Abstract
We describe the application of 16S rRNA gene sequencing in defining eight cases of bacteremia due to Haemophilus species other than Haemophilus influenzae (non-H. influenzae bacteremia) during a 7-year period. The first case of acute pyelonephritis due to Haemophilus segnis is also reported. In contrast to the extremely rare incidence of H. segnis infections reported previously, our results suggested that H. segnis is an important cause of non-H. influenzae bacteremia.
Apart from Haemophilus influenzae, other Haemophilus species, in particular H. segnis, H. haemolyticus, and H. parahaemolyticus, are considered uncommon causes of infections (14). These organisms are often fastidious and may exhibit ambiguous phenotypic profiles. Therefore, the epidemiology of Haemophilus infections not due to H. influenzae (non-H. influenzae infections) may have not been accurately defined. Comparison of the gene sequences of bacterial species has shown that the 16S rRNA gene is highly conserved within a species and among species of the same genus (18, 19). 16S rRNA gene sequences can be used to more accurately identify and classify fastidious organisms and organisms with ambiguous biochemical profiles (9, 11, 22, 24, 25, 27, 28). Recently, we reported the application of 16S rRNA gene sequencing in defining two cases of H. segnis bacteremia (10). In this study, we used 16S rRNA gene sequencing for the species level identification of non-H. influenzae blood culture isolates in a 7-year period. The clinical spectrum of diseases and outcomes for patients with non-H. influenzae bacteremia were also analyzed and are discussed.
The bacterial strains used in this study were isolates from blood cultures of patients hospitalized at the Queen Mary Hospital in Hong Kong during a 7-year period (January 1996 to December 2002). All clinical data were collected prospectively as described previously by Luk et al. (13). The BACTEC 9240 blood culture system (Becton Dickinson) was used, and all suspected colonies were identified by standard conventional biochemical methods (15). All isolates of gram-negative bacilli or coccobacilli that grew on chocolate agar but not on MacConkey agar were tested for factor X and factor V requirements (15), hemolysis on horse blood agar, CO2 enhancement of growth, catalase, and indole production and were identified by the Vitek system (NHI; bioMerieux Vitek). All isolates identified as Haemophilus species other than H. influenzae by either conventional phenotypic tests or the Vitek System (Neisseria/Haemophilus identification [NHI]) were subjected to 16S rRNA gene sequencing. Antimicrobial susceptibility was tested by the Kirby-Bauer disk diffusion method, and results were interpreted according to the NCCLS criteria (16).
Bacterial DNA extraction, PCR amplification, and DNA sequencing of the 16S rRNA genes were performed as described previously (10, 12, 26, 29). LPW55 (5′-AGTTTGATCCTGGCTCAG-3′) and LPW56 (5′-AGGCCCGGGAACGTATTCAC-3′) were used as the PCR primers, and LPW55, LPW56, LPW69 (5′-AGCACCGGCTAACTCCGT-3′), and LPW106 (5′-TAATCCTGTTTGCTCCCCAC-3′) were used as the sequencing primers. The sequences of the PCR products were compared with known 16S rRNA gene sequences in the GenBank (http://www.ncbi.nlm.nih.gov) by multiple sequence alignment using the CLUSTAL W program (23); phylogenetic tree construction was performed with PileUp, and the neighbor-joining method was performed with GrowTree (Genetics Computer Group, Inc., San Diego, Calif.).
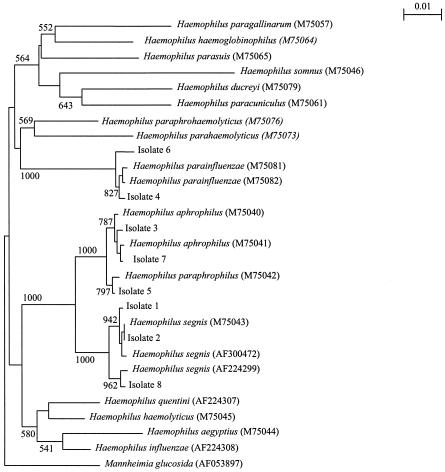
A total of 25 strains of Haemophilus spp. were isolated from the blood cultures of 25 patients during the 7-year study period. Of the 25 isolates, 17 were identified as H. influenzae and 8 were identified as other Haemophilus species by either standard phenotypic tests or the Vitek system (NHI). All of the eight non-H. influenzae isolates, including the two reported previously (10), were gram-negative coccobacilli and grew on chocolate agar to colonies of 0.5 to 1 mm in diameter after incubation for 24 h at 37°C in air with 5% CO2. Their phenotypic characteristics, identification by the Vitek System (NHI), and antibiotic susceptibilities are shown in Table 1. PCR of the 16S rRNA genes of these eight isolates showed bands at about 1,400 bp. Three isolates (isolates 1, 2, and 8) were identified by 16S rRNA gene sequencing as H. segnis, two (isolates 4 and 6) were identified as H. parainfluenzae, two (isolates 3 and 7) were identified as H. aphrophilus, and one (isolate 5) was identified as H. paraphrophilus (Fig. 1). Only one of the eight isolates (isolate 4) was identified correctly by a combination of conventional biochemical tests and the Vitek System (NHI; bioMerieux Vitek).
TABLE 1.
Patient characteristics, phenotypic characteristics, identification and antibiotic susceptibilities of the eight non-H. influenzae blood culture isolates
| Parameter | Result for patient:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Yr of isolation | 1996 | 1996 | 1998 | 1999 | 1999 | 2000 | 2000 | 2000 |
| Sex/age (yr) | F/83 | M/32 | M/62 | F/70 | M/48 | M/49 | F/65 | F/78 |
| Underlying condition(s) | Clostridium difficile colitis during the episode of bacteremia | Intravenous drug abuser | Fractured left medial malleolus due to accidental injury 1 week earlier | Diabetes mellitus, hypertension, intracerebral hemorrhage | Intravenous drug abuser, prosthetic valvular replacement, emphysema | Peptic ulcer treated several years earlier | Hypertension, diabetes mellitus, hyperlipidemia, hypertrophic obstructive cardiomyopathy | Carcinoma of cervix, vesicovaginal fistula, left ureteric obstruction |
| Diagnosis | Primary bacteremia | Bacteremic empyema | Bacteremic osteomyelitis of left distal tibia | Primary bacteremia | Infective endocarditis | Bacteremic spondylitis and discitis | Primary bacteremia | Bacteremic left pyelonephritis |
| Complication | Septic shock | None | None | None | None | None | None | Acute renal failure |
| No. of positive blood cultures | 1 | 1 | 1 | 2 | 3 | 2 | 1 | 1 |
| Type of bacteremia (concomitant isolates) | Monomicrobial | Polymicrobial (Streptococcus intermedius, Streptococcus sanguinis) | Monomicrobial | Monomicrobial | Monomicrobial | Monomicrobial | Polymicrobial (Streptococcus anginosus, Bacteroides fragilis, Eikenella corrodens) | Monomicrobial |
| X factor requirement | − | − | − | − | − | − | − | − |
| V factor requirement | + | + | − | + | + | + | − | + |
| Hemolysis of horse blood | − | − | − | − | − | + | − | − |
| CO2 enhancement of growth | − | − | − | − | − | − | + | − |
| Catalase | − | − | − | + | − | + | − | − |
| Indole production | − | − | − | − | − | − | − | − |
| Identification by Vitek system (NHI) | 95% H. influenzae VIII | 56% Actinobacillus actinomycetemcomitans, 40% Neisseria subflava | 62% H. paraphrophilus, 37% H. aphrophilus | 98% H. parainfluenzae I | 62% H. paraphrophilus, 37% H. aphrophilus | 95% H. parainfluenzae III | 67% H. ducreyi, 29% Kingella kingae | 62% H. paraphrophilus, 37% H. aphrophilus |
| Identification by 16S rRNA sequencing | H. segnis | H. segnis | H. aphrophilus | H. parainfluenzae | H. paraphrophilus | H. parainfluenzae | H. aphrophilus | H. segnis |
| Antibiotic susceptibilitya | ||||||||
| Ampicillin | S | S | S | S | S | S | S | S |
| Cefotaxime | S | S | S | S | S | S | S | S |
| Imipenem | S | S | S | S | S | S | S | S |
| Cotrimoxazole | S | S | S | S | S | M | S | S |
| Chloramphenicol | S | S | S | S | S | S | S | S |
S, sensitive; M, intermediate resistant.
FIG. 1.
Phylogenetic tree showing the relationship of the eight blood culture isolates from our patients to related species. The tree was inferred from 16S rRNA sequence data by the neighbor-joining method. Bootstrap values were calculated from 1,000 iterations. Scale bar, estimated number of substitutions per 100 bases by using the Jukes-Cantor correction. Names and accession numbers are given as cited in the GenBank database.
The clinical characteristics of the eight patients with non-H. influenzae bacteremia are summarized in Table 1. All patients had community-acquired bacteremia. With the exception of those associated with H. segnis, the clinical manifestations of Haemophilus bacteremia in our patients were compatible to those reported previously (2, 6, 17, 21). In contrast to H. influenzae bacteremia, which often affects young children and healthy adults, non-H. influenzae bacteremia was found to affect only adult patients (median age, 64 years; range, 32 to 83 years) who often had underlying diseases. Despite this, complications from non-H. influenzae bacteremia were not common. Apart from patient 1, who deteriorated rapidly after the bacteremia and died, all patients were cured with antibiotic treatment.
The present study suggests that the prevalence of H. segnis bacteremia may have been underestimated previously. Identification of H. segnis by conventional biochemical tests has been difficult (10). All three isolates in the present study could not be identified by phenotypic methods. H. segnis is phenotypically very similar to H. parainfluenzae and therefore may have been reported as H. parainfluenzae by many clinical laboratories (7). Although the numbers were small, H. segnis accounted for 37.5% (three of eight) of the cases of non-H. influenzae bacteremia in our locality, compared to 25% (two of eight) for both H. parainfluenzae and H. aphrophilus and 12.5% (one of eight) for H. paraphrophilus. This is in contrast to the rare isolation of H. segnis from blood cultures reported in the literature. Apart from the present three cases, there were only two reports of H. segnis bacteremia, one in a case of infective endocarditis and the other in a case of pancreatic abscess (1, 3). The first two cases in the present report, a case of primary bacteremia and a case of empyema thoracis, have been described previously, and the sources of the bacteremia were thought to be the gastrointestinal tract in one case and the oral cavity in the other (10). On the other hand, the last case represents the first report of pyelonephritis due to H. segnis. Since the patient had vesicovaginal fistula as a result of local radiotherapy for her carcinoma of the cervix, it is likely that the source of infection was her genital tract, which may have been transiently colonized by the bacterium. 16S rRNA gene sequencing should be used to identify more cases of H. segnis infections and better define its epidemiology, clinical spectrum, treatment, and outcome.
16S rRNA gene sequencing is the method of choice for the identification of Haemophilus species. As early as 1992, Dewhirst et al. reported the use of 16S rRNA gene sequences for phylogenetic study of Pasteurellaceae (5). Subsequently, its application for identification of isolated cases of H. parainfluenzae and H. segnis infections has been described (4, 7, 10). The present study also showed that, apart from its usefulness in identifying H. segnis, the technique is helpful for strains of other species, especially those with ambiguous biochemical profiles. One of our isolates (isolate 6) showed β-hemolysis on horse blood agar and was factor V, but not factor X, dependent. It would therefore be identified as H. parahaemolyticus by most clinical laboratories. 16S rRNA gene sequencing unambiguously identified it as H. parainfluenzae, an identification which is compatible with its other phenotypic characteristics. We speculate the isolate was a hemolytic variant of H. parainfluenzae that had acquired a hemolysin gene. Another isolate (isolate 7) also exhibited ambiguous biochemical profiles and was identified as 67% H. ducreyi by the Vitek system (NHI). It was confirmed to be H. aphrophilus only later by 16S rRNA gene sequencing. Although restriction enzyme analysis of PCR-amplified 16S rRNA genes has been used to distinguish among Actinobacillus actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus (20), such a technique has not been demonstrated to be applicable to other species, and results may be compromised by single nucleotide substitutions at restriction sites. The translation initiation factor 2 has also been shown useful for phylogenetic analysis of Haemophilus (8) and may be an alternative molecular target useful for species identification.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of blood culture isolates 1, 2, 3, 4, 5, 6, 7, and 8 have been lodged within the GenBank sequence database under accession numbers AY365447, AY365448, AY365449, AY365450, AY365451, AY365452, AY365453, and AY365454, respectively.
Acknowledgments
This work was partly supported by the University Development Fund, University Research Grant Council, and the Committee of Research and Conference Grants, The University of Hong Kong.
REFERENCES
- 1.Bangsbor, J. M., M. Tvede, and P. Skinhoj. 1988. Haemophilus segnis endocarditis. J. Infect. 16:81-85. [DOI] [PubMed] [Google Scholar]
- 2.Beauvais, C., F. Berenbaum, M. Spentchian, A. Prier, and G. Kaplan. 1992. Early diagnosis of vertebral osteomyelitis due to a rare pathogen: Haemophilus parainfluenzae. J. Rheumatol. 19:491-493. [PubMed] [Google Scholar]
- 3.Bullock, D. W., and P. G. Devitt. 1981. Pancreatic abscess and septicaemia caused by Haemophilus segnis. J. Infect. 3:82-85. [DOI] [PubMed] [Google Scholar]
- 4.Das, I., J. V. DeGiovanni, and J. Gray. 1997. Endocarditis caused by Haemophilus parainfluenzae identified by 16S ribosomal RNA sequencing. J. Clin. Pathol. 50:72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewhirst, F. E., B. J. Paster, I. Olsen, and G. J. Fraser. 1992. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J. Bacteriol. 174:2002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewire, P., B. E. McGrath, and C. Brass. 1999. Haemophilus aphrophilus osteomyelitis after dental prophylaxis. A case report. Clin. Orthop. 363:196-202. [PubMed] [Google Scholar]
- 7.Hamed, K. A., P. R. Dormitzer, C. K. Su, and D. A. Relman. 1994. Haemophilus parainfluenzae endocarditis: application of a molecular approach for identification of pathogenic bacterial species. Clin. Infect. Dis. 19:677-683. [DOI] [PubMed] [Google Scholar]
- 8.Hedegaard, J., H. Okkels, B. Bruun, M. Kilian, K. K. Mortensen, and N. Norskov-Lauritsen. 2001. Phylogeny of the genus Haemophilus as determined by comparison of partial infB sequences. Microbiology 147:2599-2609. [DOI] [PubMed] [Google Scholar]
- 9.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau, S. K. P., P. C. Y. Woo, B. Y. L. Chan, A. M. Y. Fung, T. L. Que, and K. Y. Yuen. 2002. Haemophilus segnis polymicrobial and monomicrobial bacteremia identified by 16S ribosomal RNA gene sequencing. J. Med. Microbiol. 51:635-640. [DOI] [PubMed] [Google Scholar]
- 11.Lau, S. K. P., P. C. Y. Woo, G. K. S. Woo, and K. Y. Yuen. 2002. Catheter-related Microbacterium bacteremia identified by 16S ribosomal RNA gene sequencing. J. Clin. Microbiol. 40:2681-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau, S. K., P. C. Y. Woo, H. Tse, K. W. Leung, S. S. Wong, and K. Y. Yuen. 2003. Invasive Streptococcus iniae infections outside North America. J. Clin. Microbiol. 41:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luk, W. K., S. S. Y. Wong, K. Y. Yuen, P. L. Ho, P. C. Y. Woo, R. A. Lee, and P. Y. Chau. 1998. Inpatient emergencies encountered by an infectious disease consultative service. Clin. Infect. Dis. 26:695-701. [DOI] [PubMed] [Google Scholar]
- 14.Mandell, G. L., J. E. Bennett, and R. Dolin. 2000. Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 15.Murray, P. R., E. J. Baro, M. A. Pfaller, F. C. Tenover, and. R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 16.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A8, 8th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Olk, D. G., R. J. Hamill, and R. A. Proctor. 1987. Haemophilus parainfluenzae vertebral osteomyelitis. Am. J. Med. Sci. 294:114-116. [DOI] [PubMed] [Google Scholar]
- 18.Olsen, G. J., and C. R. Woese. 1993. Ribosomal RNA: a key to phylogeny. FASEB J. 7:113-123. [DOI] [PubMed] [Google Scholar]
- 19.Olsen, G. J., R. Overbeek, N. Larsen, T. L. Marsh, M. J. McCaughey, M. A. Maciukenas, W. M. Kuan, T. J. Macke, Y. Xing, and C. R. Woese. 1992. The ribosomal database project. Nucleic Acids Res. 20:2199-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riggio, M. P., and A. Lennon. 1997. Rapid identification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, and Haemophilus paraphrophilus by restriction enzyme analysis of PCR-amplified 16S rRNA genes. J. Clin. Microbiol. 35:1630-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts, D. E. 1998. Femoral osteomyelitis after tooth extraction. Am. J. Orthop. 27:624-626. [PubMed] [Google Scholar]
- 22.. Rogall, T., T. Flohr, and E. C. Bottger. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J. Gen. Microbiol. 136:1915-1920. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, and K. Y. Yuen. 2002. Identification by 16S ribosomal RNA gene sequencing of Lactobacillus salivarius bacteremic cholecystitis. J. Clin. Microbiol. 40:265-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo, P. C. Y., A. M. Y. Fung, S. S. Y. Wong, H. W. Tsoi, and K. Y. Yuen. 2001. Isolation and characterization of a Salmonella enterica serotype Typhi variant and its clinical and public health implications. J. Clin. Microbiol. 39:1190-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo, P. C. Y., D. M. W. Tam, K. W. Leung, S. K. P. Lau, J. L. L. Teng, M. K. M. Wong, and K. Y. Yuen. 2002. Streptococcus sinensis sp. nov., a novel Streptococcus species isolated from a patient with infective endocarditis. J. Clin. Microbiol. 40:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo, P. C. Y., H. W. Tsoi, K. W. Leung, P. N. L. Lum, A. S. P. Leung, C. H. Ma, K. M. Kam, and K. Y. Yuen. 2000. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S ribosomal RNA sequencing. J. Clin. Microbiol. 38:3515-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo, P. C. Y., J. H. C. Li, W. M. Tang, and K. Y. Yuen. 2001. Acupuncture mycobacteriosis. N. Engl. J. Med. 345:842-843. [DOI] [PubMed] [Google Scholar]
- 29.Yuen, K. Y., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, M. K. M. Wong, and S. K. P. Lau. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 39:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]