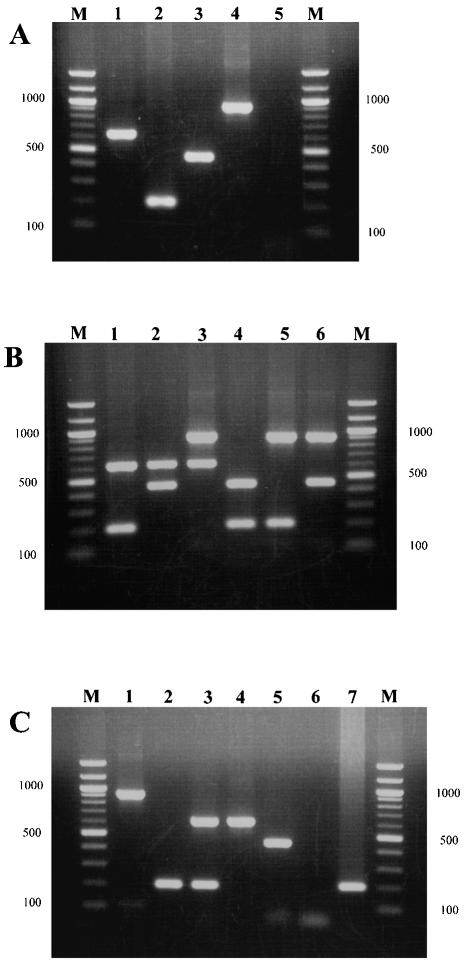
The combination of primers yielded products of 630, 204, 455, and 929 bp, lengths appropriate for Ad1, Ad2, Ad5, and Ad6 prototype strains, respectively (Fig. 1A). Mixing two different prototype DNAs produced two amplicons of different lengths, which clearly distinguished the prototypes (Fig. 1B). Twenty field isolates (Table 3) also yielded clearly visible products of the expected lengths, and one isolate (YC 85-74) showed a double band whenever tested by our assay (Fig. 1C).
FIG. 1.
Ethidium bromide-stained agarose gels showing PCR products of subgenus C Ads. Lanes M, molecular size marker (100-bp DNA ladder; New England Biolabs). (A) Subgenus C prototype (p) strains. Lane 1, Ad1p; lane 2, Ad2p; lane 3, Ad5p; lane 4, Ad6p; lane 5, negative control. (B) Mixed DNAs of subgenus C prototype strains. Lane 1, Ad1p and Ad2p; lane 2, Ad1p and Ad5p; lane 3, Ad1p and Ad6p; lane 4, Ad2p and Ad5p; lane 5, Ad2p and Ad6p; lane 6, Ad5p and Ad6p. (C) Subgenus C field isolates. Lane 1, 91-1141 (Ad6); lane 2, YC 79-57 (Ad2); lane 3, YC 85-74 (Ad1, Ad2); lane 4, YC 66-315 (Ad1); lane 5, YC 84-37 (Ad5); lane 6, negative control; lane 7, positive control (Ad2p).
TABLE 3.
Comparison of Ad serotypes determined by the fiber-based multiplex PCR assay and the hexon-based PCR-RFLP method
| Isolate | Serotype by:
|
||
|---|---|---|---|
| Original identification | Fiber-based multiplex PCR | Hexon-based PCR-RFLP | |
| YC 66-113 | Ad1 | Ad1 | Ad1 |
| YC 66-283 | Ad2 | Ad2 | Ad2 |
| YC 66-284 | Ad2 | Ad2 | Ad2 |
| YC 66-315 | Ad1 | Ad1 | Ad1 |
| YC 66-323 | Ad2 | Ad2 | Ad2 |
| YC 72-141 | Ad5 | Ad5 | Ad5 |
| YC 72-143 | Ad2 | Ad2 | Ad2 |
| YC 77-82 | Ad5 | Ad5 | Ad5 |
| YC 79-57 | Ad2 | Ad2 | Ad2 |
| YC 84-31 | Ad6 | Ad6 | Ad6 |
| YC 84-37 | Ad5 | Ad5 | Ad5 |
| YC 85-70 | Ad6 | Ad6 | Ad6 |
| YC 85-74 | Ad1, Ad2 | Ad1, Ad2 | Not typed |
| YC 85-75 | Ad1 | Ad1 | Ad1 |
| YC 85-76 | Ad1 | Ad1 | Ad1 |
| YC 85-77 | Ad1 | Ad1 | Ad1 |
| 91-1141 | Ad6 | Ad6 | Ad6 |
| 92-515T | Ad5 | Ad5 | Ad5 |
| 92-516T | Ad6 | Ad6 | Ad6 |
| 92-523T | Ad6 | Ad6 | Ad6 |
| 92-566T | Ad2 | Ad2 | Ad2 |
Specificity and limits of detection.
The specificity of the PCR was tested against DNAs from other Ad subgenera and against nonadenoviral DNA. No amplified products were detected, indicating a high specificity of the test. After serial dilution and amplification of purified Ad2 DNA, the amplicon was detected at a dilution of 1:108. This represents 38.4 fg of Ad DNA, which corresponds to 100 genome copies (data not shown).
Comparison of PCR assays.
Our PCR clearly amplified all the isolates and provided the same result (100%) as NT. However, the PCR-RFLP method was unable to detect a dual infection (Table 3).
DISCUSSION
A fiber-based PCR method for identification of subgenus C Ads (Ad1, -2, -5, and -6) has been developed. The subgenus C Ads are the most frequently isolated human Ads; they are associated with respiratory, gastrointestinal, and ocular diseases among children and are causative agents of fatal infections among immunodeficient patients (18). In routine practice, culture-NT, which requires 3 to 4 weeks for the whole procedure, is in use for the identification of Ads. In comparison, PCR-based methods do not require viable virus, and a few virus particles are enough to yield a positive result; therefore, these methods have appeared as alternatives to culture-NT during the last few years. To date, methods for type-specific identification of Ads are hexon gene based; these include type-specific PCR (22, 24), a combination of PCR and restriction enzyme analysis (26), and typing by DNA sequencing of the PCR product (29). The first method, in which type-specific primers target serotype-specific regions located in two or more of the seven hypervariable regions of the hexon, is susceptible to possible PCR failure due to differences in the target region between strains of the same serotype (8). Moreover, visual discrimination of serotypes appears to be difficult, because the amplicons of subgenus C Ads are of approximately the same length. The PCR-RFLP method targets the downstream region of the hexon gene, which is not specified as a region determining the serotype (29). Moreover, this region is highly conserved; therefore, the primers could also amplify Ads of serotypes other than the common serotypes studied, and the restriction patterns of these other serotypes may be difficult to interpret. Identification of serotypes by direct sequencing of the PCR product requires expensive instrumentation and technical skill that are beyond the scope of most diagnostic laboratories. Although dual infections are not very common, their identification is not possible by any of these methods. To address these inherent problems, an alternative method that is simple and accurate is needed.
The capsid components of Ad, the hexon and the fiber display antigenic determinant, together define the serotype specificity of Ad (20). In the present study, the fiber gene was explored as a site for type-specific identification of subgenus C Ads. In contrast to other subgenera, subgenus C is serologically distinct, and intermediate strains that arise due to homologous recombination between two parental viruses are quite uncommon; therefore, NT and HAI provide unequivocal results (5, 24). Moreover, evolution of new serotypes has not been observed so far, providing supportive evidence that this subgenus is stable in comparison to subgenera B and D. Therefore, it is reasonable to speculate that fiber-based PCR could be a reliable alternative to hexon-based PCR.
Nucleotide sequence heterogeneity is one of the prerequisites for the development of type-specific primers. Fiber genes are usually heterogeneous among subgenera and highly homologous within a subgenus (23). Our sequence analysis revealed that subgenus C fiber genes are highly heterogeneous, with 66 to 71% homology (data not shown) to each other, providing an opportunity to develop type specific primers. Type-specific primers accurately identified all the prototype strains, as well as various combinations of two prototypes, at the expected lengths. The accuracy and reproducibility of the assay was confirmed by blind evaluation of 21 field isolates previously typed by NT and/or HAI. In comparison to other PCR-based methods, our assay showed some distinct advantages: (i) it can amplify 100 genome copies of virus, indicating higher sensitivity; (ii) a single round of amplification is enough to yield a positive result; and (iii) it successfully detects dual infection. Although dual infection is not very common, this is an added benefit of our assay, particularly for patients with immunodeficiency, which allows the coexistence of different strains (16). The specificity of the assay was determined by testing Ads of other subgenera as well as nonadenoviral DNA. The lack of amplified products indicates that our assay has a high level of specificity. The minimum limit of detection of our PCR was 100 copies of viral DNA. It is not known how many copies of viral DNA are needed for clinical diseases. We were unable to evaluate our multiplex PCR for direct identification of clinical samples, due to the limited availability of suitable specimens; therefore, our assay is recommended for virus isolates. However, in this study the fiber gene of subgenus C was found to be a reliable alternative to the hexon gene for type-specific detection of Ads with high levels of sensitivity and specificity. This simple and cost-effective method has the potential to replace time-consuming techniques.
REFERENCES
- 1.Adhikary, A. K., J. Numaga, T. Kaburaki, H. Kawashima, M. Araie, Y. Ikeda, T. Ogino, E. Suzuki, H. Ushijima, A. Mukoyama, S. Matsuno, T. Inada, and N. Okabe. 2003. Genetic characterization of adenovirus type 8 isolated in Hiroshima city over a 15 year period. J. Clin. Pathol. 56:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajanowski, T., P. Wiegand, R. Cecchi, P. Pring-Akerblom, T. Adrian, G. Jorch, and B. Brinkmann. 1996. Detection and significance of adenoviruses in cases of sudden infant death. Virchows Arch. 428:113-118. [DOI] [PubMed] [Google Scholar]
- 3.Bell, J. A., and J. H. Styen. 1962. Viruses in lymph nodes of children with mesenteric adenitis and intussusception. Br. Med. J. 2:700-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, F. M., B. B. Law, W. Hamilton, and A. MacDonald. 1957. Adenovirus eye infections in Aberdeen. Lancet ii:670-673. [DOI] [PubMed] [Google Scholar]
- 5.Boursnell, M. E. G., and V. Mautner. 1981. Recombination in adenoviruses: crossover sites in intertypic recombinations are located in the regions of homology. Virology 112:198-209. [DOI] [PubMed] [Google Scholar]
- 6.Chrobozek, J., and B. Jacrot. 1987. The sequence of adenovirus fiber: similarities and differences between serotypes 2 and 5. Virology 161:549-554. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, E. J., I. A. Phillips, and E. R. Alexander. 1969. Adenovirus infection in intussusception in children in Taiwan. JAMA 208:1671-1674. [PubMed] [Google Scholar]
- 8.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of adenovirus hexon proteins revealed the location of structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1835-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elnifro, E. M., R. J. Cooper, P. E. Klapper, and A. S. Bailey. 2000. PCR and restriction endonuclease analysis for rapid identification of human adenovirus subgenera. J. Clin. Microbiol. 38:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, J. P., C. E. Hall, and M. K. Cooney. 1977. The Seattle virus watch. VII. Observation of adenovirus infection. Am. J. Epidemiol. 105:362-368. [DOI] [PubMed] [Google Scholar]
- 12.Ginsberg, H. S., R. L. Horswood, R. M. Chanock, and G. A. Prince. 1990. Role of early genes in pathogenesis of adenovirus pneumonia. Proc. Natl. Acad. Sci. USA 87:6191-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, N. M., N. G. Wrigley, W. C. Russell, S. R. Martin, and A. D. McLachlan. 1983. Evidence for a repeating cross-β sheet structure in the adenovirus fiber. EMBO J. 2:1357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hierholzer, J. C. 1973. Further subgrouping of the human adenoviruses by differential hemagglutination. J. Infect. Dis. 128:541-550. [DOI] [PubMed] [Google Scholar]
- 15.Hierholzer, J. C., Y. O. Stone, and J. R. Broderson. 1991. Antigenic relationships among the 47 human adenoviruses in reference horse antisera. Arch. Virol. 121:179-197. [DOI] [PubMed] [Google Scholar]
- 16.Hierholzer, J. C. 1992. Adenoviruses in the immunocompromised host. Clin. Microbiol. Rev. 5:262-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hierholzer, J. C. 1995. Adenoviruses, p. 169-188. In E. H. Lennette, D. A. Lennette, and E. T. Lennette (ed.), Diagnostic procedures for viral, rickettsial, and chlamydial infections, 7th ed. American Public Health Association, Washington, D.C.
- 18.Infectious Disease Surveillance Center, National Institute of Infectious Diseases. 2003. Isolation/detection of viruses from human sources, Japan. Infect. Dis. Surveillance Rep. 24:5-22. (In Japanese.) [Google Scholar]
- 19.Michaels, M. G., M. Green, E. R. Wald, and T. E. Starzl. 1992. Adenovirus infection in pediatric liver transplant recipients. J. Infect. Dis. 165:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norby, E. 1969. The structural and functional diversity of adenovirus capsid protein components. J. Gen. Virol. 5:221-236. [DOI] [PubMed] [Google Scholar]
- 21.Potter, C. W. 1964. Adenovirus infection as an aetiological factor in intussusception of infants and young children. J. Pathol. Bacteriol. 88:263-274. [PubMed] [Google Scholar]
- 22.Pring-Akerblom, P., and T. Adrian. 1994. Type- and group-specific polymerase chain reaction for adenovirus detection. Res. Virol. 145:25-35. [DOI] [PubMed] [Google Scholar]
- 23.Pring-Akerblom, P., and T. Adrian. 1995. Sequence characterization of the adenovirus 31 fiber and comparison with serotypes of subgenera A to F. Res. Virol. 146:343-354. [DOI] [PubMed] [Google Scholar]
- 24.Pring-Akerblom, P., and T. Adrian. 1997. PCR-based detection and typing of human adenoviruses in clinical samples. Res. Virol. 148:225-231. [DOI] [PubMed] [Google Scholar]
- 25.Rosen, I. 1960. A hemagglutination-inhibition technique for typing of adenoviruses. Am. J. Hyg. 71:120-128. [DOI] [PubMed] [Google Scholar]
- 26.Saitoh-Inagawa, W., A. Oshima, K. Aoki, N. Itoh, K. Isobe, E. Uchio, S. Ohno, H. Nakajima, K. Hata, and I. Hirosaki. 1996. Rapid detection of adenoviral conjunctivitis by PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:2113-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz, H., R. Wigand, and W. Heinreich. 1983. Worldwide epidemiology of adenovirus infections. Am. J. Epidemiol. 117:455-466. [DOI] [PubMed] [Google Scholar]
- 28.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2265-2300. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Takeuchi, S., N. Itoh, E. Uchio, K. Aoki, and S. Ohno. 1999. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J. Clin. Microbiol. 37:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchio, E., K. Aoki, W. Saitoh-Inagawa, N. Itoh, and S. Ohno. 1996. Rapid diagnosis of adenoviral conjunctivitis on conjunctival swab by 10-minute immunochromatography. Ophthalmology 104:1294-1299. [DOI] [PubMed] [Google Scholar]