Abstract
W5.43(194), a conserved tryptophan residue among G-protein coupled receptors (GPCRs) and cannabinoid receptors (CB), was examined in the present report for its significance in CB2 receptor ligand binding and adenylyl cyclase (AC) activity. Computer modeling postulates that this site in CB2 may be involved in the affinity of WIN55212-2 and SR144528 through aromatic contacts. In the present study, we reported that a CB2 receptor mutant, W5.43(194)Y, which had a tyrosine (Y) substitution for tryptophan (W), retained the binding affinity for CB agonist CP55940, but reduced binding affinity for CB2 agonist WIN55212-2 and inverse agonist SR144528 by 8-fold and 5-fold, respectively; the CB2 W5.43(194)F and W5.43(194)A mutations significantly affect the binding activities of CP55940, WIN55212-2 and SR144528. Furthermore, we found that agonist-mediated inhibition of the forskolin-induced cAMP production was dramatically diminished in the CB2 mutant W5.43(194)Y, whereas W5.43(194)F and W5.43(194)A mutants resulted in complete elimination of downstream signaling, suggesting that W5.43(194) was essential for the full activation of CB2. These results indicate that both aromatic interaction and hydrogen bonding are involved in ligand binding for the residue W5.43(194), and the mutations of this tryptophan site may affect the conformation of the ligand binding pocket and therefore control the active conformation of the wild type CB2 receptor. W5.43(194)Y/F/A mutations also displayed noticeable enhancement of the constitutive activation probably attributed to the receptor conformational changes resulted from the mutations.
Keywords: G-protein coupled receptors, cannabinoid receptor CB2, ligand binding and G-protein signaling, tryptophan, site-directed mutagenesis, Computer modeling
1. Introduction
CB1 and CB2 are two subtypes of cannabinoid (CB) receptors, and have been classified as Class A rhodopsin-like GPCRs. CB1 is found primarily in brain and neuronal tissue, whereas CB2 is mainly expressed in immune tissues including the spleen, tonsils and thymus [1]. Similar to CB1, activation of CB2 can generate numerous cellular responses through its signal transductions. For example, stimulation of CB2 elicits not only the activation of the mitogen-activated protein kinase [2], but also the dissociation of Gαi proteins and subsequent decrease in intracellular adenylyl cyclase activity [3]. CB2 receptor activation can also mediate endocannabinoid anandamide initiated Ca2+ signaling [4]. Importantly, CB2 has been proven to play a significant role in the regulation of the immunological functions of cannabinoid ligands. Noticeably, new anorectic anti-obesity drug Rimonabant was recently withdrawn from the European market because of concerns over suicide and depression side effects, associated with the use of this CB1 blocker. These facts render CB2 receptor as an attractive drug molecular target via developing non-psychotropic or non CB1-mediated venue to treat various human diseases, such as neuropathic pain, inflammation, osteoporosis, cancers, and autoimmune diseases as detailed in review articles [5-7].
Therefore, identification of the binding sites of cannabinoid (CB) ligands within the CB2 receptor is of great interest not only for understanding the principles that account for the interactions between the ligands and the amino acid residues, but also for the design of selective CB2 ligands. Multiple lines of evidence suggest that the ligand binding pocket of CB2 receptor, like many other GPCRs, is situated mainly within the transmembrane helices (TMH) 3, 4, 5 and 6 as well as part of the extracellular loop 2 [8]. Due to the lack of a CB2 receptor crystal structure, there is little experimental structural information for CB2 ligand-receptor recognition and activation. The combination of site-directed mutagenesis and molecular modeling provides an alternative strategy for the study of the interactions of ligands and specific sites or regions of the receptor.
Numerous CB residues have been characterized by mutational studies for ligand binding and signal transduction [7, 9]. Abood and Reggio et al have characterized aromatic CB1 microdomain in the TMH3-4-5-6, showing that F3.36, W5.43, W6.48 are important CB1 ligand interaction sites [10]. Several computational CB2 receptor models have been constructed based upon the published rhodopsin template [11-20]. While these CB2 modeling studies have contributed considerably to current understanding of ligand-receptor interactions, these models must be viewed as theoretical and tentative since cannabinoid receptors share only 20-21% identity with bovine rhodopsin [21]. There are still some important residues especially aromatic ones, either predicted by CB2 modeling, conserved in GPCRs and CB receptors, or unique to CB2 receptor, which have not been fully characterized by mutational studies.
One such residue is W5.43, an important GPCRs conserved tryptophan residue. Tryptophans are unusually abundant in membrane proteins, including cannabinoid receptors, where along with tyrosines, they tend to form aromatic clusters, which may be important for the structure and function of membrane proteins [10]. It has been reported that the mutations occurred in aromatic microdomain of CB1 (spanning from TMH3 toTMH6), such as W5.43(280)A, resulted in 16.8-fold reduction of WIN55212-2 binding and 245-fold reduction of WIN55212-2 stimulated GTPγS binding [10]. Computer modeling and docking studies [10, 11, 16, 22] predicted that the W5.43(194) of CB2 receptor might be an important residue for WIN55212-2 and SR144528 binding. However, to our knowledge, its importance has not yet been the subject of full analysis for receptor binding and G-protein signaling using mutational and computational studies.
The purpose of this study is to use site-directed mutagenesis and computer modeling in conjunction with key cannabinoid ligands as probes to clarify the importance of this aromatic residue W5.43(194) of CB2 in ligand binding and signaling. In this analysis, the conserved tryptophan residue W5.43(194) has been replaced by alanine (A), phenylalanine (F), or tyrosine (Y), corresponding to non-aromatic residue, aromatic residue, or aromatic residue with hydroxyl group, respectively. The binding affinity of CB agonists CP55940 and WIN55212-2 and CB2 inverse agonist SR144528 to the wild type (WT) and mutant CB2 receptors, along with the resultant adenylyl cyclase (AC) activity are examined. The results of receptor binding and downstream signaling upon the CB2 mutants indicate that both aromatic interaction and hydrogen bonding are involved in ligand binding for the residue W5.43(194), and the mutations of this tryptophan site may affect the conformation of the ligand binding pocket and therefore control the active conformation of the wild type CB2 receptor.
2. Materials and methods
2.1. Amino acid numbering system
The amino acid numbering system reported by Ballesteros and Weinstein [23] was adopted. In this system, the most highly conserved residue in each transmembrane helix (TMH) is assigned a position index of 0.50. This number is preceded by the TMH number and followed by the sequence position number in parentheses. All other residues in the TMH are numbered relative to this residue. For example, the most highly conserved residue in TMH4 of the human CB2 receptor is W4.50(158). The residue that immediately precedes it is M4.49(157) and the one right follows it is V4.51(159).
2.2. Site-directed mutagenesis
The tryptophan residue W5.43(194) in transmembrane helix V of CB2 receptor was replaced by alanine, tyrosine, or phenylalanine individually. The human CB2 expression plasmid pcDNA3.1+3×HA-hCB2 (Missouri S&T cDNA Resources Center) was used as a template. Mutations were introduced through overlap extension polymerase chain reaction (OE-PCR) based site-directed mutagenesis. Briefly, in each of the two separated PCR reactions, two fragments of the mutant hCB2 DNA sequence were amplified using one universal primer (T7 promoter primer or BGH reverse primer) and one mutagenic primer synthesized by Integrated DNA Technologies, Inc. (Coralville, CA). The resultant two intermediate products with terminal complementarities formed a new template DNA by duplexing in a second PCR reaction. During this so-called overlap extension, the fused product was amplified by using the universal primers (T7 promoter primer and BGH reverse primer). After digestion of the final PCR products and the plasmid pcDNA3.1+3×HA-hCB2 with the restriction endonucleasesHind III/Xho I, the mutant fragments were ligated into the expression plasmid using T4 ligase. The mutants were subsequently identified by restriction mapping and confirmed by DNA sequencing. All enzymes and reagents used for the recombinant DNA experiments were purchased from Stratagen, New England Biolabs and Invitrogen.
2.3. Plasmid DNA transfection and cell culture
HEK293 cells were maintained in a humidified atmosphere at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 4 mM glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. For stable transfection, HEK293 cells grown in a 12-well plate to 70 - 80% confluence were incubated with 1 μg of wild-type or each of the mutated human CB2 receptor cDNA plasmids in 200 μl of Opti-MEM, 1 μl of PLUS Reagent and 3 μl of Lipofectamine (Invitrogen). Five hours after transfection, the solution was removed from the plate and 0.5 ml of fresh culture medium was added. Twenty-four hours after transfection, the cells were passed into a 60-mm culture dish and 800 μg/ml of G-418 was added to select for stably transfected cells. The stably transfected HEK293 cells were then maintained in the medium containing 400 μg/ml of G-418.
2.4. Cell membrane preparation
Confluent cells were washed twice with cold phosphate-buffered saline (PBS), scraped off the tissue culture plates and then homogenized (Polytron PT1600E Homogenizer, Switzerland) in 5 ml of membrane buffer (50 mM Tris-HCl, pH 7.4, containing 5mM MgCl2, 2.5 mM EGTA and 200 mM sucrose), supplemented with 5g/mL DNase A and 0.5% protease inhibitor cocktail (no. P8340, Sigma-Aldrich). The cell lysate was centrifuged at 400 ×g for 10 min at 4 °C, and the supernatant was collected. The pellet was washed three times with 10 ml of membrane buffer containing protease inhibitors and the combined supernatant was centrifuged at 100,000 ×g for 45 min at 4 °C. The resultant membrane pellet was used immediately or stored at −80 °C. Protein concentrations were determined using Bio-Rad Protein Assay Dye Reagent (catalog #500-0006).
2.5. Ligand binding assay
Competition binding assays were performed using a previously reported method [24] with modifications in a 96-well plate format. Briefly, non-radioactive (or cold) ligand dilutions were carried out in binding buffer [50 mM Tris-HCl, pH7.4, containing 5 mM MgCl2, 2.5 mM EGTA and 0.1% (w/v) fatty acid free bovine serum albumin (no. A6003, Sigma-Aldrich)], supplemented with 10% dimethyl sulfoxide and 0.4% methyl cellulose (no. M6385, Sigma-Aldrich). Aliquots were added in 1/10 volume to [3H]CP55940 (1 nM, specific activity = 77.5 Ci/mmol; National Institute on Drug Abuse) and cell membrane protein (100g/well) in binding buffer with a final volume of 200l in each assay well. The binding reaction was performed for 90 min at 30 °C with gentle agitation. The reaction was terminated by rapid filtration through Unifilter GF/B filter plates (PerkinElmer Life and Analytical Sciences) coated with 0.05% polyethylenimine using a Unifilter Cell Harvester (PerkinElmer Life and Analytical Sciences). Unbound radioligand was removed from the filter plates by washing five times with 200l ice-cold wash buffer (50 mM Tris-HCl, pH 7.4, containing 5 mM MgCl2, 2.5 mM EGTA and 0.5% BSA). All assays were performed in duplicate or triplicate and data points presented as the mean ± S.E.M. Bound radioactivity was analyzed forKi values using nonlinear regression analysis (Graphpad Prism 5.0, GraphPad Software, San Diego, CA), withKd values for [3H]CP55940 measured from competition binding experiments using the equationKd = IC50 - L, where L is the concentration of the total radioligand [25].
2.6. cAMP assay
Total cAMP levels (intra- plus extra-cellular) were quantified according to reported method [25] with modifications using LANCE cAMP 384 kits (PerkinElmer). The assay is based on competition between a Europium-labeled cAMP trace complex and total cAMP for binding sites on cAMP-specific antibodies labeled with a fluorescent dye. The energy emitted from the Euchelate is transferred to the dye on the antibodies, which in turn generates a time-resolved fluorescent resonant energy transfer (TR-FRET) signal at 665 nm. The fluorescence intensity (665 nm) decreases in the presence of cAMP from the tested samples and resulting signals are inversely proportional to the cAMP concentration of a sample. CB2 WT or mutant transfected HEK293 cells were seeded in a 384-well white ProxiPlates with a density of 5000 cells per well in 5 μl of DMEM medium containing 1% dialysed FBS, 25 mM HEPES, 100 μg/ml Pennicilin, 100 U/ml strepmicin and 200 μg/ml of G-418. The cell culture plate was covered with another plate and incubated at 37°C in 5% CO2. After culture overnight, 2.5 μl of cAMP antibody and RO20-1724 (final 50 μM) in stimulation buffer (HBSS 1X containing 5 mM HEPES, pH 7.4, containing 0.1% BSA) was added to each assay well, followed by addition of either 2.5 μl agonist and forskolin (final 5 μM) for agonist-inhibited AC activity assay, or 2.5 μl forskolin and SR144528 (final 0.3 μM) or stimulation buffer (control) for agonist-independent AC activity assay. After the plate was incubated at room temperature for 45 min, 10 μl of detection reagent was added into each well. The plate was then incubated for 1 hr at room temperature and measured in a SpectraMax 5M plate reader (Molecular Devices, Sunnyvale, CA) with excitation at 340 nm and emission at 665 nm. Each cAMP determination was made at least three independent times, each in triplicate. EC50 values were determined by nonlinear regression, dose-response curves (GraphPad Prism 5).
2.7. Western blot analysis
Cell membrane samples (10 μg each) were prepared with 4× NuPAGE LDS Sample Buffer (106 mM Tris-HCl, 141 mM Tris base, pH 8.5, containing 2% LDS, 10% glycerol, 0.51 mM EDTA, 0.22 mM SERVA Blue G250 and 0.175 mM phenol Red)(Invitrogen NP0008), and 10× reducing agent (500 mM DTT) was added and incubated at room temperature for 15 min. SDS-PAGE were carried out on 10% NuPAGE Novex Bis-Tris gels (Invitrogen) using the NuPAGE MES SDS buffer system (50 mM MES, 50 mM Tris base, pH 7.3, containing 0.1% SDS, 1 mM EDTA). The separated proteins on the SDS-PAGE gel were transferred onto a PVDF membrane for immunodetection with a rabbit HA antibody. The transferred PVDF membrane was blocked with 5% non-fat dried milk and 0.1% (v/v) Tween 20 in PBS (PBS-T) for 1 hour and then the membrane was then incubated in diluted primary antibody for 1 hour at room temperature. The membrane was washed three times in PBS-T buffer for 10 min each and incubated in the HRP labeled secondary antibody (anti-rabbit IgG) for 1 hour at room temperature. After washing in PBS-T three times and 10 min each, the antibody-recognized protein bands were visualized using ECL Western Blotting Detection Reagents (RPN2109, GE Healthcare).
2.8. Molecular Modeling
The CB2 receptor homology model reported previously [8, 26] was used for computer molecular modeling. Docking of cannabinoid ligands CP-55940, WIN55212-2, and SR144528 as well as CB2 protein-ligand complex MD/MM studies were performed on the basis of previously published docking protocols [22] using Tripos molecular modeling packages Sybyl8.1.
3. Results
3.1. Expression of recombinant human CB2 on HEK 293 cell membrane
The expression of human CB2 wild-type (WT) receptor and mutant human CB2 receptors with residue W5.43(194) replaced by alanine (A), phenylalanine (F), or tyrosine (Y) on the stably transfected polyclonal human embryonic kidney (HEK) 293 cell lines was confirmed by Western blot (Figure 1). All the receptor variants have the same size of protein bands corresponding to 43 kDa.
Fig. 1. The expression of CB2 receptor mutants in transfected HEK293 cells was analyzed by Western blot.
From left to right: CB2 WT (wild type), W194A, W194Y, W194F.
3.2. Ligand binding of CB2 wild-type and mutant receptors
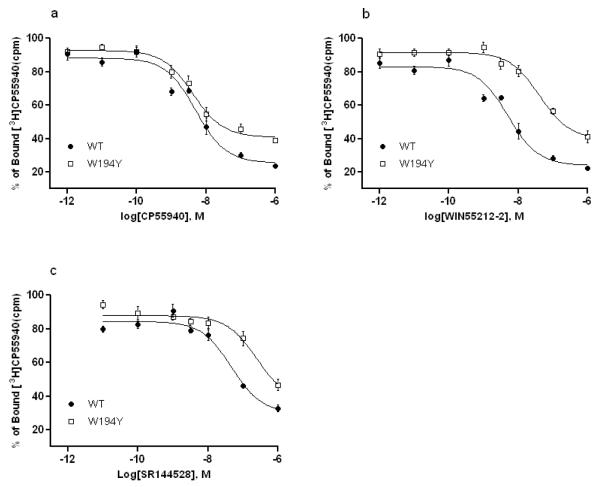
The binding affinities of the CB2 WT or mutant receptors were defined using the cannabinoid receptor agonists CP55940 and WIN55212-2 as well as the CB2 inverse agonist SR144528.Ki values (Table 1) for individual cell lines expressing CB2 WT and mutant receptors were determined in competition binding assays with [3H]CP55940 as a radioligand. The binding experiments showed that W5.43(194)Y mutation had no effect on CP55940 affinity whereas WIN55212-2 affinity was reduced up to 8-fold and SR144528 affinity reduced 5-fold comparing with the wild-type (Figure 2 a, b and c). However, W5.43(194)A and W5.43(194)F mutations resulted in a total loss of binding affinity for all tested ligands. The results were also summarized in Table 1.
Table 1.
Binding affinity of cannabinoid receptor ligands to wild type (WT) and mutant CB2 receptors stably expressed in HEK293 cells (Ki, nM)*
| CP55940 | WIN55212-2 | SR144528 | |
|---|---|---|---|
| CB2 WT | 4.11 (2.58-6.56) | 4.26 (2.89-6.29) | 38.5 (19.75-75.08) |
| W194Y | 3.21 (2.05-5.03) | 31.52 (17.11-58.08) | 191.5 (58.86-623.3) |
| W194A | ND | ND | ND |
| W194F | ND | ND | ND |
Data are the means and corresponding 95% confidence intervals of three independent experiments each performed in duplicate.
ND: binding not detected.
Fig. 2. Cannabinoid ligands (a. CP55940; b. WIN55212-2; c. SR144528) bind to CB2 receptor wild type (●) and mutant W194Y (□) using membranes prepared from transfected HEK293 cells as described.
Data are mean ± S.E.M. of three independent experiments performed in duplicate or triplicate. Curves were generated as described in Materials and Methods.
3.3. Agonist-induced inhibition of cAMP production
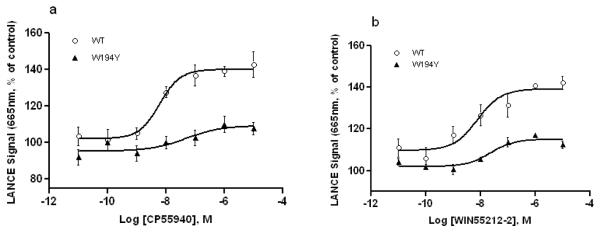
In order to characterize the importance of W5.43(194) site to CB2 receptor activity, the inhibition of CP55940 and WIN55212-2 on cAMP production stimulated by 5 μM forskolin was measured in the presence of phosphodiesterase inhibitor RO20-1724 (50 μM) using the LANCE cAMP assay with HEK293 cells expressing wild type or mutant CB2 receptors. The LANCE signal generated by fluorescence resonance energy transfer (FRET) is inversely proportional to the concentration of cAMP. Given that CB2 is a Gαi-coupled receptor, agonists will inhibit the forskolin-induced cAMP production, resulting in an enhancement of the LANCE signal as agonist concentration is increased. We clearly observed this phenomenon when CP55940 and WIN55212-2 were used for inhibiting forskolin-stimulated cAMP accumulation of the CB2 WT and W5.43(194)Y mutant. The W5.43(194)Y mutation resulted in a dramatic increase in EC50 values compared to the CB2 WT (Figure 3 a and b) (P<0.01, two-tailed t-test), indicating that the mutation W5.43(194)Y decreases agonist-induced inhibition of cAMP accumulation. These results are congruent with the ligand-receptor binding affinity data for WT and W5.43(194)Y mutant described above. The inhibition of forskolin-stimulated cAMP accumulation with the agonists was not detected for the W5.43(194)A and W5.43(194)F mutants.
Fig. 3. Comparison of CB2 WT and W194Y mutant receptors in agonist-induced inhibition of forskolin-stimulated cAMP accumulation.
EC50 values were 5.91±4.87 nM for CB2 WT and 45.48±5.86 nM for W194Y treated with CP55940 (a), 7.49±3.85 nM for CB2 WT and 21.79±1.65 nM for W194Y mutant treated with WIN55212-2 (b). Data are mean ± S.E.M. of three independent experiments in triplicate.
3.4. Constitutive activation of wild type and mutant CB2 receptors
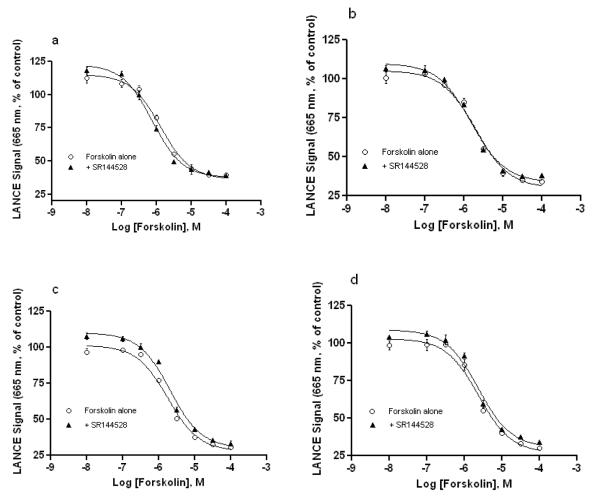
The effects of the mutations and CB2 inverse agonist SR144528 on constitutive activation of the CB2 variants were also evaluated by cAMP accumulation assay in the absence of agonist, in which the LANCE signals (inversely proportional to cAMP concentration as described earlier) were plotted as a function of forskolin concentration. As shown in Figure 4 and Table 2, treatment of the HEK293 cells stably expressing human CB2 WT receptor with CB2 inverse agonist SR144528 (0.3 μM) did not significantly increased the forskolin-stimulated cAMP accumulation with a slightly lower EC50 = 0.77 (0.59-1.0) μM, whereas treatment with 0.3 μM of CP55940 and WIN55212-2 decreased (p<0.05, two-tailed t-test) the forskolin-stimulated AC activity, showing the higher EC50 = 3.96 (3.06-5.12) μM or EC50 = 2.64 (1.93-3.62) μM, accordingly, compared to the no cannabinoid ligand treated control with EC50 = 1.32(1.0-1.74) μM.
Fig. 4. Effect of cannabinoids on forskolin-stimulated cAMP accumulation in CB2 WT transfected HEK293 cells.
HEK293 cells expressing CB2 WT was assayed for cAMP accumulation in response to a gradient concentrations forskolin (10-105 nM) alone (○) and in the presence of 0.3 μM of CP55940 (▲), WIN55212-2 (△), or SR144528 (*). The results were expressed as % of the LANCE signal representing basal cAMP accumulation, measured in the absence of forskolin. Data shown represent the mean ± S.E.M. of at least three independent experiments performed in triplicate.
Table 2.
Effects of CB2 inverse agonist SR144528 on constitutive activation of CB2 WT and mutant receptors (EC50, nM)*
| Receptor | Forskolin alone | +SR144528 | + CP55940 | + WIN55212-2 |
|---|---|---|---|---|
| Wild type | 1.32(1.0-1.74) | 0.77(0.59-1.0) | 3.96(3.06-5.12) | 2.64(1.93-3.62) |
| W194Y | 2.02 (1.5-2.73) | 1.57(1.22-2.01) | - | - |
| W194F | 1.81(1.42-2.32) | 2.09(1.62-2.71) | - | - |
| W194A | 2.42(1.73-3.39) | 2.38(1.75-3.25) | - |
HEK293 cells expressing CB2 WT, W194Y, W194A and W194F mutant receptors were assayed for cAMP accumulation in response to a gradient concentrations forskolin (10-105 nM) with or without the presence of 0.3 μM SR144528 (data from 0.3 μM CP55940 and WIN55212-2 for comparison purpose). The results were expressed as % of the LANCE signal representing basal cAMP accumulation, measured in the absence of forskolin. Data shown represent the means and corresponding 95% confidence intervals of three independent experiments performed in triplicate.
As shown in Table 2, the EC50 values of the AC activator forskolin in the HEK293 cell lines expressing CB2 W5.43(194)Y, W5.43(194)A and W5.43(194)F mutant receptors were higher than that of CB2 WT in the presence or absence of CB2 inverse agonist SR144528 (0.3 μM), indicating the cAMP levels are lower in any of the mutants than that generated in the wild type. The results showed that the effect of SR144528, a CB2 inverse agonist, on constitutive activation of W5.43(194)Y mutant was similar to that of CB2 WT receptor, and the EC50 values were lower in the presence of SR144528 than in the absence of SR144528, therein cAMP accumulation increased when CB2 WT and W5.43(194)Y were treated with SR144528. Conversely, for W5.43(194)F and W5.43(194)A mutants, the EC50 values were about same or slightly higher in the presence of SR144528 than in the absence of SR144528, indicating SR144528 has no obvious effect on the constitutive activation of W5.43(194)A and W5.43(194)F.
4. Discussion
This study provides the first experimental characterization of a cannabinoid receptor conserved residue, W5.43(194), which is located on TMH5 of CB2 receptor. Through mutational analysis, we validated the role of this aromatic residue, in the regulation of ligand-receptor binding capacity and adenylyl cyclase activity. The molecular modeling and docking studies also suggested the importance of this residue for WIN55212 binding and revealed the presence of a main aromatic interaction between the indole ring [22] or naphthyl ring [16] of WIN55212-2 with W5.43 as well as F3.36(117) and W5.46(197). Using computer modeling and docking techniques, Montero et al. [11] reported that W5.43(194) is one of the important residues involved in SR144528 binding through aromatic interaction. In the present study, we showed that replacement of this conserved tryptophan (W) residue with tyrosine (Y), another aromatic amino acid, results in a 5-fold decrease in SR144528 binding and an approximate 8-fold decrease of WIN55212-2 affinity, whereas the CP55940 affinity remained unchanged as summarized in Table 1 and Fig. 2. Our results are congruent with the computer modeling data as shown in Fig. 6 for docking of WIN55212-2 in CB2 receptor.
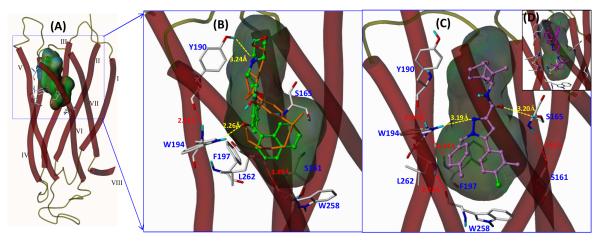
Fig. 6. 3D model of the putative interaction of cannabinoid ligands with CB2 receptor determined by Surflex-Dock and MD/MM calculations.
(A) The predicted binding pocket surrounded by known binding residues. (B) The agonists WIN55212-2 (green color) and CP55940 (orange) are located in the amphipathic CB2 receptor binding pocket. As illustrated, WIN55212-2 interacts with W194 and Y190 via hydrogen bonding and also forms significant π-π stacking interactions between the naphthyl ring and the aromatic microdomain clustered by W194, F197 and W258.(C) The CB2 antagonist SR144528 (purple) has similar π-π stacking interactions but possesses a different H-bonding interaction with Ser165. A different docking pose was also observed for SR144528 (D).
In our study, computational docking analysis using Surflex-Dock [27] and MD/MM calculations with an established CB2 receptor homology structure model [17] suggests that CB ligands bind into the amphipathic region predicted by MOLCAD solvent-accessible Connolly surface calculation, showing hydrogen bonding residues Y190 and W194, and a putative aromatic microdomain surrounded by the key residues W194, F197 and W258 [17] (Fig. 6A). As illustrated in Fig. 6, the putative binding site of CB2 receptor ligands is located adjacent to helices III, IV, V, and VI at the near extracellular side of the 7TM bundles (Fig. 6A). In this model, WIN55212-2 molecule (green color) is docked with its carbonyl group forming a hydrogen bonding interaction (2.26 Å) with the HN group at the indole ring of CB2 tryptophan W5.43(194) residue. A recent study by Sun et al. [28] also confirmed the importance of indole NH hydrogen-bonding for the folding and function of membrane spanning gramicidin channels by tryptophan to 1-methyl-tryptophan substitutions. The WIN55212-2 docking pose also reveals that its naphthalenyl group situated in the hydrophobic cavity of the amphipathic binding pocket, and the binding mode of WIN55212-2 appears to be facilitated by π-π stacking interactions (2.8 to 4.0 Å) with the illustrated residues, such as W194 and W258.
The ligand SR144528 docking pose displays a deeper binding pocket for an antagonist with hydrogen bonding (3.19 Å) with the HN group of the residue W194 indole ring and the π-π interactions (3.6 ~ 4.2 Å) between the aromatic rings of SR144528 and hydrophobic residues W194 and W258 (Fig. 6C) while possessing an additional hydrogen bonding (3.20 Å) with a known binding residue Ser165 that was reported to be important for the CB2 antagonist [29]. The computer modeling data are congruent with the present site-directed mutagenesis studies that the CB2 mutant W5.43(194)Y has lower binding affinity for WIN55212-2 and SR144528 (both with aromatic rings) in comparing with the CB2 wild-type whereas minor effect on CP55940 (with a dimethylhetyl side chain) binding affinity. CP5540 (orange color in Fig. 6B) is also docked into the predicted binding region with similar hydrogen binding patterns with W194 and Y190 whereas only its aliphatic chain interacts with the aromatic microdomain. The W5.43(194)A and W5.43(194)F mutations result in total loss of binding affinity for all tested ligands, which may attribute to the nonaromatic residue alanine (A) and non H-binding aromatic residue phenylalanine (F) as to tryptophan (W), the largest aromatic hydrophobic residue with an NH H-bonding donor.
Of course, the CB2 binding pocket was putatively predicted based on the available sited-directed mutagenesis experiments and computer modeling and will be subjected to further investigations by additional site-directed mutagenesis studies. Others also reported that CB2 double mutation K109A and S112G as well as F197V revealed that WIN55212-2 might have a different binding site with other CB ligands or the same binding site with different binding orientation [30]. In our docking studies, we also found that SR144528 may pose another docking mode, showing the similar hydrogen bonding patterns but with the ligand aromatic and bicycloheptanyl rings upside down (Fig. 6D, an insert). Thus, additional mutation studies will be needed to verify the binding regions for CB2 agonist and antagonist. In addition, the modeling data also suggested that L6.52(262) is a possible structural site on CB2 in an inactive state where its carbonyl may form an inter-helical hydrogen-bond with the indole NH of W5.43(194) with a distance of 2.03 Å, based on the reported CB2 model [11, 17], but much weaker hydrogen bonding (4.41 Å) with the phenolic hydroxyl of the W5.43(194)Y mutant. Reggio et al also reported molecular modeling data, showing V6.51, L6.52, L6.54, M6.55, L6.59 and T6.62 face into the CB2 binding pocket based on results detected using the substituted-cysteine accessibility method (SCAM) [31]. Clearly, the mutations of this W5.43(194) site to a more hydrophobic aromatic amino acid phenylalanine (F) or an aliphatic amino acid alanine (A) may disrupt these important inter-helical hydrogen-bonding interactions that is critical for stabilizing the TMH conformation, as indicated by a total loss of all tested ligand binding as shown in Table 1.
Furthermore, CB2 receptor is believed to be a Gαi coupled GPCR, where activation of CB2 receptor elicits the dissociation of Gαi proteins and a consequent inhibition of intracellular adenylyl cyclase activity [3, 32]. In our cAMP accumulation assays, W5.43(194)Y mutation severely impaired CP55940 and WIN55212-2 mediated inhibition of the forskolin-stimulated cAMP accumulation, as represented by statistically increased EC50 values. In W5.43(194)F/A mutants, agonist-dependent receptor activation was completely lost, suggesting the side chain of the conserved W5.43 site is essential for agonist binding (Table 2) and resultant conformation changes may disable G-protein activation. A similar study of CB1 receptor also reported that the mutation of W5.43 (280)A results in a significant reduction in ligand binding in the presence of WIN55212-2 and SR141716A, but not in the presence of CP55940 and anandamide, along with a reduction of WIN55212-2 induced activation by measuring GTPγS binding [10]. Therefore, we speculate that the W5.43 site of the CB2 receptor has a more critical role than the corresponding site of CB1 receptor in CB ligand binding and functional activity. Consistent with our computer modeling data, these results again suggest that in addition to hydrophobic interaction, the hydrogen bonding interaction of W5.43(194), either with the ligands or with an appropriate domain of the receptor, is important for receptor function. Such an interaction is somewhat preserved in W5.43(194)Y but not when the tryptophan residue is altered to an aliphatic amino acid alanine or to a phenylalanine, even though phenylalanine has a benzyl side chain that could contribute to aromatic interaction. Thus, the disruption of the signal transduction from extracellular stimuli to intracellular signals in the mutants W5.43(194)A/F may result from the loss of their ligand-binding property. We postulated that the hydrogen bond interaction as an additional recognition mechanism may only be required for a subset of CB2 ligands, rather than a universal recognition mechanism. For example, CP55940 binding to W149Y was similar to the CB2 WT while WIN55212-2 and SR144528 exhibited significant reduction (Table 1). The data indicated that although both tryptophan (W) and tyrosine (Y) residues have similar structures featuring both aromatic ring and hydrogen bonding, corresponding to an indole and a phenol, respectively, however, tryptophan is a non-polar hydrophobic residue while tyrosine is polar hydrophilic residue. Such difference resulted in the experimental observation that the mutation W194Y or change of the hydrophobic domain has much more impact to WIN55212-2 (with a naphthyl ring) or SR144528 (with two benzene rings) binding where less effect to CP55940 (with an alkyl chain) binding. The CB2 mutations W194A or W194F, correspondent to the non-aromatic residue (alanine, Ala or A) or non H-bonding aromatic residue (phenylanaline, Phe or F), completely lost the binding affinity for all three ligands, suggesting both aromatic ring cluster and H-bonding are essential for CB2 ligand binding. Additionally, in Table 2, all three mutations showed similar increase in EC50 (STDEV range of 0.35 to 0.78) in forskolin alone, indicating the hydrogen bonding may not be as critical as the hydrophobic aromatic interaction for in constitutive active state of the receptor. Taking these together, we argue that the combination of both aromatic interaction and hydrogen bonding interaction at this site may be important for direct ligand binding and downstream signaling; however, we cannot exclude the possibility that this amino acid could also affect the conformation of the binding pocket and therefore control the full active state of the wild type CB2 receptor.
Previous studies have indicated the constitutive activation of the CB2 receptor [33-35]. Therefore, treatment with an agonists or inverse agonists should theoretically shift the CB2 receptor to an active or inactive conformation state. Consequently, pretreatment with agonists or inverse agonists should decrease or increase the levels of forskolin-stimulated cAMP accumulation (heighten or lower LANCE signals) in the CB2 receptor, respectively. These phenomena were observed in the current study by pretreatment with CP55940, WIN55212-2 and SR144528 on the forskolin-stimulated cAMP accumulation in HEK293 cells expressing CB2 WT. Compared to the significant inhibitory effects of CP55940 and WIN55212-2 on the forskolin-stimulated AC activity, the enhancement effect of SR144528 on the forskolin-stimulated AC activity demonstrates SR144528 is an inverse agonist. As showed in Table 2 and Figure 5, all of the three CB2 receptor mutants exhibited the suppressed forskolin-stimulated AC activity (or EC50 values increase) in comparison to the wild type with or without the presence of SR144528, indicating that these mutations may cause conformational changes in the mutant receptors, which may favor their constitutive interactions with Gαi protein. The observation suggested that the W5.43(194)Y mutation induced a change in the CB2 receptor might be to mimic the effect of ligand binding and shift the receptor toward a pre-activated conformation state. Such a mutation-induced enhancement was also observed for cannabinoid receptor [36] and other GPCRs reported [37, 38]. For the W5.43(194)Y mutant, the effect of inverse agonist SR144528 on the constitutive activity is somewhat retained similar to that observed in CB2 WT. However, as a result of the mutant W5.43(194)A or W5.43(194)F, the pharmacological effect of the inverse agonist SR144528 is severely diminished, which was characterized by the similar forskolin-stimulated AC activities with or without the presence of SR144528, and is correlated with the ligand-receptor binding data.
Fig. 5. Effect of CB2 inverse agonist SR144528 on constitutive activation of CB2 WT and mutant receptors.
HEK293 cells expressing CB2 WT (a), W194Y (b), W194F(c) and W194A(d) mutant receptors were assayed for cAMP accumulation in response to a gradient concentrations forskolin (10-105 nM) in the presence (▲) or absence (○) of the inverse agonist SR144528 (0.3 μM). The results were expressed as % of the LANCE signal representing basal cAMP accumulation, measured in the absence of forskolin. Data shown represent the mean ± S.E.M. of at least three independent experiments performed in triplicate.
5. Conclusion
The present mutation study supports the hypothesis that the tryptophan residue W5.43(194) plays a key role in either ligand binding or controlling the active states of the CB2 receptor. The W5.43(194)Y mutation results in a reduction of WIN55212-2 and SR144528 binding whereas CP55940 affinity remains unchanged, and also a reduction of agonist mediated inhibition of the forskolin-induced cAMP response. The replacement of this tryptophan with the more hydrophobic aromatic residue phenylalanine or an aliphatic amino acid alanine results in completely disrupted ligand recognition and blockade of agonist-mediated inhibition of forskolin-stimulated AC activity. Based on our computer modeling studies, we therefore assume that the combination of hydrophobic aromatic and hydrogen bonding interactions might be required at this site for ligand binding and receptor signaling. W5.43(194)Y/F/A mutations have a decreased basal AC activity compared to the CB2 wild type receptor, which may be due to the inactive state of the CB2 mutant receptors possessing conformational alterations that are favored for precoupling of Gαi protein. Currently, we are conducting additional site-directed mutagenesis experiments for further investigations, including more CB2 ligands and additional residues, which will be reported elsewhere.
Acknowledgements
We thank Dr. Billy Day for reading the manuscript. We are also grateful to Missouri S&T cDNA Resources Center for providing hCB2 expression plasmid. This work was supported by NIH R01 DA025612 and CRDFA37969 grants.
Abbreviations
- AC
adenylyl cyclase
- BGH
bovine growth hormone
- cAMP
Cyclic adenosine monophosphate
- CB1
cannabinoid receptor subtype 1
- CB2
cannabinoid receptor subtype 2
- CNS
central nervous system
- DTT
dithiothreitol
- GPCR
G protein-coupled receptor
- HA
influenza hemagglutinin epitope tag
- HBSS
Hank’s balanced salt solution
- HEK293
human embryonic kidney 293 cell line
- LDS
lithium dodecyl sulfate
- TMH
transmembrane helix
- TR-FRET
time-resolved fluorescence resonance energy transfer
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Galiegue S, Mary S, Marchand J, Dussossoy D, Carriere D, Carayon P, et al. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- [2].Bouaboula M, Poinot-Chazel C, Marchand J, Canat X, Bourrie B, Rinaldi-Carmona M, et al. Signaling pathway associated with stimulation of CB2 peripheral cannabinoid receptor. Involvement of both mitogen-activated protein kinase and induction of Krox-24 expression. Eur J Biochem. 1996;237:704–11. doi: 10.1111/j.1432-1033.1996.0704p.x. [DOI] [PubMed] [Google Scholar]
- [3].Rhee MH, Bayewitch M, Avidor-Reiss T, Levy R, Vogel Z. Cannabinoid receptor activation differentially regulates the various adenylyl cyclase isozymes. J Neurochem. 1998;71:1525–34. doi: 10.1046/j.1471-4159.1998.71041525.x. [DOI] [PubMed] [Google Scholar]
- [4].Zoratti C, Kipmen-korgun D, Osibow K, Malli R, Graier WF. Anandamide initiates Ca2+ signaling via CB2 receptor linked to phospholipase C in calf pulmonary endothelial cells. Br J Pharmacol. 2003;140:1351–62. doi: 10.1038/sj.bjp.0705529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fogli S, Breschi MC. The molecular bases of cannabinoid action in cancer. Cancer Therapy. 2008;6:103–16. [Google Scholar]
- [6].Lotersztajn S, Teixeira-Clerc F, Julien B, Deveaux V, Ichigotani Y, Manin S, et al. CB2 receptors as new therapeutic targets for liver diseases. British Journal of Pharmacology. 2008;153:286–9. doi: 10.1038/sj.bjp.0707511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Raitio KH, Salo OM, Nevalainen T, Poso A, Jarvinen T. Targeting the cannabinoid CB2 receptor: mutations, modeling and development of CB2 selective ligands. Curr Med Chem. 2005;12:1217–37. doi: 10.2174/0929867053764617. [DOI] [PubMed] [Google Scholar]
- [8].Chen JZ, Xie XQ. GPCR Structure-Based Virtual Screening Approach for the CB2 Antagonist Search. J Comput Info Modeling. 2007;47:1626–37. doi: 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]
- [9].Poso A, Huffman JW. Targeting the cannabinoid CB2 receptor: modelling and structural determinants of CB2 selective ligands. Br J Pharmacol. 2008;153:335–46. doi: 10.1038/sj.bjp.0707567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McAllister SD, Hurst DP, Barnett-Norris J, Lynch D, Reggio PH, Abood ME. Structural Mimicry in Class A G Protein-coupled Receptor Rotamer Toggle Switches: The importance of the F3.36(201)/W6.48(357) interaction in cannabinoid CB1 receptor activation. Journal of Biological Chemistry. 2004;279:48024–37. doi: 10.1074/jbc.M406648200. [DOI] [PubMed] [Google Scholar]
- [11].Montero C, Campillo Nuria E, Goya P, Paez Juan A. Homology models of the cannabinoid CB1 and CB2 receptors. A docking analysis study. European journal of medicinal chemistry. 2005;40:75–83. doi: 10.1016/j.ejmech.2004.10.002. [DOI] [PubMed] [Google Scholar]
- [12].Pei Y, Mercier RW, Anday JK, Thakur GA, Zvonok AM, Hurst D, et al. Ligand-Binding Architecture of Human CB2 Cannabinoid Receptor: Evidence for Receptor Subtype-Specific Binding Motif and Modeling GPCR Activation. Chem Biol (Cambridge, MA, U S) 2008;15:1207–19. doi: 10.1016/j.chembiol.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Raduner S, Majewska A, Chen J-Z, Xie X-Q, Hamon J, Faller B, et al. Alkylamides from Echinacea Are a New Class of Cannabinomimetics: cannabinoid type 2 receptor-dependent and -independent immunomodulatory effects. Journal of Biological Chemistry. 2006;281:14192–206. doi: 10.1074/jbc.M601074200. [DOI] [PubMed] [Google Scholar]
- [14].Stern E, Muccioli Giulio G, Millet R, Goossens J-F, Farce A, Chavatte P, et al. Novel 4-oxo-1,4-dihydroquinoline-3-carboxamide derivatives as new CB2 cannabinoid receptors agonists: synthesis, pharmacological properties and molecular modeling. J Med Chem. 2006;49:70–9. doi: 10.1021/jm050467q. [DOI] [PubMed] [Google Scholar]
- [15].Salo OMH, Raitio KH, Savinainen JR, Nevalainen T, Lahtela-Kakkonen M, Laitinen JT, et al. Virtual Screening of Novel CB2 Ligands Using a Comparative Model of the Human Cannabinoid CB2 Receptor. Journal of Medicinal Chemistry. 2005;48:7166–71. doi: 10.1021/jm050565b. [DOI] [PubMed] [Google Scholar]
- [16].Tuccinardi T, Ferrarini Pier L, Manera C, Ortore G, Saccomanni G, Martinelli A. Cannabinoid CB2/CB1 selectivity. Receptor modeling and automated docking analysis. J Med Chem. 2006;49:984–94. doi: 10.1021/jm050875u. [DOI] [PubMed] [Google Scholar]
- [17].Xie XQ, Chen JZ, Billings EM. 3D structural model of the G-protein-coupled cannabinoid CB2 receptor. Proteins: Structure,Function,and Genetics. 2003;53:307–19. doi: 10.1002/prot.10511. [DOI] [PubMed] [Google Scholar]
- [18].Yates AS, Doughty SW, Kendall DA, Kellam B. Chemical modification of the naphthoyl 3-position of JWH-015: in search of a fluorescent probe to the cannabinoid CB2 receptor. Bioorg Med Chem Lett. 2005;15:3758–62. doi: 10.1016/j.bmcl.2005.05.049. [DOI] [PubMed] [Google Scholar]
- [19].Zhang R, Hurst DP, Barnett-Norris J, Reggio PH, Song Z-H. Cysteine 2.59(89) in the second transmembrane domain of human CB2 receptor is accessible within the ligand binding crevice: Evidence for possible CB2 deviation from a rhodopsin template. Molecular Pharmacology. 2005;68:69–83. doi: 10.1124/mol.104.007823. [DOI] [PubMed] [Google Scholar]
- [20].Reggio PH. Cannabinoid receptors and their ligands: Ligand-ligand and ligand-receptor modeling approaches. Handbook of Experimental Pharmacology. 2005;168:247–81. [PubMed] [Google Scholar]
- [21].Ashton JC, Wright JL, McPartland JM, Tyndall JD. Cannabinoid CB1 and CB2 receptor ligand specificity and the development of CB2-selective agonists. Curr Med Chem. 2008;15:1428–43. doi: 10.2174/092986708784567716. [DOI] [PubMed] [Google Scholar]
- [22].Song ZH, Slowey CA, Hurst DP, Reggio PH. The difference between the CB(1) and CB(2) cannabinoid receptors at position 5.46 is crucial for the selectivity of WIN55212-2 for CB(2) Mol Pharmacol. 1999;56:834–40. [PubMed] [Google Scholar]
- [23].Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosciences. 1995;25:366–428. [Google Scholar]
- [24].Lunn CA, Fine JS, Rojas-Triana A, Jackson JV, Fan X, Kung TT, et al. A novel cannabinoid peripheral cannabinoid receptor-selective inverse agonist blocks leukocyte recruitment in vivo. Journal of Pharmacology and Experimental Therapeutics. 2006;316:780–8. doi: 10.1124/jpet.105.093500. [DOI] [PubMed] [Google Scholar]
- [25].Xia M, Huang R, Guo V, Southall N, Cho M-H, Inglese J, et al. Identification of compounds that potentiate CREB signaling as possible enhancers of long-term memory. Proc Natl Acad Sci U S A. 2009;106:2412–7. doi: 10.1073/pnas.0813020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gertsch J, Leonti M, Raduner S, Racz I, Chen J-Z, Xie X-Q, et al. Beta-caryophyllene is a dietary cannabinoid. Proc Natl Acad Sci U S A. 2008;105:9099–104. doi: 10.1073/pnas.0803601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jain AN. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput-Aided Mol Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- [28].Sun H, Greathouse DV, Andersen OS, Koeppe RE., II The Preference of Tryptophan for Membrane Interfaces: insights from N-methylation of tryptophans in Gramicidin channels. J Biol Chem. 2008;283:22233–43. doi: 10.1074/jbc.M802074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gouldson P, Calandra B, Legoux P, Kerneis A, Rinaldi-Carmona M, Barth F, et al. Mutational analysis and molecular modeling of the antagonist SR 144528 binding site on the human cannabinoid CB2 receptor. European Journal of Pharmacology. 2000;401:17–25. doi: 10.1016/s0014-2999(00)00439-8. [DOI] [PubMed] [Google Scholar]
- [30].Tao Q, McAllister SD, Andreassi J, Nowell KW, Cabral GA, Hurst DP, et al. Role of a conserved lysine residue in the peripheral cannabinoid receptor (CB2): evidence for subtype specificity. Mol Pharmacol. 1999;55:605–13. [PubMed] [Google Scholar]
- [31].Nebane NM, Hurst DP, Carrasquer CA, Qiao Z, Reggio PH, Song Z-H. Residues Accessible in the Binding-Site Crevice of Transmembrane Helix 6 of the CB2 Cannabinoid Receptor. Biochemistry. 2008;47:13811–21. doi: 10.1021/bi8007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Glass M, Felder CC. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: evidence for a Gs linkage to the CB1 receptor. Journal of Neuroscience. 1997;17:5327–33. doi: 10.1523/JNEUROSCI.17-14-05327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bouaboula M, Desnoyer N, Carayon P, Combes T, Casellas P. Gi protein modulation induced by a selective inverse agonist for the peripheral cannabinoid receptor CB2: implication for intracellular signalization cross-regulation. Mol Pharmacol. 1999;55:473–80. [PubMed] [Google Scholar]
- [34].Nebane NM, Kellie B, Song Z-H. The effects of charge-neutralizing mutation D6.30N on the functions of CB1 and CB2 cannabinoid receptors. FEBS Letters. 2006;580:5392–8. doi: 10.1016/j.febslet.2006.09.001. [DOI] [PubMed] [Google Scholar]
- [35].Portier M, Rinaldi-Carmona M, Pecceu F, Combes T, Poinot-Chazel C, Calandra B, et al. SR 144528, an antagonist for the peripheral cannabinoid receptor that behaves as an inverse agonist. J Pharmacol Exp Ther. 1999;288:582–9. [PubMed] [Google Scholar]
- [36].D’Antona AM, Ahn KH, Wang L, Mierke DF, Lucas-Lenard J, Kendall DA. A cannabinoid receptor 1 mutation proximal to the DRY motif results in constitutive activity and reveals intramolecular interactions involved in receptor activation. Brain Research. 2006;1108:1–11. doi: 10.1016/j.brainres.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Leo A, Hansch C, Elkins D. Partition coefficients and their uses. Chem Rev. 1971;71:525–616. [Google Scholar]
- [38].Samama P, Cotecchia S, Costa T, Lefkowitz RJ. A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. J Biol Chem. 1993;268:4625–36. [PubMed] [Google Scholar]