Abstract
Previous studies have described IS6110-mediated polymorphism as an important driving force in Mycobacterium tuberculosis genome evolution and have provided indirect evidence for IS6110-driven deletion events. This study provides the first description of an IS6110-mediated deletion event in truly isogenic strains. We also provide further support for the hypothesis that the region from Rv1754 to Rv1765 is a hot spot for IS6110 insertion and deletion events.
The insertion sequence IS6110 has been suggested to be an important agent of Mycobacterium tuberculosis genome variation. IS6110 transposition events may disrupt open reading frames (12) or regulatory domains, and there has been much speculation regarding the possible influence of these events on strain phenotype (1, 8, 11, 19). In addition, insertion sequence-mediated deletion events have been suggested to be an important mechanism driving mycobacterial genome variation (2). Homologous recombination between directly repeated IS6110 elements has been proposed as a likely mechanism for genomic deletions in clinical isolates (5). This hypothesis is supported by the results of in silico analysis of sequences flanking the 16 IS6110 elements in the M. tuberculosis H37Rv genome to identify the deletions RvD3, RvD4, and RvD5 (2). In this study and others, the absence of 3- to 4-bp repeats immediately flanking the point of IS6110 insertion is interpreted to reflect homologous recombination events between two directly repeated IS6110 elements (2, 5). Similarly, analysis of sequences immediately flanking IS6110 elements in a highly variable 20-kb region of the chromosome provides further examples of deletions associated with the presence of IS6110 elements (10). A recent study suggested that alternative mechanisms may be involved in deletion events between inverted IS6110 elements (13).
To our knowledge, no study to date has investigated IS6110-mediated deletion in isogenic strains to directly support the hypothesis of homologous recombination. A previous report of multilesional intrapatient strain genotyping described the isolation of two isogenic strains from a single patient, one of which had a single IS6110 band polymorphism (4). From a visual perspective and in the absence of chronology, it was not clear whether this difference was due to an insertion or deletion event. In this study, we utilized the unique availability of these isogenic strains to address this question.
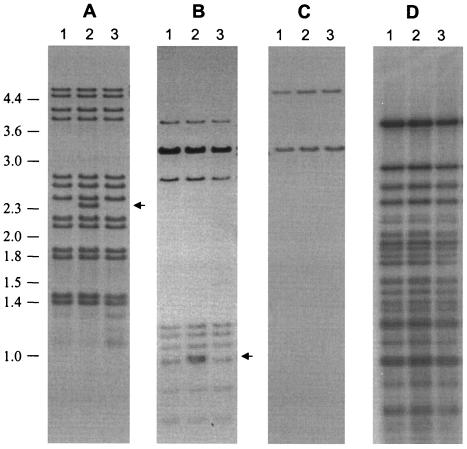
Mycobacterium tuberculosis was cultured from the cadaver of a human immunodeficiency virus-negative 41-year-old female which, on autopsy, showed pulmonary scarring and miliary pulmonary involvement (4). Extrapulmonary tuberculosis was evident in the liver; kidneys; ovaries; and hilar, mediastinal, and iliac lymph nodes. M. tuberculosis was successfully cultured from a mediastinal lymph node (culture D7030), a hilar lymph node (culture D7031), and an abdominal lymph node (culture D7033). IS-3′ DNA fingerprinting (15) showed that all cultures had identical banding patterns with the exception of D7031, which showed an additional hybridizing band at 2.4 kb (Fig. 1A), corresponding to the IS6110 insertion at position ISL7031.8, as determined by DNA sequencing (12). Similarly, the IS-5′ probe showed identical banding patterns with the exception of an additional hybridizing band at 1.0 kb (Fig. 1B), corresponding to the same insertion. DNA fingerprinting with the polymorphic GC-rich repetitive sequence probe (16) and direct repeat (DRr) probe (9) showed identical banding patterns, demonstrating that these strains were genetic variants that had evolved within this individual, possibly as the result of an IS6110-mediated event (Fig. 1C and D).
FIG. 1.
Southern blot hybridization analysis of isogenic M. tuberculosis isolates from a single patient. Genomic DNA isolated from each strain was digested with PvuII (A to C) or HinfI (D), electrophoretically fractionated in 0.8% agarose, and Southern transferred to Hybond N+. Blots were then hybridized with IS-3′ (15) (A), IS-5′ (17) (B), DRr probe (9) (C), and polymorphic GC-rich repetitive sequence probe (16) (D). Lanes: 1, D7030 from a mediastinal lymph node; 2, D7031 from a hilar lymph node; 3, D7033 from an abdominal lymph node. An additional IS6110-hybridizing band is evident in D7031 as indicated by the arrow. DNA molecular size markers are shown at left in kilobases.
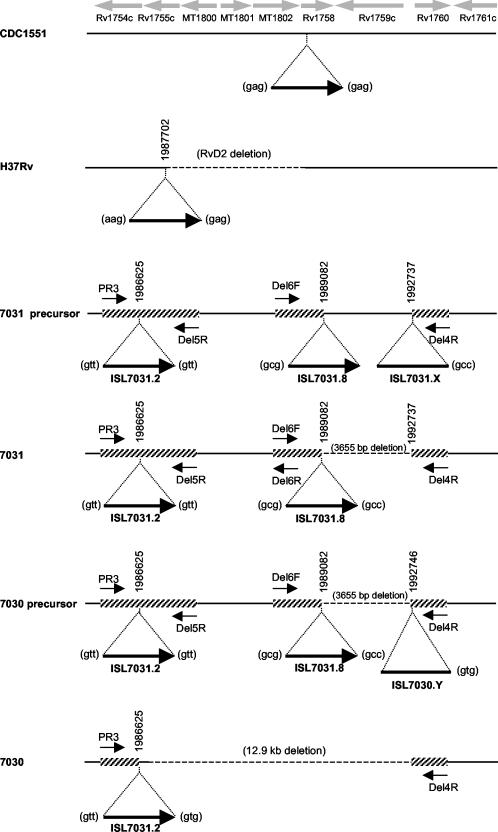
To determine whether the observed banding pattern was indeed IS6110 mediated, PCR primers were designed from the H37Rv (3) and CDC1551 (7) whole genome sequences to facilitate the amplification of the region spanning the insertions at positions ISL7031.2 and ISL7031.8 (12) (Fig. 2). PCR amplifications were performed using the Expand Long Range PCR system (Roche), as previously described (13). Amplification of D7031 with the primer combination PR3 and Del5R produced a product of 2.0 kb, while the primer combination Del4R and Del6F yielded a product of 1.9 kb. In addition, amplification with the primer combination PR3 and Del6R yielded a product of approximately 8 kb, in accordance with the 8.1-kb product that was predicted based on the CDC1551 sequence data (7). This confirmed that the RvD2 region (8) was present in D7031 and flanked by two IS6110 insertions at positions ISL7031.2 and ISL7031.8 (Fig. 2). This structure had not been previously observed, and it was in fact hypothesized that such a structure would be unstable (10). Interestingly, the Del4R and Del6F amplification product was 3.6 kb shorter than predicted (based on the CDC1551 genome sequence data), suggesting a possible deletion event. DNA sequencing of this product demonstrated the presence of an IS6110 element with the 3′ point of insertion corresponding to chromosomal position 1989082 (numbered according to H37Rv genome sequence), while the 5′ point of insertion corresponded to chromosomal position 1992737, 3,655 bp downstream (Fig. 2). The triplet repeats flanking this insertion element were not conserved. This suggests that a precursor strain existed wherein an additional IS6110 insertion (ISL7031.X [Fig. 2]) occurred downstream of the IS6110 insertion at position ISL7031.8, with subsequent recombination between the IS6110 elements leading to the deletion of the 3,655-bp intervening region. Alternatively, it is conceivable that a transpositionally mediated mechanism (14), such as deletion of target DNA on integration of ISL7031.8, may have resulted in the structure observed here, although this has not been previously reported in M. tuberculosis.
FIG. 2.
Schematic diagram of the chromosomal domain in M. tuberculosis. The IS6110 insertion elements are depicted by bold black arrows where the orientation is 3′ (tail) to 5′ (head). IS6110 insertions for which the orientation is unknown are depicted by bold black lines. The respective insertion points are indicted by vertical numbers, which relate to the whole genome sequence of H37Rv (3). Each insertion locus is numbered according to the numbering in reference 12. Open reading frames, according to the annotation of the whole genome sequences of H37Rv (3) and CDC1551 (7), are indicated by the grey arrows. Dotted lines indicate chromosomal deletions. Deletion RvD2 was assigned as described in reference 8. Thin arrows indicate the primer positions, while the hatched lines indicate regions sequenced.
PCR amplification of D7030 using the primer combination PR3 and Del5R and the primer combination Del4R and Del6F failed to generate products. Amplification using PR3 and Del4R produced a product of 2.1 kb, suggesting that the region between the IS6110 elements at position ISL7031.2 and ISL7031.8 had been deleted from strain D7030. This was confirmed by sequence analysis (Fig. 2). However, the sequence data demonstrated that this deletion event was not mediated by the recombination of these adjacent IS6110 elements. The DNA sequence of the flanking region showed that the insertion point corresponded to chromosomal position 1992746, 9 bp downstream of ISL7031.8. In addition, the 5′ triplet repeat differed from that of ISL7031.8. Based on this result, we hypothesize the existence of a second precursor strain wherein an additional IS6110 insertion (at position ISL7030.Y [Fig. 2]) occurred in the region downstream of the insertion at position ISL7031.8. Subsequently, recombination between the IS6110 elements at positions ISL7030.Y and ISL7031.2 may have mediated the deletion event, resulting in the deletion of five genes as well as the IS6110 element inserted at position ISL7031.8. Although other transpositionally mediated mechanisms may explain the structure observed here, for reasons discussed previously by Sampson et al. (13) and others (5), we favor the recombination-mediated scenario. In combination, the recombination between IS6110 elements inserted at ISL7031.X and ISL7031.8 as well as the recombination between ISL7030.Y and ISL7031.2 resulted in the overall deletion of a 12.9-kb fragment.
In summary, this study provides the first description of an IS6110-mediated deletion event in truly isogenic strains. However, as Sampson et al. have previously observed (13), the mechanism of deletion was more complex than simple homologous recombination between directly repeated IS6110 elements. From the results of this study and others (10, 13), it is evident that this chromosomal region from Rv1754 to Rv1765 has undergone a number of deletion events during its evolutionary history. The frequency at which these domains are deleted together with the fact that the strains caused disease in the human host implies that this region may not be important to the overall virulence of M. tuberculosis.
The results presented here emphasize that this region, like the DR (18) and IS6110 preferential locus (6) regions of the M. tuberculosis genome, is a hot spot for IS6110 insertions (10), which consequently drives the high rate of deletion events observed here. This suggests that regions of preferential IS6110 integration loci may be in a state of flux due to the disruptive influence of IS6110 insertion as well as IS6110-mediated deletion and thereby confirms the importance of IS6110 in defining the architecture of the M. tuberculosis chromosome. If preferential integration loci are also preferential deletion regions (13), these findings suggest that some caution should be exercised when reconstructing evolutionary histories of strains based on deletion mapping. Depending on the sensitivity of the detection method, deletions offset by only a few base pairs may not be distinguished from one another, thereby leading to an overestimation of relatedness. In addition, deletions in different regions may be more or less informative, depending on whether the region is a deletion hot spot or not. Together, these factors may lead to inaccurate measures of phylogenetic distances.
Acknowledgments
We thank the GlaxoSmithkline Action TB Initiative for supporting this study.
We are grateful to D. G. du Plessis for providing the mycobacterial strains.
REFERENCES
- 1.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 2.Brosch, R., W. J. Philipp, E. Stavropoulos, M. J. Colston, S. T. Cole, and S. V. Gordon. 1999. Genomic analysis reveals variation between Mycobacterium tuberculosis H37Rv and the attenuated M. tuberculosis H37Ra strain. Infect. Immun. 67:5768-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 4.du Plessis, D. G., R. Warren, M. Richardson, J. J. Joubert, and P. D. van Helden. 2001. Demonstration of reinfection and reactivation in HIV-negative autopsied cases of secondary tuberculosis: multilesional genotyping of Mycobacterium tuberculosis utilizing IS6110 and other repetitive element-based DNA fingerprinting. Tuberculosis 81:211-220. [DOI] [PubMed] [Google Scholar]
- 5.Fang, Z., C. Doig, D. T. Kenna, N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang, Z., and K. J. Forbes. 1997. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J. Clin. Microbiol. 35:479-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, J. W. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 9.Hermans, P. W., D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho, T. B., B. D. Robertson, G. M. Taylor, R. J. Shaw, and D. B. Young. 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sampson, S. L., R. M. Warren, M. Richardson, G. D. van der Spuy, and P. D. van Helden. 1999. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuber. Lung Dis. 79:349-359. [DOI] [PubMed] [Google Scholar]
- 13.Sampson, S. L., R. M. Warren, M. Richardson, T. C. Victor, A. M. Jordaan, G. D. van der Spuy, and P. D. van Helden. 2003. IS6110-mediated deletion polymorphism in the direct repeat region of clinical isolates of Mycobacterium tuberculosis. J. Bacteriol. 185:2856-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherratt, D. 1989. Tn3 and related transposable elements: site-specific recombination and transposition., p. 163-184. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. ASM Press, Washington, D.C.
- 15.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren, R., M. Richardson, S. Sampson, J. H. Hauman, N. Beyers, P. R. Donald, and P. D. van Helden. 1996. Genotyping of Mycobacterium tuberculosis with additional markers enhances accuracy in epidemiological studies. J. Clin. Microbiol. 34:2219-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warren, R. M., M. Richardson, S. L. Sampson, G. D. van der Spuy, W. Bourn, J. H. Hauman, H. Heersma, W. Hide, N. Beyers, and P. D. van Helden. 2001. Molecular evolution of Mycobacterium tuberculosis: phylogenetic reconstruction of clonal expansion. Tuberculosis 81:291-302. [DOI] [PubMed] [Google Scholar]
- 18.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zumarraga, M., F. Bigi, A. Alito, M. I. Romano, and A. Cataldi. 1999. A 12.7 kb fragment of the Mycobacterium tuberculosis genome is not present in Mycobacterium bovis. Microbiology 145:893-897. [DOI] [PubMed] [Google Scholar]