Summary
Numerous computational methodologies have been developed to facilitate the process of drug discovery. Broadly, they can be classified into ligand-based approaches, which are solely based on the calculation of the molecular properties of compounds, and structure-based approaches, which are based on the study of the interactions between compounds and their target proteins. This chapter deals with two major categories of ligand-based and structure-based methods for the prediction of biological activities of chemical compounds, namely quantitative structure-activity relationship (QSAR) analysis and docking-based scoring. QSAR methods are endowed with robustness and good ranking ability when applied to the prediction of the activity of closely related analogs; however, their great dependence on training sets significantly limits their applicability to the evaluation of diverse compounds. Instead, docking-based scoring, although not very effective in ranking active compounds on the basis of their affinities or potencies, offer the great advantage of not depending on training sets and have proven to be suitable tools for the distinction of active from inactive compounds, thus providing feasible platforms for virtual screening campaigns. Here, we describe the basic principles underlying the prediction of biological activities on the basis of QSAR and docking-based scoring, as well as a method to combine two or more individual predictions into a consensus model. Finally, we describe an example that illustrates the applicability of QSAR and molecular docking to G protein-coupled receptor (GPCR) projects.
Keywords: G protein-coupled receptors (GPCRs), Ligand-based drug discovery, Structure-based drug discovery, Molecular Docking, Quantitative structure-activity relationships (QSAR), Comparative molecular field analysis (CoMFA), Comparative molecular similarity index analysis (CoMSIA), Multiple linear regression (MLR), Partial least square (PLS) regression.
1. Introduction
The discovery of new drugs involves the expenditure of large amounts of money and manpower. Introducing a compound into clinical trials typically entails the scouting and biological evaluation of a large set of diverse molecules. This lengthy process can now be assisted and accelerated through the integration of a computer-aided drug discovery (CADD) strategy that helps the selection of candidate compounds, provides mechanistic hypotheses on their mode of action, and facilitates their development. Notably, CADD is a rapidly growing field and has already experienced a significant advancement since the early days, thanks to the efforts that academic researchers and pharmaceutical companies are putting into the development of new and improved computational methods and to the rapid technological improvement of computers (1-2).
CADD strategies can be broadly categorized into ligand-based and/or structure-based approaches (2-3). The former methods rely on the analysis of molecular properties of known ligands without taking into account explicitly the interactions of the ligands with their target protein. Clearly, ligand-based methodologies can only be applied when known ligands exist. Structure-based approaches, instead, are based on the direct calculation of protein-ligand interactions and can be applied only when the structure of the target protein has either been solved experimentally or generated through computational modeling.
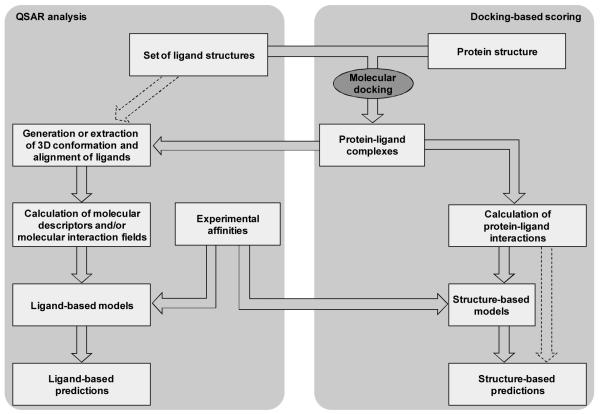
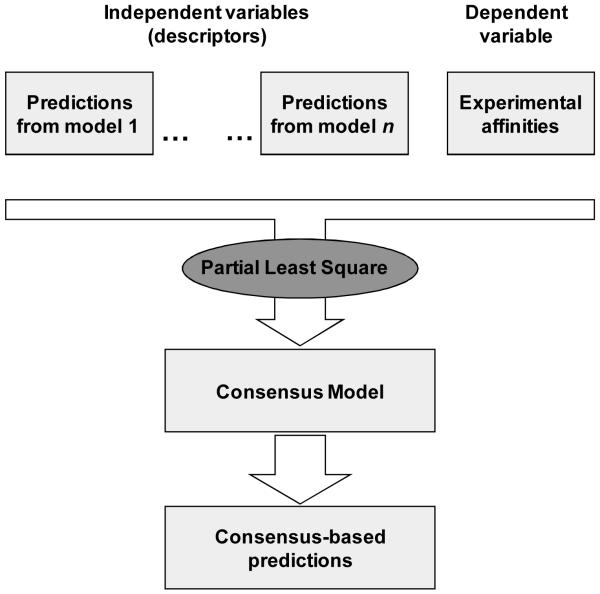
In this chapter, after a brief introduction to two specific categories of ligand-based and structure-based CADD approaches to the prediction of biological activities of chemicals, namely quantitative structure-activity relationship (QSAR) analysis and docking-based scoring, we describe the various phases necessary for their implementation (see Figure 1). We also describe how to generate consensus models that combine the predictions of two or more individual models (see Figure 2). Moreover, we illustrate the application of QSAR and molecular docking to the prediction of the activity of ligands of G protein-coupled receptors (GPCRs), a superfamily of proteins that, in light of their vast physiological and pathophysiological implications, are among the most pursued targets for pharmacological intervention (4). In particular, we present a case study that deals with the prediction of the activity of ligands for the β2-adrenergic receptor (5).
Figure 1.
Flowchart for the construction of ligand-based and structure-based models. According to this scheme, molecular docking plays a key role at the basis of both approaches. Alternatively, as indicated by the dashed arrow on the left side of the figure, a pure ligand-based approach can be adopted, in which the conformation and the alignment of the ligands are derived exclusively from their molecular features. Additionally, the scheme also illustrate that, when a training set of ligands with known activity is available, this can be used to train structure-based scoring functions. Alternatively, as indicated by the dashed arrows on the right side of the figure, a pure structure-based approach can be adopted in which prepackaged scoring functions are applied without the need for the use of a training set.
Figure 2.
Flowchart for the construction of a consensus model based on the combination, through PLS regression, of the predictions derived from n individual models.
1.1. QSAR
QSAR methods encompass a number of ligand-based analyses designed to correlate biological activities with molecular properties calculated using two-dimensional (2D) or three-dimensional (3D) ligand structures (6-7). QSAR analyses can only be conducted when a set of ligands with known biological activities, known as a training set, is available. Statistical models linking biological activities to molecular properties are built on the basis of such training sets and subsequently applied to the prediction of the activity of novel compounds. In the field of GPCRs, biological activity data have been published for ligands of numerous receptors and can be utilized to generate training sets. For this reason and because of the paucity of information on the 3D structure of GPCRs that, up until recently, has characterized the superfamily, QSAR has been extensively applied to the prediction of the activity of GPCR ligands (3). However, for orphan or less studied receptors, the absence or the paucity of known ligands may prevent or seriously hinder the application of ligand-based modeling.
QSAR analyses require the calculation of molecular descriptors that reflect the topology or the physicochemical properties of molecules. Once such descriptors have been calculated for the whole dataset, the correlation between descriptors and experimental activities is studied through statistical analyses, such as linear regression, multiple linear regression (MLR), or partial least square (PLS) regression.
In 2D-QSAR, molecules are described through properties calculated on the basis of their 2D topology. Instead, 3D-QSAR analyses are based on molecular properties that depend on the 3D structure of the molecules. For the calculation of some of these properties, models of the bioactive 3D conformation of the ligands are sufficient. For others, instead, a 3D alignment of the bioactive conformation of all the ligands is also necessary. In a pure ligand-based modeling approach, 3D alignments are generated simply by superimposing the ligands on the basis of their common features. However, more effectively, 3D alignments can be obtained through molecular docking, with a strategy that combines structure-based and ligand-based modeling (see Figure 1).
Within 3D QSAR methodologies, it is worth mentioning two techniques that have been among the most widely applied to the prediction of the activity of GPCR ligands (2), namely Comparative Molecular Field Analysis (CoMFA) (8) and Comparative Molecular Similarity Index Analysis (CoMSIA) (9). CoMFA and CoMSIA are based on the representation of ligands through molecular fields measured in the space that surrounds them. In particular, molecular fields are sampled at each point of a 3D lattice in which the aligned ligands are immersed and used as descriptors in a subsequent QSAR analysis. Due to the high number of descriptors that CoMFA and CoMSIA entail, a fundamental factor that contributed to their development has been the introduction of the PLS regression technique. This statistical method combines characteristics from principal component analysis (PCA) and MLR and reduces the dimensionality of the independent variables into fewer orthogonal components, thus allowing the conduction of regression analyses even when the number of independent variables is very high (10-11).
Recently, alignment independent 3D-QSAR analyses have also been developed and applied to study of GPCR ligands, for instance the autocorrelation of molecular electrostatic potential approach devised by Moro and coworkers (12-13) and the grid-independent descriptors (GRIND) approach devised by Clementi, Cruciani and coworkers (14-15).
QSAR models are very dependent on the nature of the training set. They are endowed with high predictive power when applied to compounds structurally related to those included in the training set, but perform poorly when applied to structurally diverse molecules. Thus, to ensure a robust predictive power, their use should be circumscribed to the analysis of compounds with molecular characteristics well represented within the training set.
1.2. Docking-based scoring
Predictions of biological activities based on docking scores do not require a training set of ligands with known activities. However, an absolute condition for the molecular docking experiments that lie at their foundation and the subsequent calculation of the protein-ligand interactions is that the structure of the target protein be known, possibly experimentally. For a long time, their applicability to the discovery and the development of GPCR ligands has been hampered, although not precluded, by the paucity of structural knowledge that has characterized the superfamily. In particular, for years, rhodopsin has been the only receptor with experimentally derived 3D information and has served as a prototype for the study of the whole GPCR superfamily (16). In recent years, however, breakthroughs in GPCR crystallography led to the solution of additional crystal structures, while many more are expected to be solved in the near future (17). Notably, besides their obvious direct application to structure-based CADD, these crystal structures will also provide an increasingly solid platform for the construction of homology models for the members of the superfamily that have yet to be experimentally elucidated (18-20).
The most accurate structure-based methodologies to rank the binding affinity of a set of given ligands for a target protein are those that rely on first-principle methods for the calculation of their free binding energy, for instance through free energy perturbations (FEP) or thermodynamic integration (TI) (21-22). However, these techniques are time-consuming and require significant effort for the preparation and optimization of the system to allow high-throughput application. Moreover, they are best suited for the analysis of closely related compounds. For these reasons, in the context of molecular docking, compounds are usually ranked through simpler and faster scoring functions, broadly classifiable into force field-based, empirical or knowledge-based methods (3). Scoring functions are notorious for yielding scores that lack fine correlation with experimental affinities, even when the calculations are based on geometrically accurate complexes (23). Nevertheless, they have been shown capable of effectively distinguishing between active and inactive compounds (1). Accordingly, many studies have demonstrated the applicability of molecular docking methods to virtual screening campaigns, including those targeting GPCRs, especially when the structure of the receptor is known crystallographically (5, 24-28). Particularly noteworthy are recent studies that reported the discovery, in high yields, of novel ligands of the β2-adrenergic and the adenosine A2A receptors through docking-based virtual screening targeting the crystal structures of the two receptors (24-27). Moreover, sufficiently accurate in silico models of GPCRs, although outperformed by crystal structures, have been shown to be effectively applicable to virtual screening not only in controlled studies but also through real-life quests for new ligands (25-26, 28, 33-37).
2. Materials
2.1. Software
Molecular descriptors can be calculated using a plethora of dedicated software, including, but certainly not limited to, DRAGON, CODESSA, MOE, the Schrödinger package, and ICM (38-42). Alternatively, they can be measured experimentally. CoMFA and CoMSIA calculations are implemented in SYBYL (43). Molecular docking experiments can be carried out by means of a variety of modeling software, including, but certainly not limited to, MOE, the Schrödinger package, SYBYL, GOLD, and ICM (40-44). Some modeling packages, including, but certainly not limited to, the Schrödinger package, MOE, SYBYL, and ICM, allow directly performing statistical analyses. Specialized software packages for statistical analyses are also available, including, but certainly not limited to, R (45) and STATISTICA (46).
2.2. Skills
Molecular modeling software can run with a variety of operating systems, including various implementations of Unix and Linux. Some software can be operated through a graphic user interface (GUI). However, most software can also (or exclusively) be operated through command-line or through scripts written by the user. For these reasons, some knowledge of the Unix/Linux operating systems as well as the ability of writing scripts and interacting with software through command-line is advisable.
2.3. Data
2.3.1. Structure and biological activity of ligands
Sets of known ligands, to be used as training and/or test sets, can be compiled using in-house data or data retrieved from the literature (see Note 1). Instead sets of novel potential ligands, to be evaluated in silico prior to their experimental testing, can be designed through computer-aided or classic medicinal chemistry approaches.
The 3D structures of the retrieved ligands can either be sketched within the chosen molecular modeling package or, if available, downloaded from databases such as PubChem (pubchem.ncbi.nlm.nih.gov). Particular attention to the chirality of the compounds is necessary. Once drawn, the structures should be saved in a format readable by the software chosen for the docking or the QSAR analysis. Some programs require one file per ligand; others allow the use of one file enlisting all the ligands.
For training and test sets, but not for novel putative ligands, biological activities - preferably binding affinities - need to be collected, either from the literature or from in-house data, and properly codified. If the experimental measurements were conducted on different species, it is advisable to verify the presence of sufficient similarity in the amino acid sequence of the target protein across species, especially within the binding cavity. If there are discrepancies between the activities reported for a compound by different articles, the data can either be excluded or an average value can be considered. Of note, information on GPCR ligands, with references to the relevant literature, is available through the GLIDA database (pharminfo.pharm.kyoto-u.ac.jp/services/glida) (47).
For the statistical analyses, a spreadsheet must be compiled, in the format required by the chosen software, listing the biological activity of the ligands along with the values of the molecular descriptors associated to them and/or their docking scores. Several software packages allow the calculation of the descriptors and also the subsequent statistical analyses. Usually, these packages require saving structures and biological activities in a single spreadsheet file.
2.3.2. Protein Structure
A file with the 3D coordinates of the target protein is required in order to perform docking experiments. For proteins that have been solved experimentally, such files can be downloaded from the Protein Data Bank (www.rcsb.org). Alternatively, homology models of the receptor can be constructed (18-20).
3. Methods
3.1 Preparation of the ligands
Known or candidate ligands, collected or designed as indicated in section 2.3.1, need to be subjected to a careful preparation procedure in order to: add hydrogen atoms, if these are not present; generate all ionization and tautomeric states available to the molecules within a certain pH range; generate all possible stereoisomers by varying the configuration of all the chiral centers in a combinatorial manner (unless the dataset contains molecules with known, specified chiralities); minimize the energy of the ligands through a molecular mechanics engine.
For pure ligand-based QSAR analyses, the most probable ionization and tautomeric state of each compound can be chosen on the basis of energetic considerations. Instead, for docking-based calculations, the ionization and tautomeric state favored by the receptor can be identified for each compound on the basis of the docking scores. All the other states can then be discarded, leading to a dataset containing only a single instance for each molecule.
3.2 Preparation of the protein
Crystal structures downloaded from the Protein Data Bank and homology models must be carefully inspected and processed in order to: add hydrogen atoms, if they are not present; optimize the geometry and interaction network of the hydrogen atoms; ensure that bond orders are properly assigned; ensure that disulfide bridges are properly connected; delete water molecules, if so desired; and add capping groups to truncated termini (see Note 2). Most docking packages offer automated procedures that help carry out these operations in an automated manner.
3.3 Generation of protein-ligand complexes trough molecular docking
As illustrated in Figure 1, besides being a fundamental step in the calculation of structure-based scores that reflect protein-ligand interactions, molecular docking is also our method of choice for the generation of the structural alignments of ligands necessary for most 3D-QSAR analyses, such as CoMFA and CoMSIA (5, 48).
During the docking procedure, typically, a variety of ligand conformations and orientations are sampled within the target protein by means of specific algorithms. Most docking software requires the user to specify a region of the protein within which to confine the docking of the ligands. Knowledge of the biology of the target is fundamental for a correct identification of the binding site. When data are not available, all the cavities present within a protein and on its surface should be explored. Target proteins are usually treated as rigid molecules, mostly to reduce calculation times. Most software packages, however, allow granting some flexibility to the receptor, if so desired, although usually with a considerable increase of the computational demand. Most docking programs allow usage of constraints on protein-ligand interactions derived from experimental data. For example, required hydrogen-bonds or hydrophobic interactions may be specified, or the occupation of a particular region of the binding pocket may be enforced.
3.4 Prediction of biological activities through QSAR analysis
After the collection of the compounds and, where necessary, their alignment, QSAR analyses can be conducted through the steps described in the following paragraphs.
3.4.1. Calculation of the independent variables: QSAR based on molecular descriptors
Different numerical values associated with each molecule, known as descriptors, can be either calculated trough a variety of existing software packages or measured experimentally (see Note 3) – for a non-comprehensive list of programs that can calculate molecular descriptors see the Materials section. While for the calculation of 2D descriptors only the topology of the ligands is required, the calculation of 3D descriptors requires also the bioactive conformation and, in some instances, the 3D alignment of the ligands. As mentioned, our method of choice for the derivation of bioactive conformations and structural alignments is molecular docking. Alternatively, ligand-based conformational analyses and 3D superimpositions can be applied. Most of the software allows including the descriptors in the same spreadsheet that contains the structures and biological activities of the compounds.
3.4.2. Calculation of the independent variables: CoMFA and CoMSIA
CoMFA (8) and CoMSIA (9) are 3D-QSAR analyses implemented in SYBYL (43). Once the molecules have been aligned, our protocol for the development of CoMFA and CoMSIA models proceeds as follows: Gasteiger-Hückel charges are calculated for all the compounds; a 3D cubic lattice is defined, extending 4 Å over the aligned molecules in all directions, with a spacing of 1 Å and a probe atom consisting of an sp3 hybridized carbon (c.3) with a charge of +1.0; two molecular interaction fields (steric and electrostatic) are calculated for CoMFA studies, using a distance dependent dielectric and a cut-off of 30.0 kcal/mol for the calculation of the Coulombic electrostatic energy; five fields (steric, electrostatic, H-bond donor, H-bond acceptor, hydrophobic) are calculated for CoMSIA studies, using the standard parameters.
3.4.3. Statistical analysis
The relationships between the descriptors and the experimental activity can be studied through various statistical analyses, for instance MLR or PLS regression. Several statistical parameters can then be employed to assess the quality of the model, the most prominent of which are the square of the correlation coefficient (r2) and the root mean square error (RMSE) (see Note 4) of the predictions. For CoMFA and CoMSIA studies, PLS regression analyses are directly carried out within SYBYL (43), using 4-6 components as independent variables and the experimental pIC50 or pKi values of the molecules as a dependent variable.
3.4.4. Validation of the model
Models can be validated through the use of cross-validation techniques. Among these, one of the most widespread is the leave-one-out test, which involves taking one molecule out of the training set and predicting its activity on the basis of a model trained with the remaining molecules. The operation is repeated until all the molecules, in turn, have been taken out of the training set one by one. The most used parameter to assess the quality of a leave-one-out cross-validation model is the cross-validated r2, or q2 – see Golbraikh and Tropsha for further details and caveats (49). Moreover, and most effectively, models can be validated through the evaluation of the activity of a test set, i.e. an additional set of molecules not included in the training set.
3.4.5. Prediction of the activity of novel compounds
Once a QSAR model has been built and validated, it can be used to predict the activity of newly designed compounds on the basis of the molecular properties associated with them. Following these initial predictions, the compounds can be further modified in order to improve their predicted activity. Eventually, all the designed compounds are ranked on the basis of the QSAR predictions and selected for experimental testing.
3.5 Prediction of biological activities through docking-based scoring
In molecular docking, the generated protein-ligand complexes are scored through specific scoring functions, which are based on different principles and endowed with different levels of accuracy (2) (see Note 5). Parallel docking calculations can be run combining various docking algorithms and scoring functions. If experimentally solved protein-ligand complexes are available, they may be conveniently used as controls to assess the accuracy of the docking poses and choose the algorithm the most suitable algorithm to work with a given target.
Docking-based scoring does not require training sets and can be directly used to rank the relative predicted affinity of a set of compounds. The numeric values of the docking scores do not represent free binding energies or affinities and can only be used to estimate the affinity of a compound relativity others. However, if a set of known ligands is available, this can be used as a training set in order to correlate docking scores and the experimental affinities through linear regression analysis. The affinity of novel ligands can then be inferred on the basis of the calculated relationship between docking scores and experimental affinity values. Just as in ligand-based QSAR, the quality of the models can be assessed on the basis of statistical parameters such as r2 and RMSE of calculated and experimental affinities. Cross-validation and external validation techniques can then be used to validate the models prior to their application to the prediction of the affinity of novel ligands. Moreover, when a training set is available, it can also be used to infer adhoc “trained” scoring functions through regression analysis, cherry-picking and combining the components that better correlate with the experimental affinities. However, such ad hoc scoring functions incur the risk of being predictive only within the training set for which they were generated, and deserve a careful and extensive validation through external test sets in order to assess their applicability.
As a caveat, it is worth mentioning that docking scores are rather crude and hence not very effective in the fine ranking of active compounds on the basis of their affinities or potencies. Instead, they are usually suited for distinguishing active from inactive compounds. If a set of known ligands is available, this ability can be monitored through controlled experiments in which the ligands are docked at the target protein together with a large set of decoy compounds. The receiver operating characteristic (ROC) curves obtained in these pilot screenings can be conveniently used to optimize the docking protocol and select the most appropriate scoring functions for a given target.
3.3. Consensus Models
Our method for the construction of consensus models is based on a PLS regression in which the experimental activities are used as the dependent variable, while the activities predicted through different individual models are used as independent variables (see Figure 2) (5, 48). In particular, the independent variables are converted into one or more components through PLS (see Note 6) and then the regression analysis is performed. As usual, the quality of the model can be monitored on the basis of its r2 and RMSE values. Moreover, cross-validations and validations with external test sets can be performed.
Consensus modeling can be applied to combine different ligand-based models, different structure-based models, or even ligand-based and structure-based models together. As mentioned in the introduction, since our consensus models originate from PLS regressions, a training set is necessary for their construction, even if the individual components are exclusively structure-based models.
4. Notes
The selection of the ligands is a very important step in the construction of a QSAR model. To generate a broadly applicable model, it is fundamental to collect a representative set of diverse ligands.
If the protein structure is truncated, i.e. misses a few residues at the N-terminus or the C-terminus, N-acetyl and N-methylamide groups are often used to cap the first solved N-terminal and last solved C-terminal residue, respectively. This operation prevents the first and last solved residues from being unnaturally electrically charged.
In order to avoid overfitting, the model must not be based on an excessive number of descriptors (independent variables). A commonly observed rule prescribes the use of no more than one independent variable per 10 observations (50-51). For instance, if the training set contains 100 compounds, no more than 10 descriptors should be used. Validating the model through an external test set, i.e. a set of compounds that have not been used to generate the model, is also a good practice to assess the quality of the model, since overfitted models, although very good within training sets, usually perform poorly with external test sets.
Many statistical analyses require the variables to follow a normal distribution. Thus, it is recommended that the activity of compounds included in QSAR studies be expressed in logarithmic form, since the use of logarithmic values may help normalizing the distribution of a variable. For instance, the expression of affinities as pKi (-log Ki) rather than Ki values is preferable. Furthermore, in order to ensure a correct interpretation of QSAR equations and an immediate perception of the weight of each descriptor on the basis of the value of its coefficient, it is also important that the descriptors be standardized. One way of doing this is subtracting to the value of a descriptor calculated for a particular molecule the mean value of the descriptor and dividing the resulting number by the standard deviation of the descriptor (52).
The computational time required to score a set of ligands with a scoring function is usually directly correlated to the accuracy of the scoring function. The fastest, less accurate, functions are intended to be used when docking a very high number of molecules; on the contrary, the most accurate and slowest functions are intended to be used when docking a smaller number of molecules.
The number of components to be used in a PLS regression should be determined in each case, trying to keep it to a minimum and terminating the introduction of additional components when the last added component barely adds anything to the explanation of the variance of the data.
Acknowledgment
This research was supported by the Intramural Research Program of the NIH, NIDDK.
References
- 1.Congreve M, Marshall F. The impact of GPCR structures on pharmacology and structure-based drug design. Br. J. Pharmacol. 2010;159:986–996. doi: 10.1111/j.1476-5381.2009.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vilar S, Cozza G, Moro S. Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery. Curr. Top. Med. Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 3.Costanzi S, Tikhonova IG, Harden TK, Jacobson KA. Ligand and structure-based methodologies for the prediction of the activity of G protein-coupled receptor ligands. J. Comput. Aided Mol. Des. 2009;23:747–754. doi: 10.1007/s10822-008-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 5.Vilar S, Karpiak J, Costanzi S. Ligand and structure-based models for the prediction of ligand-receptor affinities and virtual screenings: Development and application to the β2-adrenergic receptor. J. Comput. Chem. 2010;31:707–720. doi: 10.1002/jcc.21346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potemkin V, Grishina M. Principles for 3D/4D QSAR classification of drugs. Drug Discov. Today. 2008;13:952–959. doi: 10.1016/j.drudis.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Estrada E. How the parts organize in the whole? A top-down view of molecular descriptors and properties for QSAR and drug design. Mini-Rev. Med. Chem. 2008;8:213–221. doi: 10.2174/138955708783744128. [DOI] [PubMed] [Google Scholar]
- 8.Cramer RD, Patterson DE, Bunce JD. Comparative Molecular-Field Analysis (CoMFA) .1. Effect of Shape on Binding of Steroids to Carrier Proteins. J. Am. Chem. Soc. 1988;110:5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 9.Klebe G, Abraham U, Mietzner T. Molecular Similarity Indexes in A Comparative-Analysis (CoMSIA) of Drug Molecules to Correlate and Predict Their Biological-Activity. J. Med. Chem. 1994;37:4130–4146. doi: 10.1021/jm00050a010. [DOI] [PubMed] [Google Scholar]
- 10.Wold S, Albano C, Dunn WJ, Esbensen K, Hellberg S, Johansson E, et al. Modeling data tables by principal components and PLS-class patterns and quantitative predictive relations. Analusis. 1984;12:477–485. [Google Scholar]
- 11.Geladi P, Kowalski BR. Partial least-squares regression: a tutorial. Anal. Chim. Acta. 1986;185:1–17. [Google Scholar]
- 12.Moro S, Bacilieri M, Ferrari C, Spalluto G. Autocorrelation of molecular electrostatic potential surface properties combined with partial least squares analysis as alternative attractive tool to generate ligand-based 3D-QSARs. Curr. Drug Discov. Technol. 2005;2:13–21. doi: 10.2174/1570163053175439. [DOI] [PubMed] [Google Scholar]
- 13.Moro S, Bacilieri M, Cacciari B, Spalluto G. Autocorrelation of molecular electrostatic potential surface properties combined with partial least squares analysis as new strategy for the prediction of the activity of human A3 adenosine receptor antagonists. J. Med. Chem. 2005;48:5698–5704. doi: 10.1021/jm0502440. [DOI] [PubMed] [Google Scholar]
- 14.Pastor M, Cruciani G, McLay I, Pickett S, Clementi S. GRid-INdependent descriptors (GRIND): a novel class of alignment-independent three-dimensional molecular descriptors. J. Med. Chem. 2000;43:3233–3243. doi: 10.1021/jm000941m. [DOI] [PubMed] [Google Scholar]
- 15.Benedetti P, Mannhold R, Cruciani G, Ottaviani G. GRIND/ALMOND investigations on CysLT1 receptor antagonists of the quinolinyl(bridged)aryl type. Bioorg. Med. Chem. 2004;12:3607–3617. doi: 10.1016/j.bmc.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Costanzi S, Siegel J, Tikhonova IG, Jacobson KA. Rhodopsin and the others: a historical perspective on structural studies of G protein-coupled receptor. Curr. Pharm. Des. 2009;15:3994–4002. doi: 10.2174/138161209789824795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson MA, Stevens RC. Discovery of new GPCR biology: one receptor structure at a time. Structure. 2009;17:8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costanzi S. On the applicability of GPCR homology models to computer-aided drug discovery: a comparison between in silico and crystal structures of the beta2-adrenergic receptor. J. Med. Chem. 2008;51:2907–2914. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michino M, Abola E, GPCR Dock 2008 participants. Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat. Rev. Drug Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costanzi S. Modeling G protein-coupled receptors: a concrete possibility. Chimica Oggi-Chemistry Today. 2010;28:26–31. [PMC free article] [PubMed] [Google Scholar]
- 21.Chipot C, Rozanska X, Dixit SB. Can free energy calculations be fast and accurate at the same time? Binding of low-affinity, non-peptide inhibitors to the SH2 domain of the src protein. J. Comput. Aided Mol. Des. 2005;19:765–770. doi: 10.1007/s10822-005-9021-3. [DOI] [PubMed] [Google Scholar]
- 22.Foloppe N, Hubbard R. Towards predictive ligand design with free-energy based computational methods? Curr. Med. Chem. 2006;13:3583–3608. doi: 10.2174/092986706779026165. [DOI] [PubMed] [Google Scholar]
- 23.Warren GL, Andrews C, Capelli AM, Clarke B, LaLonde J, Lambert MH, et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 24.de Graaf C, Rognan D. Selective structure-based virtual screening for full and partial agonists of the β2-adrenergic receptor. J. Med. Chem. 2008;51:4978–4985. doi: 10.1021/jm800710x. [DOI] [PubMed] [Google Scholar]
- 25.Reynolds KA, Katritch V, Abagyan R. Identifying conformational changes of the β2-adrenoceptor that enable accurate prediction of ligand/receptor interactions and screening for GPCR modulators. J. Comput. Aided Mol. Des. 2009;23:273–288. doi: 10.1007/s10822-008-9257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katritch V, Rueda M, Lam PC, Yeager M, Abagyan R. GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2A receptor complex. Proteins. 2010;78:197–211. doi: 10.1002/prot.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya S, Vaidehi N. Computational mapping of the conformational transitions in agonist selective pathways of a G-protein coupled receptor. J. Am. Chem. Soc. 2010;132:5205–5214. doi: 10.1021/ja910700y. [DOI] [PubMed] [Google Scholar]
- 28.Vilar S, Ferino G, Sharangdhar SP, Berk B, Cavasotto CN, Costanzi S. Docking-based virtual screening for ligands of G protein-coupled receptors: not only crystal structures but also in silico models. Submitted for publication. 2010 doi: 10.1016/j.jmgm.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topiol S, Sabio M. Use of the X-ray structure of the β2-adrenergic receptor for drug discovery. Bioorg. Med. Chem. Lett. 2008;18:1598–1602. doi: 10.1016/j.bmcl.2008.01.063. [DOI] [PubMed] [Google Scholar]
- 30.Kolb P, Rosenbaum DM, Irwin JJ, Fung JJ, Kobilka BK, Shoichet BK. Structure-based discovery of β2-adrenergic receptor ligands. Proc. Natl. Acad. Sci. U.S.A. 2009;106:6843–6848. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlsson J, Yoo L, Gao ZG, Irwin JJ, Shoichet BK, Jacobson KA. Structure-based discovery of A2A adenosine receptor ligands. J. Med. Chem. 2010;53:3748–3755. doi: 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, Yeager M, et al. Structure-based discovery of novel chemotypes for adenosine A2A receptor antagonists. J. Med. Chem. 2010;53:1799–1809. doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaidehi N, Schlyer S, Trabanino RJ, Floriano WB, Abrol R, Sharma S, et al. Predictions of CCR1 chemokine receptor structure and BX 471 antagonist binding followed by experimental validation. J. Biol. Chem. 2006;281:27613–27620. doi: 10.1074/jbc.M601389200. [DOI] [PubMed] [Google Scholar]
- 34.Engel S, Skoumbourdis AP, Childress J, Neumann S, Deschamps JR, Thomas CJ, et al. A virtual screen for diverse ligands: Discovery of selective G protein-coupled receptor antagonists. J. Am. Chem. Soc. 2008;130:5115–5123. doi: 10.1021/ja077620l. [DOI] [PubMed] [Google Scholar]
- 35.Tikhonova IG, Sum CS, Neumann S, Engel S, Raaka BM, Costanzi S, et al. Discovery of novel agonists and antagonists of the free fatty acid receptor 1 (FFAR1) using virtual screening. J. Med. Chem. 2008;51:625–633. doi: 10.1021/jm7012425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavasotto CN, Orry AJ, Murgolo NJ, Czarniecki MF, Kocsi SA, et al. Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J. Med. Chem. 2008;51:581–588. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharya S, Subramanian G, Hall S, Lin J, Laoui A, Vaidehi N. Allosteric antagonist binding sites in class B GPCRs: corticotropin receptor 1. J. Comput. Aided Mol. Des. 2010;24:659–674. doi: 10.1007/s10822-010-9364-2. [DOI] [PubMed] [Google Scholar]
- 38.Dragon, Talete, SRL E-Dragon, Virtual Computational Chemistry Laboratory. www.talete.mi.it www.vcclab.org
- 39.The CODESSA PRO project www.codessa-pro.com.
- 40.MOE . Chemical Computing Group, Inc.; www.chemcomp.com. [Google Scholar]
- 41.Schrödinger LLC. www.schrodinger.com.
- 42.ICM, MolSoft, LLC www.molsoft.com.
- 43.SYBYL . Tripos, Inc.; www.tripos.com. [Google Scholar]
- 44.GOLD . Cambridge Crystallographic Data Centre; www.ccdc.cam.ac.uk/products/life_sciences/gold. [Google Scholar]
- 45.The R project for statistical computing www.r-project.org.
- 46.STATISTICA . StatSoft, Inc.; www.statsoft.com. [Google Scholar]
- 47.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, et al. GLIDA: GPCR--ligand database for chemical genomics drug discovery--database and tools update. Nucleic Acids Res. 2008;36:D907–912. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costanzi S, Tikhonova IG, Ohno M, Roh EJ, Joshi BV, Colson AO, et al. P2Y1 antagonists: Combining receptor-based modeling and QSAR for a quantitative prediction of the biological activity based on consensus scoring. J. Med. Chem. 2007;50:3229–3241. doi: 10.1021/jm0700971. [DOI] [PubMed] [Google Scholar]
- 49.Golbraikh A, Tropsha A. Beware of q2! J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 50.Normolle D, Ruffin MT, Brenner D. Design of early validation trials of biomarkers. Cancer Inform. 2005;1:25–31. [PMC free article] [PubMed] [Google Scholar]
- 51.Concato J, Peduzzi P, Holford TR, Feinstein AR. Importance of events per independent variable in proportional hazards analysis I. Background, goals, and general strategy. J. Clin. Epidemiol. 1995;48:1495–1501. doi: 10.1016/0895-4356(95)00510-2. [DOI] [PubMed] [Google Scholar]
- 52.Hill T, Lewicki P. STATISTICS Methods and Applications. StatSoft; Tulsa: 2006. [Google Scholar]