Abstract
A total of 202 methicillin-resistant Staphylococcus aureus (MRSA) single-patient isolates recovered between January and June 1998 in two hospitals in Miami, Florida, were characterized by a combination of several molecular typing techniques: multilocus sequence typing, spaA typing, pulsed-field gel electrophoresis, and determination of the structure of the SCCmec element. The overwhelming majority of the isolates—187of 202, or 93%—belonged to one of three internationally spread epidemic clones which were identified on the basis of their multilocus sequence type (ST) as E-MRSA-16 (ST36), the New York clone V (ST8), and the New York/Japan clone (ST5; SCCmec II) and its single- and double-locus variants. The rest of the isolates (15 of 202, or 7%) were more genetically diverse and were each recovered from a few patients only. Of the 23 MRSA strains isolated from confirmed human immunodeficiency virus-positive patients, as many as 17 (or 70%) belonged to a single ST8 clone carrying SCCmec type IV. The data provide further evidence for the conclusion of earlier studies that most MRSA disease in hospitals is caused by relatively few pandemic clones.
The introduction of molecular typing techniques into epidemiological investigations has provided powerful new tools for tracking the origin and routes of dissemination of methicillin-resistant Staphylococcus aureus (MRSA) strains not only among patients in a hospital but also among hospitals in different countries or even across different continents. One of the interesting conclusions that have emerged from large international surveillance studies using such molecular typing techniques is that a relatively few MRSA clones have been responsible for a disproportionately large fraction of all MRSA disease worldwide (8, 16). In addition these studies have also indicated a considerable degree of geographic specificity in the spread of various MRSA lineages. For instance, the Iberian clone of MRSA was found to be the dominant clone in hospitals in Southern Europe (16), while the clone referred to as E-MRSA-16 was most frequently recovered in hospitals in the United Kingdom (11), Canada (24), and Mexico (2). In surveillance of MRSA disease in 12 New York City hospitals in 1996 (18), as many as 42% of all MRSA isolates belonged to the New York/Japan clone. In a follow-up study conducted at 29 hospitals and/or health centers in the tristate area of New Jersey, Pennsylvania, and Connecticut in 1998 (19), the same clonal type was represented in 92% of the MRSA isolates from the participating centers in Pennsylvania, 65% of MRSA isolates from New Jersey, and 39% of MRSA isolates from Connecticut. The purpose of the study described here was to sample hospitals in the United States at a geographic site distant from the Northeast. Two hospitals, both located in Miami, Florida, were selected for such a study, and the basic observations are described in this communication.
MATERIALS AND METHODS
The first of the two sites selected for the study was The Jackson Memorial Hospital (hospital C), a 1,550-bed hospital that serves the population of Miami-Dade County. Services include general acute care, a trauma center, and multiorgan transplantation programs. The second site was the Miami Veterans Affairs Medical Center (hospital D), a 220-bed acute care facility serving the veterans of southeastern Florida. Both hospitals are closely affiliated with the University of Miami School of Medicine. During the calendar year of 1998, the frequency of methicillin resistance among S. aureus isolates was 38% in hospital C and about 50% in hospital D.
Of the MRSA isolates recovered in these two hospitals between 1 June and 23 December 1998, a total of 202 isolates were available for molecular typing: 113 from hospital C and 89 from hospital D. Information was obtained on the source of each isolates (blood, the respiratory tract, wounds, urine, or other [including the nose, stool, and skin]), whether the isolate was collected within the first 72 h of hospitalization (community acquired) or after 72 h of hospitalization (nosocomial), and whether the isolate was from a patient with human immunodeficiency virus infection (if that information was available). The major clinical sources of the MRSA isolates were blood (n = 32), the respiratory tract (n = 53), wounds (n = 25), urine (n = 35), and other sources (n = 57). The great majority of isolates were of nosocomial origin. Species identification and antibiograms were performed at the clinical microbiology laboratories of the hospitals. Cultures were frozen in tryptic soy broth containing 10% glycerol (Difco, Detroit, Mich.) and were stored at −70°C until transfer to the Laboratory of Microbiology at The Rockefeller University, New York, N.Y., for molecular typing.
Table 1 lists the MRSA isolates according to their molecular types and antibiotypes. Multilocus sequence typing (MLST) (6, 7), determination of the spaA type (15, 22), and pulsed-field gel electrophoresis (PFGE) of SmaI-digested chromosomal DNA (5) were performed by published methods. A multiplex PCR technique was used to identify the type of the mec element (SCCmec type) carried by the bacterium (17).
TABLE 1.
Molecular types and antibiotypes of MRSA isolates recovered at two Miami hospitals
| STa | MLST | Trivial name | spaA type | SCCmecb | PFGE typec | Typical antibiogramd | No. of isolates in:
|
|
|---|---|---|---|---|---|---|---|---|
| Hospital C | Hospital D | |||||||
| 36 | 2, 2, 2, 2, 3, 3, 2 | EMRSA-16 | WGKAKAOMQQQ | II | A1 | P, OX, CI, CM, E, G | 41 | 23 |
| 30 | 2, 2, 2, 2, 6, 3, 2 | EMRSA-16 (SLV) | WGKAKAOMQQ | II | A2 | P, OX, E | 1 | 0 |
| 5 | 1, 4, 1, 4, 12, 1, 10 | NY/JP | TJMBMDMGMK | II | ND | P, OX, CI, CM, E | 11 | 12 |
| 5 | 1, 4, 1, 4, 12, 1, 10 | NY/JP | TJMGMK | II | B2 | P, OX, CI, CM, E | 1 | 0 |
| 5 | 1, 4, 1, 4, 12, 1, 10 | NY/JP | TMBMAMGMK | II | ND | P, OX, CI, CM, E, G | 0 | 1 |
| 105 | 1, 4, 1, 4, 12, 1, 28 | NY/JP (SLV) | TJMBMDMGMK | II | B1 | P, OX, CI, CM, E | 11 | 5 |
| 105 | 1, 4, 1, 4, 12, 1, 28 | NY/JP (SLV) | TJMGM | II | ND | P, OX, CI, CM, E, G | 0 | 1 |
| 231 | 1, 34, 1, 4, 12, 1, 28 | NY/JP (DLV) | TJMBMDMGMK | II | B3 | P, OX, CI, CM, E | 5 | 1 |
| 83 | 1, 4, 1, 4, 12, 1, 1 | NY/JP (SLV) | TJMBMDMGMK | II | B4 | P, OX, CI, CM, E, G | 0 | 1 |
| 8 | 3, 3, 1, 1, 4, 4, 3 | NYV | YHGCMBQBLO | IV | C | P, OX, CI, CM, E, G, RI, T, TMP/SXT | 25 | 40 |
| 8 | 3, 3, 1, 1, 4, 4, 3 | NYV | YHB2CMBQBLO | IV | ND | P, OX, CI, T, TMX/SXT | 1 | 0 |
| 8 | 3, 3, 1, 1, 4, 4, 3 | NYV | YC2CMBQBLO | IV | ND | P, OX, CI, T | 0 | 1 |
| 8 | 3, 3, 1, 1, 4, 4, 3 | NYV | YHHGCMBQBLO | IV | ND | P, OX, CI, CL, CM, E, G, TMP/SXT | 0 | 1 |
| 8 | 3, 3, 1, 1, 4, 4, 3 | NYV | YHGFMBQBLO | I | ND | P, OX, CI, CL, CM, E, G | 5 | 0 |
| 239 | 2, 3, 1, 1, 4, 4, 3 | Brazil | WGKAOMQ | III | D1 | P, OX, CI, CM, E, G, RI, T | 1 | 0 |
| 239 | 2, 3, 1, 1, 4, 4, 3 | Brazil | XKAOMQ | III | D2 | P, OX, CI, CM, E, G, T, TMP/SXT | 1 | 0 |
| 241 | 2, 3, 1, 1, 4, 4, 30 | Brazil (SLV) | WGKAOMQ | IIIA | ND | P, OX, CI, CM, E, G, T, TMP/SXT | 1 | 0 |
| 72 | 1, 4, 1, 8, 4, 4, 3 | None | UJGFMDMGMGM | IV | E | P, OX, E, G | 1 | 0 |
| 107 | 1, 4, 2, 8, 4, 4, 3 | ST72 (SLV) | UJGFGMDMGMGM | IV | E | P, OX, CM, E, G | 1 | 0 |
| 9 | 3, 3, 1, 1, 1, 1, 10 | None | UKGJB | NT | F1 | NA | 1 | 0 |
| 9 | 3, 3, 1, 1, 1, 1, 10 | None | H2GJAABB | NT | F2 | P, OX, CL, CM, E, T | 1 | 0 |
| 15 | 13, 13, 1, 1, 12, 11, 13 | None | UJGBBBGGJA | NT | G | P, OX, T | 1 | 0 |
| 97 | 3, 1, 1, 1, 1, 5, 3 | None | UJFMBBBPB | IV | I | P, OX, CL, T | 1 | 0 |
| 45 | 10, 14, 8, 6, 10, 3, 2 | Community aquired | A2KBEMBKB | II | L1 | P, OX, CI, CM, E | 2 | 0 |
| 45 | 10, 14, 8, 6, 10, 3, 2 | Community aquired | A2AKEMBKB | II | L2 | P, OX, CI, CM, E | 0 | 1 |
| 45 | 10, 14, 8, 6, 10, 3, 2 | Community aquired | XKABMB | IV | M | OX, CI, CM, E | 0 | 2 |
| 106 | 3, 1, 14, 15, 11, 43, 3 | None | ZDGMDMGM | NT | K | P, OX, CL, CM, T | 1 | 0 |
ST, sequence type.
NT, nontypeable.
ND, not done.
P, penicillin; OX, oxacillin; CI, clindamycin; CM, chloramphenicol; E, erythromycin; G, gentamicin; RI, rifampin; T, tetracycline; TMP/SXT, trimethoprim-sulfamethoxazole.
RESULTS AND DISCUSSION
We chose a combination of several molecular typing techniques for characterization of the MRSA isolates recovered in the two Miami hospitals, since these methods can complement one another both in terms of expedience and in terms of the type of information they provide. While superior in resolving power, PFGE may blur evolutionary relationships among strains which may be revealed by MLST, particularly when the structure of the SCCmec type is also determined. Experience with the relatively simple sequence-based spaA typing technique, recently introduced, indicates that this method can frequently predict the MLST type (14, 16). While short-term epidemiological investigations need high-resolution tracking techniques such as PFGE, additional characterization of strains by MLST and other sequence-based techniques allows one to recognize issues that transcend the geographic boundaries of a hospital and permits insights into the global epidemiology and population biology of S. aureus as well. From this perspective, the molecular typing data generated in the two Miami hospitals allow one to draw several conclusions.
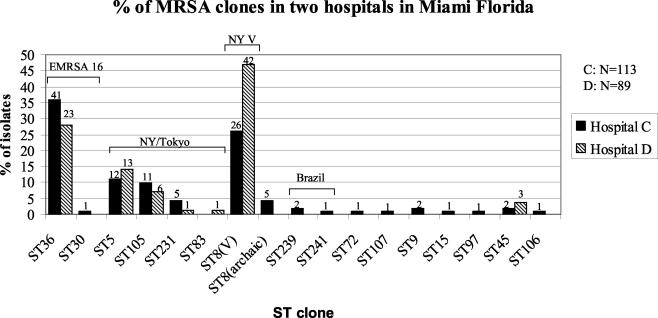
The most frequent MRSA clone in hospital C and the second most frequent clone in hospital D was E-MRSA-16 (ST36) (Fig. 1). All MRSA isolates belonging to this sequence type in both hospitals were homogeneous: they had identical MLSTs, carried SCCmec type II, and showed a uniform PFGE pattern as well as an invariant spaA type (Fig. 2). E-MRSA-16 is widespread in the United Kingdom (11) and has also been detected in Greece (3), Scandinavian countries (20, 21), Switzerland and Belgium (12), Mexico (2), and Canada (24). Our identification of this clone in the two Miami hospitals represents the first detection of E-MRSA-16 in the United States. The uniformity of MRSA isolates belonging to this clone may be interpreted as an indication that E-MRSA-16 has arrived relatively recently at the Miami hospitals.
FIG. 1.
Molecular types of MRSA isolates recovered in two Miami hospitals. MRSA isolates recovered in hospitals C and D were characterized by MLST, and the frequencies of the various sequence types (ST) are plotted as percentages of the total number of MRSA isolates. The number of strains with the indicated genotype at the indicated hospital is given above each bar.
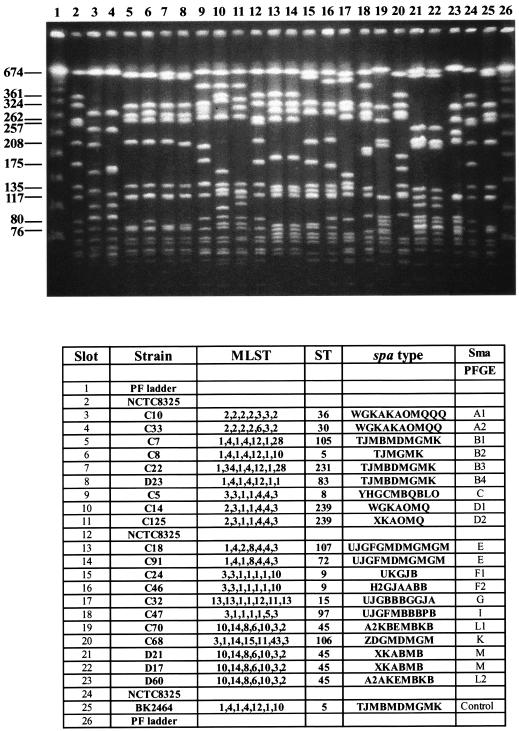
FIG. 2.
(Top) PFGE patterns of the various sequence types of MRSA isolates recovered in the two Miami hospitals. Chromosomal DNAs were isolated, restricted by SmaI, and separated by PFGE. (Bottom) The various strains used as sources of the chromosomal DNA are identified by their MLST, sequence type (ST), and spaA type. The hospital source of each isolate is indicated by the letter C or D in the strain designation; strains NTCT8325 and BK2464 are included for comparison.
The second most frequent MRSA clone was the New York clone V (14), which accounted for 47% of all MRSA isolates in hospital D and was the second most frequent clone (36%) in hospital C (Fig. 1). This clone was first identified as an epidemic MRSA clone in a surveillance study in New York, N.Y., where it was associated with SCCmec IV and was assigned the multilocus sequence type ST8 (14). However, the ST8 genetic background was proposed to represent the predicted ancestor of the very first European MRSA strain (8). Furthermore, a single-locus variant of ST8, MRSA clonal type ST247, also known as the Iberian clone (6), is one of the most widely spread MRSA clones in Southern and Western Europe (16). In the two Miami hospitals, most of the strains belonging to ST8 (68 of 73) carried SCCmec type IV but 5 strains of this sequence type were associated with SCCmec type I, which was shown to be present in the first MRSA isolates from the United Kingdom and Denmark (6, 8). Variation in the SCCmec types associated with this genetic background has been demonstrated previously (8).
MRSA strains belonging to the New York clone V and carrying SCCmec type IV were recovered with relatively high frequency (70%) from human immunodeficiency virus-positive patients in the two Miami hospitals, and a similar association was noted in a surveillance study in New York (18, 19). The significance of these observations is not clear at present. Unlike that of SCCmec type I, the structure of SCCmec type IV lacks the pls gene, which may be involved in colonization by S. aureus (16).
In contrast to the MRSA isolates belonging to the E-MRSA-16 clone, MRSA isolates belonging to the New York clone V showed variations in spaA type (Fig. 2).
The third most frequent MRSA clone in both Miami hospitals was the New York/Japan clone (multilocus sequence type ST5), together with its single-locus variants ST105 and ST83 and its double-locus variant ST231 (Fig. 1). All these strains carried SCCmec type II. MRSA isolates belonging to this clone are most frequent in the northeastern United States (7, 18, 19), Canada (24), and Japan (1). All MRSA isolates identified so far in the United States and Japan with reduced susceptibility to vancomycin (so-called VISA strains) belong to this clone (8, 23). Variations both in spaA type and in PFGE pattern were detected among representatives of this clone (Fig. 2).
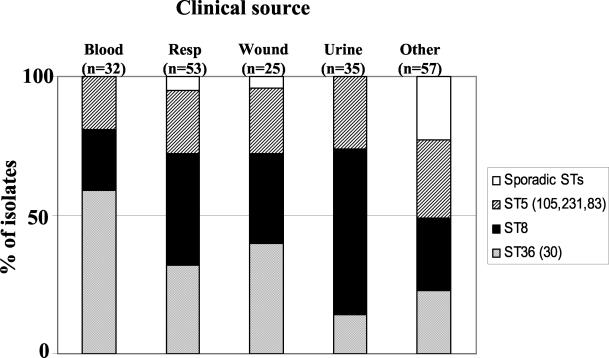
The capacity of each of these three major MRSA clonal types to cause staphylococcal disease is documented by their frequent recovery from all major clinical sites (Fig. 3).
FIG. 3.
Recovery of the three major MRSA clonal types and sporadic MRSA sequence types (STs) from various clinical sites of disease in the two Miami hospitals. The frequencies of association of the MRSA clonal types ST36 (light shading), ST8 (solid), and ST5 (105, 231, 83) (dark shading) and of sporadic STs (open) with the different clinical sites of disease are shown. Resp, respiratory tract.
In contrast to the majority of MRSA isolates represented by the three dominant clones, the rest of the isolates (sporadic MRSA isolates; n =15) were more genetically diverse: they included 9 different sequence types in association with several SCCmec types and 12 distinct PFGE patterns. Three of these genetically diverse isolates were representatives of the Brazilian MRSA clone (ST239 and ST241)—a major internationally spread MRSA lineage (4, 16). An additional group of five MRSA isolates belonged to ST45—a clone widely spread both as methicillin-susceptible S. aureus (MSSA) and as MRSA in Western Europe, Scandinavia, Canada, and the northeastern United States (8; www.mlst.net). Two MRSA isolates, with ST107 (SCCmec type IV) and ST106 (SCCmec type unknown), represent novel genetic backgrounds not described previously. One isolate, with ST72 (SCCmec type IV), has previously been described in Spain (17) and Cuba (www.mlst.net). Two isolates belonging to ST9 (SCCmec type unknown) have been reported in the United Kingdom but only as MSSA (www.mlst.net). A single MRSA isolate of ST15 (SCCmec type unknown) and another MRSA isolate belonging to ST97 (SCCmec type IV) have been reported previously from several countries, but only as MSSA strains (www.mlst.net). The SCCmec types that gave no PCR products, namely, those identified in ST106, ST9, and ST15 MRSA strains, are likely to represent new structural types or structural rearrangements of the mec element (10). It is also interesting that three of the genetic backgrounds identified in MRSA isolates from the Miami hospitals, namely, ST9, ST15, and ST97, were reported only as backgrounds of MSSA strains until now. The fact that these genetic backgrounds were identified in MRSA strains in the Miami hospitals is consistent with the notion of the relatively frequent acquisition of the SCCmec element by S. aureus (9, 16). It is noteworthy that most of these 15 genetically more diverse isolates were associated with hospital C, the patient population of which—in contrast to that of hospital D—is more likely to represent a geographically diverse group (from international patient referrals, immigrants, and tourists visiting the Miami area) that may have been the source of these diverse MRSA isolates. Thus, at least some of the sporadic MRSA isolates identified in hospital C may not have a strictly nosocomial origin. It is interesting in this respect that a few of the sporadic isolates belonging to ST45 had the genetic background also seen in some so-called community-acquired MRSA isolates (13).
With the continued and increasing mobility of human populations through international travel, it is expected that the geographic boundaries of dominance among MRSA clones will become blurred in time, and the appearance of the genetically diverse sporadic isolates in hospital C may represent this trend. Why such a large proportion of MRSA disease is caused by so few clonal types of bacteria still remains a puzzle, but one component of the extensive geographic spread of these few pandemic MRSA clones may be related to a combination of particular genetic traits in the genetic background of the bacteria (16).
Acknowledgments
Partial support for these investigations came from a grant from the U.S. Public Health Service (1 RO1 AI45738) and from the Palm Beach Institute of Infectious Diseases (to Istvan Krisko).
We thank Carmen Bermudez for assistance with this project.
REFERENCES
- 1.Aires de Sousa, M., H. de Lencastre, I. Santos Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6:253-258. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., M. Miragaia, I. Santos Sanches, S. Ávila, I. Adamson, S. T. Casagrande, M. C. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America, 1996-1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., C. Bartzavali, I. Spiliopoulou, I. Santos Sanches, M. I. Crisostomo, and H. de Lencastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41:2027-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aires de Sousa, M., M. I. Crisóstomo, I. Santos Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, and Multilaboratory Project Collaborators. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob, Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore, P. C., and J. A. Lindsay. 2002. Molecular characterization of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 51:516-521. [DOI] [PubMed] [Google Scholar]
- 12.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Lijequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis for epidemiological typing of methicillin-resistant Staphylococcus aureus by consensus in 10 European centers and its use to plot the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira, D. C., I. Crisostomo, I. Santos-Sanches, P. Major, C. R. Alves, M. Aires-de-Sousa, M. K. Thege, and H. de Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob, Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, and the MRSA Collaborative Study Group. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in twelve New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 19.Roberts, R. B., M. Chung, H. de Lencastre, J. Hargrave, A. Tomasz, and the Tri-State MRSA Collaborative Study Group. 2000. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey and Pennsylvania. Microb. Drug Resist. 6:245-251. [DOI] [PubMed] [Google Scholar]
- 20.Salmenlinna, S., O. Lyytikainen, and J. Vuopio-Varkila. 2002. Community-acquired methicillin-resistant Staphylococcus aureus, Finland. Emerg. Infect. Dis. 8:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeberg, S., L. Larsson, C. Welinder-Olsson, T. Sandberg, E. Skyman, B. Bresky, A. Lindqvist, and M. van Raalte. 2002. How an outbreak of MRSA in Gothenburg was eliminated: by strict hygienic routines and a massive control-culture program. Lakartidningen 99:3198-3204. [PubMed] [Google Scholar]
- 22.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 24.Simor, A. E., M. Ofner-Agostini, E. Bryce, A. McGeer, S. Paton, M. R. Mulvey, and the Canadian Hospital Epidemiology Committee and Canadian Nosocomial Infection Surveillance Program, Health Canada. 2002. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: results of 5 years of National Surveillance, 1995-1999. J. Infect. Dis. 186:652-660. [DOI] [PubMed] [Google Scholar]