Abstract
A major hurdle for molecular mechanistic studies of many proteins is the lack of a general method for fluorescent labeling with high efficiency, specificity, and speed. By incorporating an aldehyde motif genetically into a protein and improving the labeling kinetics substantially under mild conditions, we achieved fast, site-specific labeling of a protein with ~100% efficiency while maintaining the biological function. We demonstrate that an aldehyde-tagged protein can be specifically labeled in cell extracts without protein purification and then can be used in single-molecule pull-down analysis. We further show the unique power of our method in a series of single-molecule studies on the transient interactions and switching between two quantitatively labeled DNA polymerases on their processivity factor.
Introduction
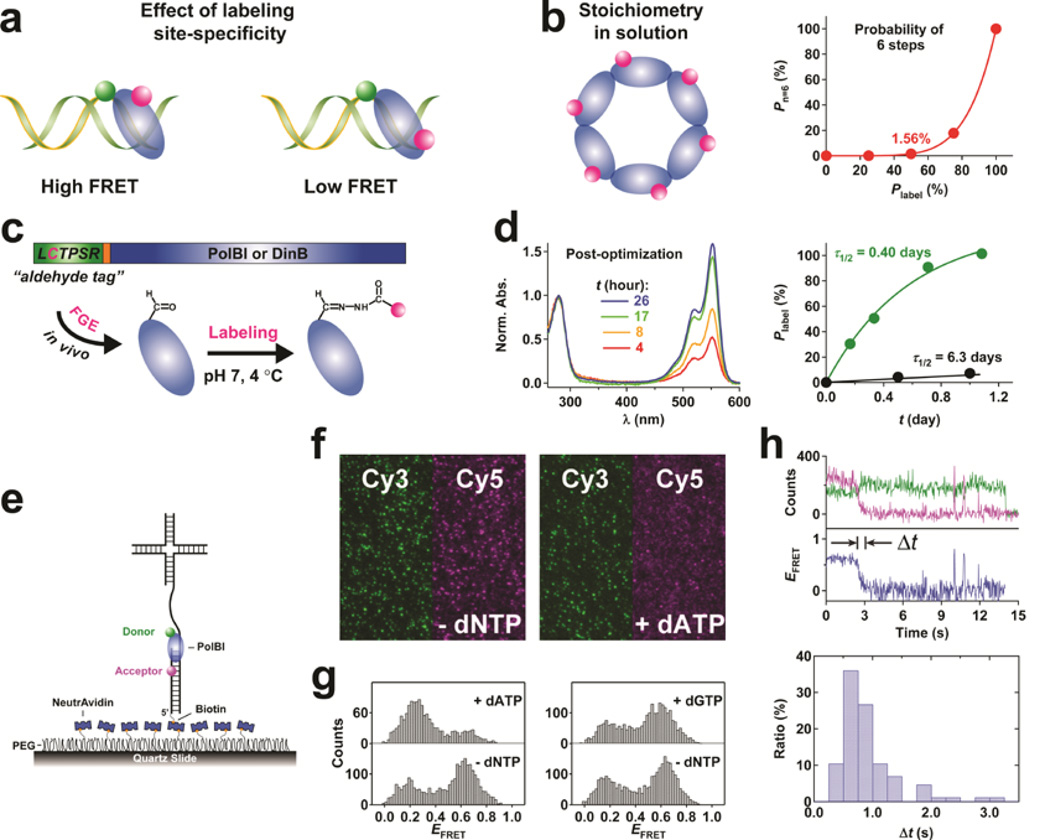
In recent years, single-molecule techniques have become standard tools for studying complex biological problems.1–3 Often a prerequisite for these studies is the fluorescent labeling of a protein at one location with high efficiency. Poor site-specificity introduces undesirable heterogeneity into the problem whereas low labeling efficiency limits the data throughput and makes certain experiments challenging. For example, when determining the solution stoichiometry for a hexameric complex, only 1.56% of the molecules will show all six photobleaching steps if the labeling efficiency is 50%. Protein labeling has been a major hurdle for many mechanistic studies at the ensemble or single-molecule level due to the lack of a generally applicable method. Current methods work by cysteine-specific chemistry, N-terminal transamination,4 expressed protein ligation,5 unnatural amino acid incorporation,6 installation of a peptide such as tetracysteine7 or polyhistidine,8 etc.; however, they suffer from common problems including poor site-specificity, location requirement, byproduct formation, limited commercial availability, unpredictable protein yield, low labeling efficiency, or limited fluorophore choice.
Given the limitations of existing techniques, we took a different approach by employing the genetically-encoded aldehyde tag, LCTPSR,9 that can be fused to a protein. The cysteine in this motif is converted in vivo into formylglycine by co-expressed formylglycine-generating enzyme (FGE).9 The aldehyde group in this residue then serves as an exclusive target for labeling with commercially available cyanine hydrazides even in the presence of other cysteines on the protein. This seemingly straightforward strategy however comes associated with several practical challenges. First, the condition reported for aldehyde labeling9 is harsh for many proteins. Second, fast and quantitative labeling is yet to be demonstrated for any protein. Third, the cysteine-to-formylglycine conversion may not be complete. Despite the importance of these issues, none has been addressed in the literature thus far.9,10 Moreover, labeling site-specificity and fluorophore linkage stability may present further challenges.
Here we establish a simple and robust method for labeling a protein at one location with ~100% efficiency rapidly under physiologically relevant conditions. We demonstrate the labeling of two different DNA polymerases with different fluorophores that allowed us to detect the transient and dynamic interactions between the two polymerases on a shared processivity factor. We also show that an aldehyde-tagged protein can be labeled efficiently and specifically within cell extracts and can then be used for single-molecule pull-down analysis.11–14
Online Methods
Plasmid Preparation
All oligonucleotides used in this study were purchased from IDT. For the purpose of cloning, the two NcoI sites found in PolBI’s gene were removed using a QuikChange Multi site-directed mutagenesis kit (Agilent).
The sequence of N-terminal aldehyde tag was designed into the primer ald6N-forward with an NcoI site at the 5’ end. This primer was used in a PCR along with a reverse primer containing a NotI or SalI site at the 5’ end. Purified PCR product was tailed at both 3’ ends with a single adenine using Taq DNA polymerase (NEB), followed by ligation into pGEM-T Easy vector (Promega) using T4 DNA ligase (Promega). After transformation with the ligation product, JM109 cells (Promega) were cultured at 37 °C for 1 h, spread on a LB-ampicillin plate, and grown at 37 °C overnight. Colonies containing the gene for Ald6N-PolBI or -DinB were cultured in LB containing 100 µg/ml ampicillin at 37 °C overnight. Plasmid DNA was extracted using QIAprep Spin Miniprep kit (Qiagen). Restriction digestion and sequencing confirmed the gene for Ald6N-PolBI or -DinB in the plasmid obtained. This plasmid and the one containing pET-28a(+) vector were digested with restriction enzymes at 37 °C for 3 h. Purified Ald6N-PolBI or -DinB insert and pET-28a(+) vector were ligated using T4 DNA ligase at 16 °C overnight. The final plasmid was obtained and validated by the same procedure above.
DH5α cells transformed with the FGE plasmid in pBAD/Myc-His A vector were obtained from Addgene (plasmid 16132). Purified plasmid was prepared as described above.
Protein Expression and Purification
After co-transformation with the FGE and Ald6N-PolBI or -DinB plasmids, BL21(DE3) cells (Agilent) were cultured at 37 °C for 1 h, spread on a LB-ampicillin-kanamycin plate, and grown at 37 °C overnight. Single colonies were cultured in 10 ml LB with 100 µg/ml ampicillin and 30 µg/ml kanamycin at 37 °C for 6 h, followed by dilution into 1 l LB containing the same antibiotics. When OD600 reached 0.3, expression of FGE was turned on by adding L-(+)-arabinose to 0.2%. Expression of Ald6N-PolBI or -DinB was induced 30 min later by adding IPTG to 0.1 mM, followed by lowering the temperature to 16 °C. Cells were harvested 16 h later by centrifugation at 7,000 × g and then resuspended in the lysis buffer at pH 7.0 containing 50 mM sodium phosphate and 300 mM NaCl, followed by homogenization using French press. After centrifugation at 9,200 × rpm, the supernatant was collected, mixed with TALON metal affinity resin (Clontech), and incubated at 4 °C for 1 h. After washing with a second buffer containing 10 mM imidazole in addition, proteins were eluted off the resin using a third buffer containing 150 mM imidazole.
Proteins were exchanged into buffer A at pH 8.5 containing 50 mM Tris by using Amicon Ultra-15 centrifugal filter unit (Millipore), and separated on a Mono Q 5/50 GL column using ÄKTA FPLC (GE Healthcare) and a 50 ml gradient from 0 to 50% buffer B containing 1 M NaCl in addition at a flow rate of 1 ml/min. Desired fractions were exchanged into the gel filtration buffer at pH 8.5 containing 50 mM Tris, 150 mM NaCl and 0.5 mM DTT, and further separated on a HiLoad 16/60 Superdex 200 column (GE Healthcare). Purified protein was exchanged into the storage buffer containing 10% (v/v) glycerol in addition.
M. acetivorans RFC and PCNA were expressed in E. coli and purified as described.15
Labeling Aldehyde-Tagged Proteins
Ald6N-PolBI or -DinB was exchanged into a pH 7.0 labeling buffer using Amicon Ultra-4 or −0.5 centrifugal filter unit (Millipore). This buffer contains 50 mM potassium phosphate, 100 mM KCl and 1 mM DTT (pre-optimization), or identical ingredients but each with a concentration five times higher (post-optimization). After buffer exchange, protein was mixed with dried Cy3HZ or Cy5HZ (GE Healthcare). For the quantitative labeling established in this work, we used either 100 µg dye in 3 µl reactions or 1 mg dye in 30 µl.
To characterize the labeling kinetics, we withdrew at each time point an aliquot of 0.5–1 µl from the 3 µl mixture. Unincorporated free dye was removed by passing the sample through two Micro Bio-Spin columns (Bio-Rad). A UV-Vis spectrum was taken for each sample using a Cary 50 spectrophotometer (Varian), from which absorbance was obtained for the label and protein. A correction factor of 0.08 or 0.017 was used for Cy3 or Cy5 to account for their absorption at 280 nm. Published molar extinction coefficients of Cy3 and Cy5,31 150,000 and 250,000 M−1 cm−1, were used. Note that covalent conjugation of these dyes to protein led to a small but detectable red-shift in their absorption maxima and, possibly, a minor change in extinction coefficient; however, this was not considered in our calculation. For Ald6N-PolBI and -DinB, the extinction coefficients calculated by the ProtParam program (ExPASy), 83,500 and 28,500 M−1 cm−1, were used. In principle, the values obtained experimentally (Supplementary Fig. 7), 81,600 and 30,850 M−1 cm−1, should be used instead. That is, all values of Plabel (t) reported in this work should be corrected by −2.3% and 8.2% for PolBI and DinB, respectively.
To label Ald6N-PolBI at the N-terminal α-amine,21 we exchanged the protein into the labeling buffer above, mixed it with 2 µg dried Cy3 NHS-ester (GE Healthcare), and incubated at room temperature for 30 min and 4 °C overnight.
To characterize the stability of the hydrazone linker between the label and protein, we exchanged Cy3HZ-labeled PolBI into the pH 8.5 storage buffer and kept it in the dark at 4 °C. After 1, 2, and 4 days, aliquots were withdrawn from the sample and free dye was removed by using one Micro Bio-Spin column.
Analysis of Aldehyde-Tagged Protein’s Labeling Kinetics
The labeling of aldehyde-tagged protein with cyanine hydrazide and the reverse reaction of hydrolysis can be described by
| (1) |
where A, B, and AB are the unlabeled protein, dye, and labeled protein, respectively. The rate equation is
| (2) |
where k2 and k−1 are rate constants for the forward and reverse reaction, respectively. When B is in large excess, [B]t is essentially a constant, making the forward reaction pseudo first-order. Then,
| (3) |
where k1' = k2[B]0 is the rate constant for the pseudo first-order reaction. Solving (3) gives
| (4) |
The labeling efficiency at time t is
| (5) |
The labeling kinetics is governed by the apparent rate constant kapp that equals k1' + k−1. The time to reach half of the maximal labeling efficiency, τ1/2, is ln(2)/kapp, while the maximal labeling efficiency, Plabel(t→∞), is k1'/kapp. When k1' ≫ k−1, Plabel(t) becomes 1 – exp (−k1't) and Plabel(t→∞) should reach 100%.
Hydrophobic Interaction Chromatography
Partially labeled Ald6N-DinB was diluted into buffer A at pH 8.0, containing 50 mM sodium phosphate and 1 M (NH4)2SO4, and separated on a TSKgel Phenyl-5PW column (Tosoh Bioscience) at 4 °C using ÄKTApurifier (GE Healthcare) and a 20-column-volume gradient from 0 to 100% buffer B containing no (NH4)2SO4 at a flow rate of 1 ml/min.
DNA Preparation
27-nt DNA D27-5Am with an amino-C6-dT modification at the 5th base from the 3’ end and a biotin at the 5’ end was mixed with 200 µg dried Cy5 NHS-ester (GE Healthcare) in a pH 8.5 buffer containing 100 mM sodium tetraborate and incubated at room temperature overnight. Free dye was removed by using two Micro Bio-Spin columns. 22-nt strand J7b+4Am with an amino-C6-dT modification at the 4th base from the 5’ end was labeled the same way. The labeling efficiency was ~100% for both strands.
Cy5-labeled D27-5Am was mixed in a 1.5:1:2:2:2 molar ratio with 69-nt strand HJG47A-T20 and three 22-nt strands J7b, J7h, and J7r, respectively. Unlabeled D27-5Am was mixed in the same ratio with these strands. Cy5-labeled J7b+4Am was mixed in a 2:1.5:1:2:2 ratio with D27-5Am, HJG47A-T20, J7h, and J7r. After heat denaturation at 90 °C for 1 min and slow cooling to room temperature, annealed DNA was separated on an 8% native polyacrylamide gel. 27-nt strand D27-5Cy5 internally labeled with Cy5 was mixed in a 1:2 ratio with 47- or 69-nt strand D27-T20 or −42, and the partial duplex DNA desired was obtained without gel purification.
Ensemble Polymerase Assay
A mixture of DNA, polymerase and dNTPs was prepared at a concentration of 0.1, 0.5 and 250 µM, respectively, in a pH 8.8 buffer, containing 20 mM Tris, 2 mM β-mercaptoethanol, 5 mM MgCl2 and 0.1 mg/ml BSA. After 10 min of incubation at 37 °C, the reaction was stopped by dilution in a 1:1 volume ratio into a solution containing 98% formamide and 1 mM EDTA. After heat denaturation at 95 °C for 5 min, reaction product was examined using a 10% TBE-Urea gel (Bio-Rad). The gel was then imaged using an ImageQuant LAS 4000 system (GE Healthcare).
Single-Molecule Polymerase Assay
Quartz slide and cover glass were subjected to PEGylation before assembly into an imaging chamber.32 Cy5-labeled DNA containing a (dT)20 template and a four-way junction was immobilized on the surface through biotin-NeutrAvidin (Thermo) linkage. Cy3HZ-labeled PolBI was then added at a concentration of 0.5–1 nM in a pH 8.0 imaging buffer containing 25 mM Tris, 5 mM MgCl2, 0.8% (w/v) dextrose, 2 mM Trolox (Aldrich), 0.04 mg/ml catalase (Sigma) and 1.0 mg/ml glucose oxidase (Sigma).32 After 5 min of incubation at room temperature, unbound protein was removed. 1 mM dATP or dGTP was then added at pH 8.8 into the chamber and incubated for 1 min prior to imaging. In the case of DinB, Cy3HZ-labeled polymerase was added at 1–2 nM after incubating the DNA with 20 nM RFC, 1 mM ATP and 5 nM PCNA.
Imaging was performed with the prism-type total internal reflection microscopy.19,32 A 532 nm Excelsior laser (Spectra Physics) was used to excite Cy3 on the labeled polymerase. Emission from both Cy3 and Cy5 was collected through a 60×, 1.2 NA, water immersion objective (Olympus), separated with a 630dcxr dichroic beam splitter (Chroma), and detected using an iXon DU-897 EMCCD camera (Andor). Individual molecules were identified using a custom program written in IDL.
Restrictive Proteolysis
3.1 µL labeled polymerase was incubated with 0.5 U restriction-grade thrombin (EMD) at room temperature overnight. After a 1:1 dilution into the Laemmli sample buffer (Bio-Rad) and heat denaturation at 95 °C for 5 min, the sample was examined by SDS-PAGE on a 4–20% Tris-HCl gel (Bio-Rad). Unless noted, Precision Plus Protein Kaleidoscope Standards (Bio-Rad) were used as molecular weight markers. The gel was imaged for fluorescence using the imager mentioned before. After staining with Bio-Safe Coomassie (Bio-Rad), this gel was imaged again under white light illumination.
Measurement of Protein Extinction Coefficient
Aliquot of aldehyde-tagged polymerase was diluted into either the native storage buffer, or a pH 6.5 denaturing buffer containing 20 mM sodium phosphate and 6.0 M guanidine hydrochloride.33 The extinction coefficient of a protein in the storage buffer, εnat(λmax), is
| (1) |
where Anat(λmax) and Adenat(280) are the peak absorbance under native condition and 280 nm absorbance under denaturing condition, respectively, and εdenat(280) is the protein’s extinction coefficient at 280 nm under denaturing condition (Supplementary Fig. 7). εdenat(280) is obtained by
| (2) |
where cW, cY, cC’ are the number of tryptophans, tyrosines, and cystines in the protein, and εW, εY, εC are the extinction coefficient of corresponding model compounds at 280 nm under denaturing condition.33 The extinction coefficient of Ald6N-PolBI and -DinB under native condition was thus determined to be 81,300–81,900 and 30,700–31,000 M−1 cm−1, respectively.
High-Resolution Mass Spectrometry
Aldehyde-tagged polymerase was mixed in a 50:1 weight ratio with sequencing-grade modified trypsin (Promega) in a pH 7.8 buffer containing 100 mM ammonium bicarbonate and incubated at 37 °C overnight. The product was separated on a nanoLC-2D system (Eksigent) at a flow rate of 300 nl/min, using a gradient from 95% A containing 5% acetonitrile and 0.2% formic acid and 5% buffer B containing 95% acetonitrile and 0.2% formic acid, to 55% A and 45% B in 50 min, and then to 15% A and 85% B in 10 min.
Sample eluted off the RPLC was electrosprayed into an 11-tesla LTQ-FT Ultra hybrid mass spectrometer (Thermo). For MS/MS analysis, a data-dependent, top 3 strategy with a 10 m/z isolation window was used. MS1 was performed in the 350–1,300 m/z scan range at 50,000 resolving power with a FT-ICR cell. MS2 was performed at 25,000 resolving power after collision-induced dissociation. Data were analyzed with the ProSightPC 2.0 SP1 software (Thermo).34
Labeling Ald6N-DinB in Cell Extracts and Pull-down
BL21(DE3) cells co-transformed with the Ald6N-DinB and FGE plasmids were cultured in 1 l LB as described in Protein Expression and Purification. The same procedure was carried out for the controls, except that cells were transformed with either plasmid alone and then cultured in the presence of a single antibiotic. Cells were harvested 16 h later and resuspended in a 1:4.2 weight-to-volume ratio in the labeling buffer mentioned before, followed by French press. After centrifugation, the supernatant was collected and a cocktail of protease inhibitors containing 500 µM AEBSF, 150 nM aprotinin, 1 µM E-64 and 1 µM leupeptin (Research Products International) was added to the lysate. Small molecules were removed from the extract by centrifugation at 14,000 × g for six times using Amicon Ultra-0.5 centrifugal filter unit with Ultracel-10 membrane (Millipore). 3 µl extract was then mixed with 100 µg dried Cy3HZ and incubated at 4 °C for one day, followed by the removal of free dye.
Labeled cell extract was diluted in a 1:1 ratio into the Laemmli sample buffer, heat denatured at 95 °C for 5 min, and examined by SDS-PAGE on a 4–20% Tris-HCl gel. For the purpose of calibration, samples of ~92% labeled pure DinB at various concentrations were run on the same gel. The gel was first imaged for Cy3 fluorescence; after staining with Bio-Safe Coomassie, it was imaged again under white light illumination. The density of Cy3 fluorescence, ICy3, was plotted against that of Coomassie stain, ICoomassie, for each calibration sample’s DinB band (C) and these data were fitted to a straight line. The slope, ΔICy3(C)/ΔICoomassie(C), and the labeling efficiency of the calibration samples were used to estimate the labeling efficiency of DinB in the cell extract (E),
| (3) |
In the single-molecule pull-down assay, diluted cell extract containing ~1 nM Cy3HZ-labeled DinB was added to surface-immobilized DNA in complex with PCNA (see Single-Molecule Polymerase Assay). After 5 min of incubation at room temperature, DinB was pulled down from the mixture of cellular proteins and other macromolecules, followed by removing unbound molecules. As a control, the imaging buffer was added in the place of RFC-ATP-PCNA mixture prior to pulling down DinB.
Imaging Transient Interactions and Switching between Polymerases
The DNA mentioned in DNA Preparation containing a (dT)20 template and Cy5-labeled four-way junction was immobilized in the imaging chamber. Cy3HZ-labeled PolBI was added at 1 nM, followed by removing unbound protein 5 min later using the pH 8.0 imaging buffer.
To observe the interactions and switching between different DNA polymerases, the DNA described in DNA Preparation containing a (dT)20 template and an unlabeled four-way junction was immobilized on the surface and incubated with a mixture of 20 nM RFC, 1 mM ATP and 5 nM PCNA for 5 min. Cy5HZ-labeled DinB was then added at a concentration of 5 nM, followed by the addition of 1 nM Cy3HZ-labeled PolBI. A movie was recorded during the flow of PolBI. Alternatively, Cy5HZ-labeled PolBI was added first at a concentration of 2 nM, followed by the addition of 1 nM Cy3HZ-labeled DinB. A similar procedure was carried out for the controls, except that the imaging buffer was added in the place of RFC-ATP-PCNA mixture, Cy5-labeled polymerase, or Cy3-labeled polymerase, respectively.
Results
Quantitative Labeling of Aldehyde-Tagged Proteins
We constructed plasmids for the archaeal DNA polymerases PolBI15 and DinB16 from Methanosarcina acetivorans with an aldehyde tag positioned at the N terminus followed by a polyhistidine tag and thrombin cleavage site, and co-expressed the protein with FGE in E. coli. PolBI and DinB have 10 and 4 native cysteines respectively, making it difficult to engineer ‘cysteine-light’ mutant rationally in the absence of structural information. Purified PolBI or DinB was exchanged into the buffer reported in the literature for labeling: pH 5.5 with 1% SDS, and incubated at 37 °C for 2 hours.9 After this treatment, both polymerases lost the primer-extension activity (Supplementary Note 1). For this reason, we attempted to use a much milder condition: pH 7.0, with no detergent, at 4 °C. However, even with excess Cy3 hydrazide (Cy3HZ), only ~5% of DinB and ~40% of PolBI were labeled after 4.5 and 9.7 days of incubation, respectively (Supplementary Note 1). By measuring the labeling kinetics of Ald6N-PolBI, we determined that the time to reach half of the maximal labeling efficiency, τ1/2, was 6.3 ± 0.6 days for the condition above.
After characterizing the effects of temperature, Cy3HZ concentration or [Cy3HZ], pH, and the reported catalyst aniline17 on the labeling kinetics and efficiency (Supplementary Note 1), we focused on increasing [Cy3HZ] while keeping the temperature at 4 °C and pH at 7.0. Owing to the high water solubility of Cy3, a concentration of 75.6 mM can be obtained for Cy3HZ, which allowed us to achieve ~100% labeling efficiency for Ald6N-PolBI at 4 °C and pH 7.0 (Fig. 1a). τ1/2 was also shortened dramatically from 6.3 to 0.4 days. Despite the use of high dye concentration, the overall contribution of nonspecific dye association with protein to the labeling efficiency observed was minimal as shown using a protein without the aldehyde tag (Supplementary Note 2).
Figure 1.
Development and validation of the quantitative, fluorescent labeling of an aldehyde-tagged protein. (a) Quantitative, fluorescent labeling of Ald6N-PolBI at 4 °C with 75.6 mM Cy3HZ in a pH 7.0 buffer containing 250 mM potassium phosphate, 500 mM KCl and 5 mM DTT (green). The labeling result before optimization is shown for comparison (black). (b) Schematic for the single-molecule polymerase assay. The DNA is labeled with a FRET acceptor (Cy5, magenta) at near the primer-template junction, while PolBI is labeled with a FRET donor (Cy3, green). (c) Total internal reflection microscopy images of the Cy3 and Cy5 channels taken before (left, –dNTP) and after (right, +dATP) adding dATP to PolBI and DNA immobilized as shown in b. (d) FRET efficiency histograms before (bottom) and after (top) adding dATP (left) or dGTP (right) to PolBI and DNA. (e) Donor (green) and acceptor (magenta) intensity and FRET efficiency (blue) traces of a molecule during replication (top, dATP added at ~2.5 s) and replication time (Δt) histogram (bottom).
Intact Protein Function after Quantitative Labeling
To confirm the biological function of labeled protein, we used standard ensemble biochemical assays for DNA polymerases and observed identical primer-extension and strand-displacement activities of unlabeled and Cy3HZ-labeled PolBI (Supplementary Note 3). We also examined DNA binding and replication using single-molecule fluorescence resonance energy transfer (FRET).18,19 Cy3HZ-labeled PolBI exhibited robust binding to a Cy5-labeled DNA immobilized on the surface (Fig. 1b,c). Adding dATP led to a substantial decrease in the FRET efficiency, EFRET = ICy5/(ICy3+ICy5), as a result of template-dependent replication (Fig. 1c,d). Real-time single-molecule FRET trajectories showed a gradual FRET decrease during the reaction and fitting the replication time histogram to a Gaussian distribution gave a replication rate of 34 ± 13 nt/s (Fig. 1e), which agrees well with the value of ~23 nt/s estimated from bulk primer-extension experiments15 and is close to the value of ~17 nt/s reported for a closely related member of the PolB family from Thermococcus litoralis.20 As a control, adding an incorrect dNTP gave no detectable change in EFRET (Fig. 1d and Supplementary Fig. 1). These single-molecule data further demonstrate that Ald6N-PolBI is functional after labeling.
Site-Specificity, Linker Stability, General Applicability
Through incubation with restriction-grade thrombin, Cy3HZ-labeled Ald6N-PolBI was cleaved at the thrombin site between the aldehyde tag and main protein (see Supplementary Note 4 for amino acid maps), producing a small N-terminal fragment that was fluorescent and a large C-terminal fragment that was nonfluorescent (Fig. 2a and see Supplementary Fig. 2 for the case of Ald6N-DinB). These results demonstrate that only the N-terminal region of PolBI or DinB containing the aldehyde tag was labeled despite the high concentration of dye used. In comparison, a widely used method that targets the N-terminal α-amine at pH 7.0 (Supplementary Fig. 3)21 by utilizing the difference in pKa and, thus, reactivity between this group and the ε-amine of lysines did not result in N-terminal specific labeling, as evaluated by the same restrictive proteolysis assay (Supplementary Note 5).
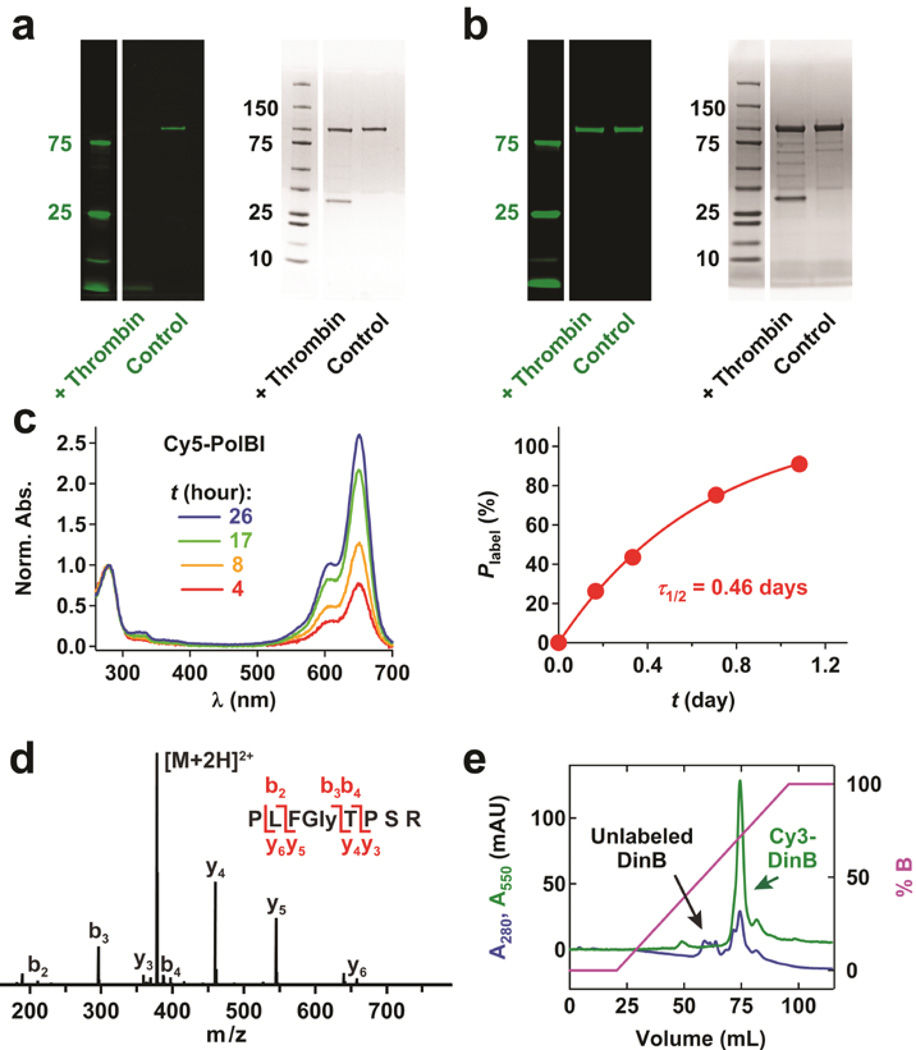
Figure 2.
General applicability of labeling an aldehyde-tagged protein. (a) Fluorescence (left) and white-light (right) images of a gel for Cy3HZ-labeled Ald6N-PolBI after incubation with thrombin (+Thrombin) or no thrombin (Control). The nonfluorescent band between 25 and 37 kDa in thrombin-treated sample is from thrombin. (b) Normalized UV-Vis spectra (left) and the labeling kinetics (right) of Ald6N-PolBI incubated with 40.5 mM Cy5HZ under the condition shown in Fig. 1a. (c) Detection of the fragment ions from an Ald6N-PolBI’s N-terminal peptide using high-resolution mass spectrometry. (d) Separation of Cy3-labeled DinB from unlabeled protein in a partially labeled sample of Ald6N-DinB using hydrophobic interaction chromatography.
We tested the hydrolytic stability of the hydrazone linker formed between the fluorophore and protein and observed only a negligible loss of the fluorophore from protein after several days of incubation under standard polymerase storage conditions, indicating good stability of the hydrazone linker therein (Supplementary Fig. 4).
Besides Cy3, the labeling of Ald6N-PolBI with Cy5 hydrazide (Cy5HZ) was also quantitative under similar conditions (Fig. 2b), allowing us to label two DNA polymerases with different fluorophores and to observe their transient interactions as discussed later. Such quantitative labeling can probably be extended to other fluorophores such as Alexa555 and 647, because many fluorophores are commercially available in aldehyde-reactive forms and are highly water soluble.
The procedure optimized for PolBI also improved the labeling efficiency of DinB from ~5 to 60%, but not quantitative yet (Supplementary Note 6). Unlike Ald6N-PolBI, which exhibits a complete cysteine-to-formylglycine conversion in vivo (Fig. 2c), Ald6N-DinB shows only incomplete aldehyde biosynthesis, limiting the maximal labeling efficiency achievable. By employing hydrophobic interaction chromatography,22 we separated Cy3HZ-labeled Ald6N-DinB from unlabeled protein (Fig. 2d), thereby obtaining ~100% labeled protein.
Labeling a Protein in Cell Extracts and Pull-down
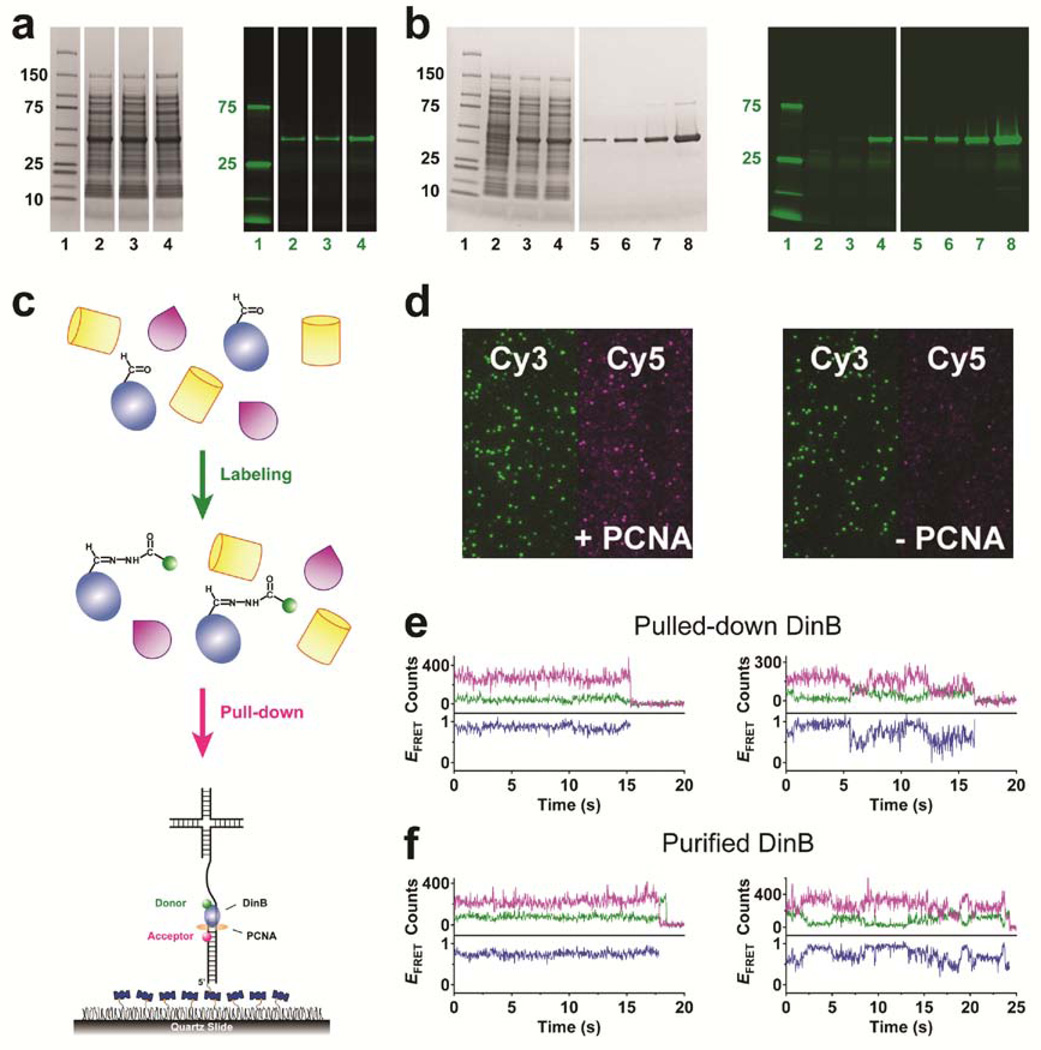
Because cellular proteins rarely possess aldehyde functionality, it should be possible to label an aldehyde-tagged protein specifically even without purification. We therefore labeled Ald6N-DinB in E. coli extracts using the same procedure. As expected, crude lysate shows the presence of many proteins; however, only DinB exhibits substantial labeling in this large pool of proteins (Fig. 3a), demonstrating high target-specificity. As a control, we examined the labeling of E. coli extract with FGE or DinB expressed alone and observed no appreciable labeling of any protein in either case (Fig. 3b). These results confirm the specificity of our method, and show that the background, FGE-like activity in E. coli10 is much weaker than that of co-expressed FGE.
Figure 3.
Specific and efficient labeling of unpurified Ald6N-DinB in cell extracts and single-molecule pull-down. (a) White-light (left) and fluorescence (right) images of a gel for the extracts of E. coli co-expressing Ald6N-DinB and FGE and labeled at 4 °C with 100 µg Cy3HZ in 3 µL for one day, with no other treatment (2nd lane), with the addition of a protease inhibitor cocktail (3rd lane), or with both the addition of protease inhibitors and the removal of small, exchangeable molecules (4th lane). (b) White-light (left) and fluorescence (right) images of a gel for the extracts of E. coli expressing FGE (2nd lane) or DinB (3rd lane) alone, or both simultaneously (4th lane), all labeled as in a (4th lane), and ~92% labeled purified DinB at various concentrations used for calibration (5th–8th lanes). (c) Schematic for the labeling and pull-down of Ald6N-DinB (blue) from a cell extract, using a surface-immobilized, Cy5-labeled DNA in complex with PCNA (orange). (d) Total internal reflection microscopy image showing the binding of DinB from cell extract to DNA in the presence of PCNA (left), and the control in the absence of PCNA (right). (e) Representative donor (green) and acceptor (magenta) intensity and FRET efficiency (blue) traces of individual molecules obtained by direct pull-down. (f) Representative data obtained with purified DinB.
We also examined the effects of adding protease inhibitors and removing small molecules. Adding a protease inhibitor cocktail to the extract limited proteolysis, with no substantial impact on the labeling efficiency of target protein (Fig. 3a). Removing small, exchangeable molecules led to a substantial improvement in the labeling efficiency, likely due to the exclusion of molecules containing a carbonyl group such as pyruvate (Fig. 3a). We estimated the labeling efficiency to be 70–75% after one day of incubation at 4 °C (Supplementary Fig. 5).
After labeling, the cell extract containing labeled DinB was subjected to single-molecule pull-down.11 A Cy5-labeled DNA was immobilized on the surface followed by the loading of polymerase processivity factor PCNA16 (Fig. 3c). After adding an extract containing 1 nM Cy3HZ-labeled DinB and a brief incubation, binding of DinB from the extract to PCNA-DNA was detected through single-molecule FRET; by contrast, only nonspecific binding was observed without PCNA (Fig. 3d). These data closely resemble those obtained with purified DinB and such similarity is also seen in the FRET traces of individual molecules (Fig. 3e,f). The capacity to label a protein in cell extracts should allow examination of protein function under near-native conditions, where proteins maintain their native post-translational modifications and physiological partners.11
Observation of Transient Interactions between Polymerases
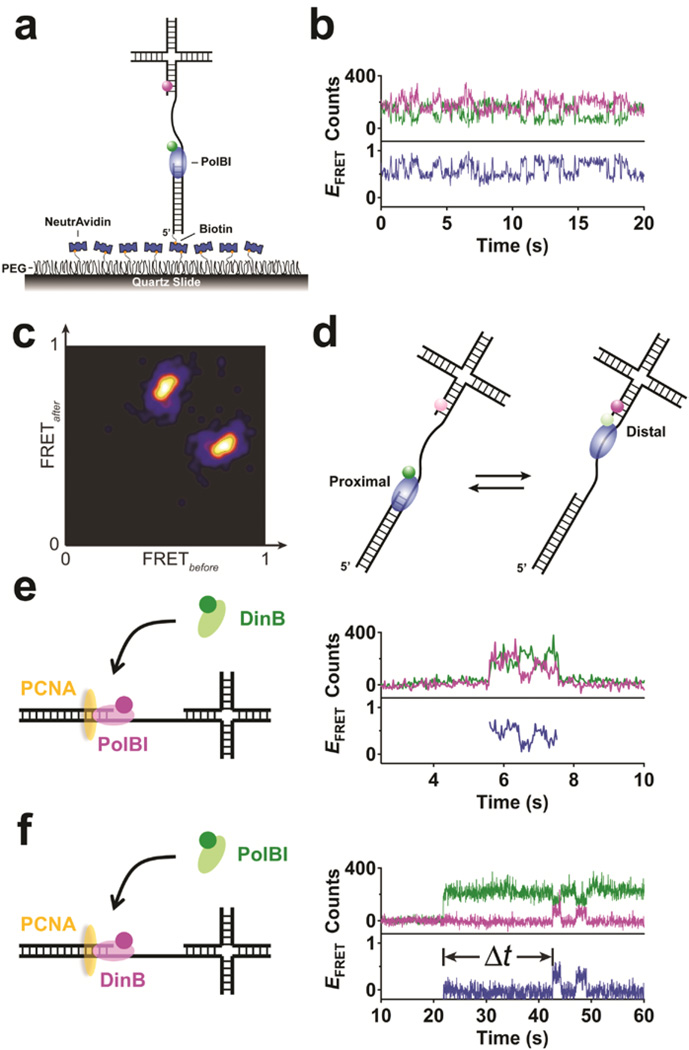
Using quantitatively labeled PolBI and DinB, we carried out a single-molecule study of transient interactions and switching between these two polymerases on PCNA. This is relevant to the process of translesion DNA synthesis, in which the actions of a high-fidelity replicative polymerase (PolBI) and an error-prone translesion polymerase (DinB) need to be coordinated.23–25 First, we examined the dynamic interaction between PolBI and DNA alone, without DinB and PCNA (Fig. 4a). Using Cy3HZ-labeled PolBI and a DNA labeled with Cy5 at a four-way junction away from the 3’ end of the primer, we observed frequent FRET fluctuations between two well-defined states at 0.5 and 0.8 (Fig. 4b,c). This observation, along with the results obtained using several related DNA constructs and with multi-color FRET26 (not shown) indicated that PolBI can shuttle repetitively on a single-stranded DNA between the proximal and distal positions (Fig. 4d).
Figure 4.
Real-time observation of the transient interactions and switching between PolBI and DinB. (a) Schematic for the study of dynamics in the interaction between PolBI and DNA. The DNA is labeled with a FRET acceptor (magenta) at the four-way junction, while PolBI is labeled with a FRET donor (green). (b) Donor (green) and acceptor (magenta) intensity and FRET efficiency (blue) traces of the molecule in a. (c) Transition density plot depicting the FRET transitions in b. (d) Schematic for the shuttling of PolBI on single-stranded DNA between the proximal and distal positions. (e) Schematic for the study of dynamic interaction between free DinB (green) and DNA-bound PolBI (magenta) mediated by PCNA (orange) and single-molecule FRET traces. (f) Schematic for the study of interaction between free PolBI (green) and DNA-bound DinB (magenta) in the presence of PCNA (orange) and single-molecule FRET traces.
Next, we examined the “toolbelt” model, in which more than one polymerase can bind to a PCNA simultaneously.25 This proposal has been difficult to validate due to the involvement of highly transient and dynamic interactions. Using quantitatively labeled PolBI and DinB, we captured relevant events in real time. When free Cy3-DinB was added to Cy5-PolBI already on PCNA-DNA, a brief and highly dynamic interaction was observed between the two polymerases as FRET fluctuations (Fig. 4e), and such an interaction requires PCNA. The interaction between DNA-bound Cy5-DinB and free Cy3-PolBI also requires PCNA but a long delay was observed for many molecules between PolBI binding and the appearance of appreciable FRET between PolBI and DinB, suggesting long-lived colocalization of both polymerases on the same DNA (Fig. 4f). Combined with PolBI shuttling, our data suggest that the two proteins can bind to the same DNA, one on PCNA and the other off PCNA, until making a direct contact. A more in-depth examination of this and other possible models (Supplementary Fig. 6) will be reported elsewhere.
These data are the first single-molecule observation of two DNA polymerases interacting and switching in real time. This was made possible by our labeling method that ensures minimal sample heterogeneity, adequate data throughput, and preservation of biological function.
Discussion
Though we only demonstrated labeling at the N terminus of a protein in this work, previous studies suggest this method should be applicable to internal locations as well as the C terminus. The formylglycine motif in sulfatases from which the aldehyde tag was originally identified is always located 50–80 amino acids away from the N terminus, suggesting that co-expressed FGE can modify the aldehyde tag in an internal location. Indeed, we made a preliminary observation of specific labeling of a helicase with the aldehyde tag inserted into an internal loop. In addition, the aldehyde formation at the C-terminus had 99% and 45–69% efficiency for maltose-binding protein expressed in E. coli9 and the Fc region of Immunoglobulin G expressed in CHO cells,10 respectively.
Another protein labeling technique capable of achieving high efficiency and specificity is the one based on the ybbR tag developed by the Walsh group.27 This method27,28 utilizes an enzymatic reaction catalyzed by Sfp or AcpS phosphopantetheinyl transferase to attach a small molecule such as a fluorescent dye in a synthetic CoA conjugate to a specific serine in a 11–12 amino acid peptide identified by phage display.27,28 In comparison to the aldehyde-tag method which can utilize most cyanine, Alexa and ATTO dyes, the ybbR-tag method is currently limited to Dy-547, Dy-647 and ATTO488 only. In addition, the ybbR-tag method involves the expression and purification of Sfp or AcpS, and necessitates purification of the target protein to remove the enzyme after labeling. The aldehyde tag is slightly shorter than the ybbR tag (6 versus 11–12 residues) and the ybbR-tag method yielded only 17% labeling efficiency in an earlier study,29. With further optimization, the ybbR-tag method may complement the aldehyde tag method, for example in applications requiring orthogonal labeling chemistries.
Our aldehyde-based labeling method shouldn’t be limited to proteins produced in bacteria. Proteins expressed in eukaryotic cells could also be labeled except that an FGE compatible with the host should be used. Co-expression of a target protein with human FGE in mammalian cells has been demonstrated.10 In principle, this strategy is applicable to proteins expressed in insect cells as well using human or fruit fly (CG7049, D. melanogaster) FGE and the MultiBac vector.30 Besides purified proteins, an unpurified protein can be labeled in cell extracts directly and then pulled down from the mixture in a physiological form with appropriate posttranslational modifications or in a complex with cellular partners.11–14 The use of a short aldehyde tag and small cyanine dye makes this labeling scheme more advantageous than existing methods using a fluorescent protein or labeled antibody for imaging, both of which are much larger in size and may not accurately reflect the physiological function, complex stoichiometry, in vivo modification, and interaction with cellular partners associated with a protein of interest. Note that it may be difficult to apply this method to systems where genetic manipulation is cumbersome, such as Xenopus extracts.
By using the aldehyde-based labeling method, a large number of proteins can now be examined directly. One good example is the five-subunit, clamp loader complex RFC, which has been difficult to label using conventional methods. This complex consists of three components named RFCL, RFCS1 and RFCS2 in a 1:3:1 ratio, containing 16 cysteines and 132 lysines in total. We have achieved labeling of this protein complex specifically at the N terminus of RFCL with ≥78% efficiency (unpublished observations). From the moderate-sized DinB to the larger PolBI and the five-subunit RFC complex with a molecular weight of 43.8, 108.2 and 222.2 kDa, respectively, the range of proteins we have successfully labeled implies that those even larger in size, such as the ribosome, may be tackled the same way.
Supplementary Material
Acknowledgments
We thank C. Bertozzi (University of California, Berkeley) for providing the plasmid DNA for FGE through Addgene, J. Fei for the suggestion of using hydrophobic interaction chromatography, and K. Ragunathan for a critical reading of the manuscript. This work was supported by the US National Institutes of Health grant GM065367 (to T.H.) and the US National Science Foundation grant MCB-0238451 (to I.C.).
Footnotes
Author Contributions. X.S. and T.H. conceived the study; X.S. designed the experiments; X.S., Y.J., L.L., C.L. and C.W. performed the experiments and analyzed the data; X.S., I.C. and T.H. wrote the manuscript.
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 2.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha T, Tinnefeld P. Photophysics of Fluorescent Probes for Single-Molecule Biophysics and Super-Resolution Imaging. Annu. Rev. Phys. Chem. 2012;63:1–23. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. N-terminal protein modification through a biomimetic transamination reaction. Angew. Chem. Int. Ed. Engl. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 5.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams SR, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 8.Lata S, Gavutis M, Tampe R, Piehler J. Specific and stable fluorescence labeling of histidine-tagged proteins for dissecting multi-protein complex formation. J. Am. Chem. Soc. 2006;128:2365–2372. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]
- 9.Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat. Chem. Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 10.Wu P, et al. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc. Natl. Acad. Sci. USA. 2009;106:3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeom KH, et al. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep. 2011;12:690–696. doi: 10.1038/embor.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YH, et al. Biochemical and mutational analyses of a unique clamp loader complex in the archaeon Methanosarcina acetivorans. J. Biol. Chem. 2005;280:41852–41863. doi: 10.1074/jbc.M508684200. [DOI] [PubMed] [Google Scholar]
- 16.Lin LJ, et al. Molecular analyses of an unusual translesion DNA polymerase from Methanosarcina acetivorans C2A. J. Mol. Biol. 2010;397:13–30. doi: 10.1016/j.jmb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Dirksen A, Hackeng TM, Dawson PE. Nucleophilic catalysis of oxime ligation. Angew. Chem. Int. Ed. Engl. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 18.Ha T, et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong H, Kucera RB, Jack WE. Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement, and exonuclease activities. J. Biol. Chem. 1993;268:1965–1975. [PubMed] [Google Scholar]
- 21.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 22.Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL., Jr Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat. Struct. Mol. Biol. 2009;16:861–868. doi: 10.1038/nsmb.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, et al. Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin J, et al. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl. Acad. Sci. USA. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, et al. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem. Biol. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 29.Lee G, Yoo J, Leslie BJ, Ha T. Single-molecule analysis reveals three phases of DNA degradation by an exonuclease. Nat. Chem. Biol. 2011;7:367–374. doi: 10.1038/nchembio.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat. Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 31.Berlier JE, et al. Quantitative comparison of long-wavelength Alexa Fluor dyes to Cy dyes: fluorescence of the dyes and their bioconjugates. J. Histochem. Cytochem. 2003;51:1699–1712. doi: 10.1177/002215540305101214. [DOI] [PubMed] [Google Scholar]
- 32.Shi X, Lim J, Ha T. Acidification of the oxygen scavenging system in single-molecule fluorescence studies: in situ sensing with a ratiometric dual-emission probe. Anal. Chem. 2010;82:6132–6138. doi: 10.1021/ac1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 34.Lee JE, et al. A robust two-dimensional separation for top-down tandem mass spectrometry of the low-mass proteome. J. Am. Soc. Mass. Spectrom. 2009;20:2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.