Abstract
Abnormalities of prefrontal cortical function are prominent features of schizophrenia and have been associated with genetic risk, suggesting that susceptibility genes for schizophrenia may impact on the molecular mechanisms of prefrontal function. A potential susceptibility mechanism involves regulation of prefrontal dopamine, which modulates the response of prefrontal neurons during working memory. We examined the relationship of a common functional polymorphism (Val108/158 Met) in the catechol-O-methyltransferase (COMT) gene, which accounts for a 4-fold variation in enzyme activity and dopamine catabolism, with both prefrontally mediated cognition and prefrontal cortical physiology. In 175 patients with schizophrenia, 219 unaffected siblings, and 55 controls, COMT genotype was related in allele dosage fashion to performance on the Wisconsin Card Sorting Test of executive cognition and explained 4% of variance (P = 0.001) in frequency of perseverative errors. Consistent with other evidence that dopamine enhances prefrontal neuronal function, the load of the low-activity Met allele predicted enhanced cognitive performance. We then examined the effect of COMT genotype on prefrontal physiology during a working memory task in three separate subgroups (n = 11–16) assayed with functional MRI. Met allele load consistently predicted a more efficient physiological response in prefrontal cortex. Finally, in a family-based association analysis of 104 trios, we found a significant increase in transmission of the Val allele to the schizophrenic offspring. These data suggest that the COMT Val allele, because it increases prefrontal dopamine catabolism, impairs prefrontal cognition and physiology, and by this mechanism slightly increases risk for schizophrenia.
Schizophrenia is a complex genetic disorder characterized by chronic psychosis, cognitive impairment, and functional disability. Linkage studies have implicated several possible susceptibility loci, including regions on chromosomes 1q, 6p, 8p, 13q, and 22q (1–3). Attempts to replicate these findings have met with limited success, perhaps due to the weak effects of susceptibility loci and limited power of linkage (4, 5). Of genes mapped to 22q11, a common functional polymorphism of catechol-O-methyltransferase (COMT), a methylation enzyme that metabolizes released dopamine (6), has been a popular candidate because of the long hypothesized role of dopamine in schizophrenia (7). Although two family-based association studies using the transmission disequilibrium test (TDT) have provided evidence for a role of COMT in schizophrenia (8–10), several small case-control association studies of COMT alleles have been negative, and it has been unclear how either protein variation would increase risk for schizophrenia (11, 12).
One approach that may improve power to find genes for complex disorders is to target biological traits found in ill subjects and their unaffected relatives, so-called intermediate phenotypes, rather than clinical diagnosis (13, 14). Such traits may be more directly related to the biological effects of susceptibility genes. Abnormal function of the prefrontal cortex, a cardinal aspect of schizophrenia, also may represent an intermediate phenotype related to genetic risk for schizophrenia (15, 16). Stable deficits in cognitive functions referable to the dorsolateral prefrontal cortex and cortical physiological abnormalities during performance of such tasks have been consistently reported in studies of schizophrenia (17–22). Recent evidence indicates that healthy siblings of patients, including monozygotic cotwins, show similar cognitive and physiological abnormalities (14–16, 22, 24).¶
Prefrontal deficits also are appealing phenotypes for genetic studies because the molecular mechanisms underlying such deficits have been sufficiently clarified to permit an hypothesis-driven test of candidate functional polymorphisms (25, 26). Electrophysiological studies in primates (27, 28) and rodents (29), and neuroimaging studies in humans (30, 31), have shown that dopamine plays an important role in modulating the activity of prefrontal circuitry during performance of working memory tasks. Although there are many proteins involved in the biological actions of dopamine, COMT, because it metabolizes released dopamine, may be an important factor during such prefrontally mediated tasks. Despite COMT's widespread distribution in nondopaminergic neurons and glia, pharmacological studies have shown that catabolic flux of synaptic dopamine through the COMT pathway is characteristic of the prefrontal cortex in contrast to the striatum (32). Studies of COMT knockout mice, similarly, have demonstrated that dopamine levels are increased only in prefrontal cortex (33) and, remarkably, that memory performance is enhanced.‖ This regionally selective effect of COMT may be because, in contrast to striatum, in prefrontal cortex dopamine transporters are expressed in low abundance and not within synapses (34, 35). As a consequence, released synaptic dopamine appears to be inactivated by diffusion, receptor internalization, and COMT degradation. These findings strongly support the notion that variation in COMT activity may have neurobiological effects specific to the prefrontal cortex.
The COMT gene contains an evolutionarily recent G to A missense mutation that translates into a substitution of Met for Val at codon 108/158 (Val108/158 Met) (GenBank accession no. Z26491). The enzyme containing Met is unstable at 37°C and has 1/4 of the activity of the enzyme containing Val (36). The alleles are codominant, as heterozygous individuals have enzyme activity that is midway between homozygote individuals (6). Thus, genetically determined variations in COMT activity might affect prefrontal cortical activity, especially during executive and working memory tasks. We hypothesized that the high-activity Val allele, because it leads to increased dopamine catabolism, would be associated with relatively compromised prefrontal function, and, by virtue of this effect, would increase risk for schizophrenia.
To test these hypotheses, we studied prefrontal executive cognition and physiology in control subjects, patients with schizophrenia, and their unaffected siblings. To measure executive cognition and working memory, we used the Wisconsin Card Sorting Test (WCST). Deficits in WCST performance are enduring and core features of schizophrenia and predict long term-disability, independent of other cognitive deficits (17, 21); healthy siblings of patients with schizophrenia also perform abnormally on it (24, 37). Functional neuroimaging studies have found that the WCST activates the dorsolateral prefrontal cortex (17, 38) and that dopamimetic drugs improve performance on this task in patients with schizophrenia and enhance the signal to noise of the prefrontal physiological response (30, 31). We hypothesized, therefore, that COMT genotype would affect WCST performance and that Val/Val individuals would have the poorest performance.
To assay prefrontal physiology, we used functional MRI (fMRI) while subjects performed the N-back task. This task has been shown to activate dorsolateral prefrontal cortex as well as a distributed cortical working memory network (20, 39). In studies of patients with schizophrenia who perform relatively well on the N-back and similar tasks, fMRI activation of dorsolateral prefrontal cortex is “inefficient,” i.e., there is excessive activity for a given level of performance (19, 20). Similar fMRI results have been described in their unaffected siblings,¶ suggesting that inefficient prefrontal information processing is related to genetic risk for schizophrenia. Using the N-back fMRI paradigm, Mattay et al. recently reported analogous inefficiency in hypodopaminergic patients with Parkinson's disease.** In contrast, the efficiency of the N-back fMRI response in dorsolateral prefrontal cortex is enhanced by the dopamimetic drug, amphetamine, in healthy individuals whose performance remains stable (40). Thus, deviations of prefrontal physiology can be appreciated with this in vivo fMRI assay even if there is compensation at the level of performance accuracy, and changes in cortical dopaminergic function impact on physiological efficiency during this task. We hypothesized, therefore, that COMT genotype would affect the efficiency of the prefrontal fMRI response during this task and predicted an allele dosage relationship with activation, with Val/Val individuals being least efficient.
Methods
Subjects and Cognitive Testing.
Subjects were recruited from local and national sources as volunteers for the Clinical Brain Disorders Branch/National Institute of Mental Health sibling study, as described (41). Briefly, all participants gave written informed consent of an Institutional Review Board-approved protocol. Most families had two eligible full siblings (at least one of whom met DSM-IV criteria for schizophrenia or schizoaffective disorder, depressed subtype). All subjects had to be from 18 to 60 years of age, above 70 in premorbid IQ, and able to give informed consent. Applicants with significant medical problems, history of head trauma, alcohol or drug abuse within the last 6 months were excluded. All subjects were medically screened and interviewed by a research psychiatrist using the Structured Clinical Interview (42).
To reduce the possibility of artifactual association due to ethnic stratification, the final sample included only individuals of European ancestry born and educated in the U.S. This sample included 175 patients with schizophrenia, 219 healthy siblings, and 55 control subjects.
Subjects performed the WCST. Perseverative errors was used as a dependent measure because it is thought to best reflect prefrontal function. Scores were transformed to t scores and normalized for age and education based on population means, a routine convention (43). Thus, better performance is reflected in a higher t score. IQ (from the Wechsler Adult Intelligence Scale, revised edition, or WAIS-R) and reading comprehension (using the Wide Range Achievement Test, WRAT, a measure of premorbid IQ) also were collected (44).
Neuroimaging.
Two cohorts of siblings (all nonsmokers) and one cohort of probands were randomly selected, based on scanner availability. Blood oxygen level-dependent fMRI was performed while subjects took the two-back and zero-back versions of the N-back task (20). In contrast to the WCST, the N-back is a relatively simple working memory task more suitable for fMRI.
The N-back task was presented via a fiber-optic goggle system and responses were recorded via a pneumatic button box. Stimuli were displayed randomly at a rate of 1.8 per sec. All subjects were first trained to maximal performance. The first group of unaffected siblings (n = 16) and the group of patients with schizophrenia (n = 11) were studied with an echo planar imaging blood oxygen level-dependent fMRI sequence at 1.5 Tesla (20). The second sibling group (n = 11) was studied by using a more rapid scanning pulse sequence, fast spiral imaging also at 1.5 Tesla (45).
Whole brain echo planar imaging data were collected in a modified block design with pseudorandomized intermixing of zero-back and two-back working memory tasks. Fast spiral imaging data were collected by using a simple block design alternating between zero-back and two-back (16 sec/task epoch) occurring during one 256-sec run. All fMRI data were reconstructed, registered, linear detrended, globally normalized, and then smoothed (10 mm Gaussian kernel) before analysis within statistical parametric mapping (SPM) (46). All data were rigorously screened for artifacts as described (20). Individual data from 18 task epochs were collapsed as adjusted means and then entered into a general linear model within SPM96 (for cohort 1) or SPM 99 (for cohort 2) (Wellcome Department of Cognitive Neurology, London). We first estimated parameters that reflected activation as a contrast between the two-back task and the zero-back task. These parameter estimates were then entered into a second analysis to test inferences about differential activations among the three genotype groups. This analysis is formally identical to a random effects analysis where the subject effect is a random effect. Because we had an anatomically specified hypothesis about prefrontal activation, we used an uncorrected threshold of P = 0.005 (voxelwise) to identify these regionally specific differences. The resultant statistical maps then were rendered onto a three-dimensional standard brain.
Genetic Analysis.
Blood was collected from all subjects as well as all available parents of patients with schizophrenia. DNA was extracted by using standard methods. DNA from 104 pairs of parents were available for the final analysis. COMT Val108/158 Met genotype was determined as a restriction fragment length polymorphism after PCR amplification and digestion with NlaIII, similar to a previously described method (47) (details available on request).
To address at a genomic level the issue of potential population admixture, 19 unlinked, short tandem repeat markers, all with heterozygosities >65%, were genotyped by using PCR and gel analysis as described (48) in selected subjects (details available on request). The markers were: D1S1612, D1S1678, D2S1356, D4S1280, D5S1471, D6S1006, D7S2847, D17S1308, D18S843, D18S535, D19S714, D20S604, D20S477, D20S481, D21S1437, D21S1446, D22S445, SLC6A3 3′ untranslated region VNTR (GenBank accession no. 162767), and the (TAA) repeat in locus HSMHC3A5 (GenBank accession no. U89335).
Statistical Analyses.
Between groups comparisons of demographic data were performed by using paired or unpaired t tests or χ2, as appropriate. To avoid lack of independence among family members, we used one randomly selected sib per family for comparisons with the control group. The effects of COMT genotype were analyzed several ways. First, groups were compared by using standard parametric techniques (case/control comparisons). Second, to avoid spurious results due to admixture, we used TDT (49, 50), which are family-based methods that substantially sacrifice power.
The effect of COMT genotype on WCST performance was assessed by using
two case-control analyses: (i) ANOVA and (ii)
multiple regression. With ANOVA, we first included all subjects.
Because this assumes independence of individuals, we also report ANOVA
results including only patients and controls. Second, using multiple
regression we tested the hypothesis that the number of Met alleles was
parametrically related to enhanced performance (patients and controls
only); diagnostic group was included as the only additional independent
variable. Next, we performed a family-based test to examine the effect
of COMT genotype on WCST performance (quantitative sib TDT; ref. 50).
Subsequently, we examined whether admixture was present in Val/Val
and Met/Met groups for patients and controls (Fig.
1) by comparing allele frequencies of 19
unlinked polymorphic genetic markers using an overall
χ as described by Pritchard and Rosenberg (51).
as described by Pritchard and Rosenberg (51).
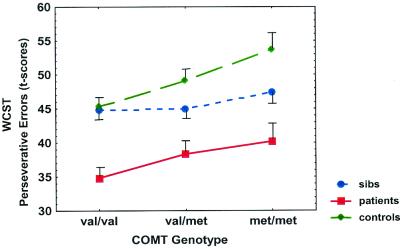
Figure 1.
WCST perseverative error t scores (± SE) by genotype for each group (population mean = 50, SD = 10, lower scores indicate worse performance). Main effect of genotype: F = 4.93, df = 2,224, P = 0.008.
The effect of COMT genotype on risk for schizophrenia was analyzed by using both case-control and family-based methods. The case-control analysis was a comparison of allele frequencies. The family-based analysis used the TDT (49). A critical issue in assessing the significance of association with phenotypic measures is the likelihood of type I errors. Many genes and phenotypes can be evaluated for schizophrenia and ultimately may be examined in this dataset, but Bonferroni correction for all possible combinations that ultimately may be performed seems overly stringent. The approach here was to selectively analyze a single candidate functional polymorphism, chosen for its biological effect, against a target phenotype likely impacted by this biological effect. Given the role of COMT in prefrontal dopamine metabolism, and the role of dopamine in prefrontal function and working memory, the prior probability of this gene modifying prefrontal function may be high relative to other polymorphisms and phenotypes.
Results
Demographic data are presented in Table 1. Briefly, siblings and controls were well matched on age, gender, education, IQ, and WRAT. There was no difference between patients receiving typical and atypical neuroleptic treatment on any cognitive variable. History of alcohol abuse and dependence did not affect any cognitive measure in this study, most likely because subjects with recent or prolonged abuse or dependence were excluded (41).
Table 1.
Demographics
| Variable | Patients (n = 175) | Siblings (n = 219) | Controls (n = 55) |
|---|---|---|---|
| Age | 36.1 (8.5) | 35.6 (8.8) | 33.9 (9.2) |
| Gender (M/F) | 138/37*† | 97/122 | 23/32 |
| Education years | 13.7*† (2.1) | 15.5 (2.5) | 15.7 (2.5) |
| WRAT | 102.0*† (12.1) | 106.3 (11.2) | 107.3 (11.4) |
| IQ | 92.8*† (13.1) | 107.4 (10.6) | 109.1 (11.5) |
| WCST perseverative errors | 37.6*† (12.6) | 45.2 (9.5)* | 49.4 (9.0) |
Means ± SD.
Significantly different compared to controls (P < 0.05).
Significantly different compared to siblings (P < 0.05).
Patients and siblings scored significantly worse on the WCST compared with the control group (Table 1), as reported (24, 37) (F = 29.6, df = 2,440, P < 0.00001). An ANOVA for all groups revealed a significant effect of COMT genotype on WCST performance (F = 6.00, df = 2,440, P = 0.003) with no group by genotype interaction (F = 1.40, df = 4,440, P = 0.23, Fig. 1). A second ANOVA including only patients and controls also detected a significant effect of genotype (F = 4.93, df = 2,224, P = 0.008). Post hoc analysis showed that subjects with the Val/Val genotype performed worse than those with the Val/Met and Met/Met genotypes (P < 0.002). In contrast, no genotype effect was seen on tasks of general academic ability, e.g., WRAT reading scores or IQ, and no differences were seen between genotype groups in other demographic measures (Table 2).
Table 2.
Demographics by genotype for patients and controls
| Patients
|
Controls
|
|||||
|---|---|---|---|---|---|---|
| Val/Val | Val/Met | Met/Met | Val/Val | Val/Met | Met/Met | |
| Age | 37.1 (8.3) | 35.7 (8.1) | 35.1 (8.3) | 34.5 (10.5) | 33.7 (10.0) | 34.2 (9.5) |
| Gender (M/F) | 49/13 | 68/17 | 21/7 | 6/9 | 10/20 | 7/3 |
| Education years | 13.9 (2.0) | 13.6 (2.0) | 13.5 (2.6) | 16.3 (2.5) | 15.8 (2.3) | 15.8 (2.6) |
| WRAT | 102.1 (10.7) | 102.4 (11.4) | 100.9 (13.4) | 108.0 (9.1) | 106.8 (10.6) | 107.4 (6.0) |
| IQ | 89.9 (13.7) | 94.3 (12.0) | 94.5 (12.6) | 111.5 (8.7) | 107.3 (9.2) | 110.4 (8.8) |
Means ± SD. Within each group (patients or controls), there is no significant difference between genotype for any variable.
Using multiple regression, the number of Met alleles was parametrically related to perseverative errors t scores [r2 = 0.041, t(228) = 3.29, P = 0.001]. COMT genotype accounted for 4.1% of the variance in performance. Because prior reports have found an effect of gender on COMT expression in animal models (33), we added gender into both the ANOVA and multiple regression analyses. There was no effect of gender or gender by genotype interaction. To exclude other possible spurious effects, we added diagnosis, age, gender, and education to a stepwise multiple regression analysis. This resulted in a small decrease in the r2 for the COMT effect but its significance at entry remained high (increment in adjusted r2 = 0.024, P = 0.003). Using the family-based quantitative sib TDT, a trend was seen for a COMT genotype effect on WCST performance (F = 2.36, df = 2,159, P < 0.10). Using 19 polymorphic genetic markers, no evidence for population stratification was found between Val/Val and Met/Met groups in patients or controls (omnibus χ2 = 113.5, df = 112 P = 0.44).
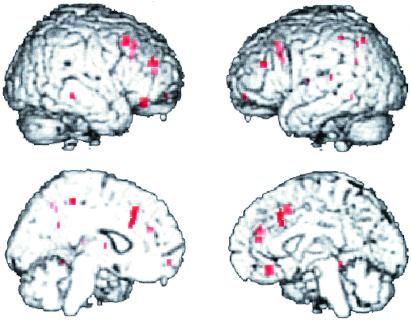
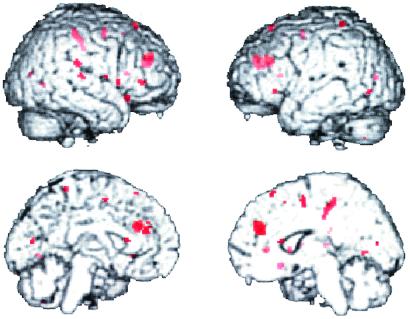
Figs. 2 and 3 show the effect of COMT allele load on the fMRI response during the two-back version of the N-back task in the two groups of siblings. The first group (Fig. 2) consisted of five Met/Met individuals, six Val/Met individuals, and five Val/Val individuals. The genotype subgroups used did not differ in mean age, gender, education, handedness, or performance accuracy. The second group (Fig. 3) consisted of three Met/Met, five Met/Val, and three Val/Val individuals; these genotype subgroups did not differ significantly in age, education, gender, handedness, or performance accuracy. In both groups, locales in dorsolateral prefrontal and cingulate cortices show the predicted genotype effects, with Val/Val individuals having the greatest response (i.e., being least efficient), followed by Val/Met and then Met/Met individuals. Similar results were seen in the patient group as well (data not shown).
Figure 2.
Effect of COMT genotype on fMRI activation during the two-back working memory task. Regions showing a significant effect of genotype on fMRI activation (voxelwise P < 0.005) are in red (shown clockwise from upper left in right lateral, left lateral, right medial, and left medial views, respectively). In dorsolateral prefrontal cortex (e.g., Brodmann area 46; x = 58, y = 32, z = 12; cluster size = 47; Z = 2.55) and anterior cingulate (e.g., Brodmann 32; x = 6, y = 60, z = 8; cluster size = 77; Z = 2.36), Val/Val individuals showed a greater fMRI response (and by inference, greater inefficiency, as performance is similar) than Val/Met individuals who have greater activation than Met/Met individuals. Post hoc analysis of genotype group contrasts confirmed these significant relationships in dorsolateral prefrontal and cingulate cortices across all groups.
Figure 3.
Effect of COMT genotype on fMRI activation during the two-back working memory task in a second group of subjects. Again, Val/Val individuals showed greater activation (and by inference greater inefficiency) than Val/Met individuals who showed less efficiency than Met/Met individuals in the dorsal prefrontal cortex and several other locales.
We next addressed the possibility that in the 104 family trios, the COMT Val allele is a risk factor for schizophrenia, per se. A total of 126 transmissions were counted from heterozygous parents to probands. The Val allele was transmitted 75 times, compared with 51 transmissions of the Met allele. These proportions are different from that predicted by random assortment (χ2 = 4.57; P = 0.03) and indicate that the COMT Val allele is weakly associated with schizophrenia. The odds ratio for the Val/Val genotype is 1.5. Unaffected siblings (n = 117) had 77 Val transmissions and 87 Met transmissions, indicating that meiotic segregation distortion is not present. Monte Carlo simulation of 10,000 TDT replicates confirmed that our result would occur at the P < 0.04 level of significance. In the case-control analysis, no significant differences in allele (χ2 = 0.92; df = 1; P = 0.34) or genotype (χ2 = 1.25; df = 2, P = 0.54) frequencies were seen comparing patients and controls (Table 3), similar to most (11, 52–54), but not all (55, 56) earlier case-control studies.
Table 3.
Distribution of genotypes and alleles
| Genotype | Patients (n = 175) | Sibs (n = 219) | Controls (n = 55) |
|---|---|---|---|
| Val/Val | 62 (35%) | 69 (31%) | 15 (27%) |
| Val/Met | 85 (49%) | 114 (51%) | 30 (55%) |
| Met/Met | 28 (16%) | 39 (18%) | 10 (18%) |
| Frequency Val | 0.60 ± 0.03 | 0.57 ± 0.02 | 0.54 ± 0.03 |
| Frequency Met | 0.40 ± 0.03 | 0.43 ± 0.02 | 0.46 ± 0.03 |
Frequency is ± SE.
Discussion
We report several convergent findings that implicate an effect of COMT genotype on prefrontal cortical function and, as a result, on increased risk for schizophrenia. First, COMT genotype is specifically associated with level of performance on a neuropsychological test of executive cognition that is related to function of prefrontal cortex, but not with general intelligence. This effect of COMT is independent of psychiatric diagnosis and explains 4.1% of the variance on the WCST. The high-activity Val allele is associated with a reduction in performance compared with the Met allele. Second, Val allele load is related to reduced “efficiency” of the physiologic response in the dorsolateral prefrontal cortex during performance of a simple working memory task in three cohorts studied with fMRI. Neural net modeling of the effects of dopamine on working memory circuits predicted that reductions in synaptic dopamine would reduce signal-to-noise ratios, thus reducing efficiency (57). This prediction recently was confirmed in an fMRI study of patients with Parkinson's disease.** It is also consistent with the effect of the Val allele observed in our fMRI data. These convergent findings suggest that the COMT Val allele, presumably by compromising the postsynaptic impact of the evoked dopamine response, may reduce signal to noise in prefrontal neurons and thereby alter working memory function. Third, the Val allele is transmitted slightly more often (P < 0.04) to probands with schizophrenia. The association of the Val allele with schizophrenia suggests that this allele, by virtue of its physiological effect on prefrontal information processing, increases susceptibility to schizophrenia.
This proposed genetic/neurophysiological mechanism is consistent with prior studies of the neurobiology of schizophrenia. As described above, deficits in prefrontal function are core manifestations of schizophrenia and are related to genetic risk for schizophrenia (15, 16, 24). Neuroimaging and postmortem studies have found evidence of reduced dopaminergic innervation of dorsolateral prefrontal cortex in patients with schizophrenia (17, 58, 59). Thus, the COMT Val allele, by imposing an additional adverse load specifically on prefrontal function, might add to or interact with other causes of prefrontal malfunction in those at risk for schizophrenia and thereby increase their susceptibility. However, the effect of COMT genotype on prefrontal function is small; indeed, it was not significant in the cohort of siblings. This latter negative finding could be due to siblings being a mixed group, in terms of other genetic risk and protective factors. Val/Val siblings who have no psychiatric disorder, for example, could have protective factors positively affecting prefrontal cortical function, otherwise they might themselves have schizophrenia.
With an odds ratio of 1.5, the effect of the Val/Val genotype acting alone on diagnosis is weak. Indeed, a 4.1% variation in prefrontal function by itself may not pose much of a risk for behavioral decompensation. This risk, however, represents an average effect across many individuals. The effect of COMT genotype within any particular individual could be large or small, depending on a variety of background factors. Thus, a gene such as COMT could have an important clinical effect in combination with other genes and environmental factors and could be of value in identifying such factors, especially if their effects are nonadditive. Nevertheless, it seems possible or even likely that most susceptibility genes for schizophrenia will either have a relatively low genotypic relative risk or will be very uncommon in the general patient population and affect only a small portion of patients (5, 60).
Although our results offer a mechanism for how the Val allele might increase susceptibility for schizophrenia, the results of genetic studies, including this one, showing linkage or association between COMT and schizophrenia are, at best, weak. Linkage studies generally have found logarithm of odds scores of 2 or less for markers near 22q11, the chromosomal region containing the COMT gene (3, 61, 62) (see ref. 60 for review). Of previously published TDT-based association studies, one found a significant relationship between schizophrenia and Val transmissions (9); a second also reported an excess (22 vs. 13, χ2 = 2.31, P = 0.13) of Val transmissions (8). In an expanded sample of 198 trios, Li et al. (10) performed a haplotype analysis and again showed a significant association with the Val allele and schizophrenia. Although TDT analyses have been uniformly positive, the results of case control association studies (including our own), which have generally used small sample sizes (relative to those needed to detect a weak genetic effect), have been negative in most (11, 52–54), but not all (55) cases. These negative results are not unexpected, given the lack of power in these studies to detect alleles of minor effect.
Population stratification artifacts are an important consideration in genetic case control analyses and might be an occult factor in our genetic effect on prefrontal function. COMT Val/Met allele frequencies differ across some ethnic groups, although this is probably not the case for the western European populations represented in our study (12). Given that the predicted COMT effect on WCST performance was seen in two unrelated samples (patients and controls) and the predicted effect on cortical physiology was found in three samples, similar stratification would have to be common to all these cohorts and both phenotypes. Furthermore, the genetically distinct subpopulations would have to differ only on prefrontal measures and not on general intelligence, because genotype groups did not differ on other cognitive tests. Nevertheless, we also used two methods to test whether admixture might account for our genetic effect on cognition, a family-based analysis (50), and genomic controls (51). The quantitative sibling TDT used with the WCST data was not significant, although with a trend P value of <0.10, but this is a random effects model with limited degrees of freedom. Using 19 unlinked polymorphic genetic markers, we found no genetic evidence for stratification. The family-based TDT, which found a weakly significant association with schizophrenia, also controls for stratification (49).
A second possible artifact to consider is that the COMT Val/Met polymorphism is not the causative locus but is in linkage disequilibrium with another mutation. We suggest that, given (i) the strong impact of the COMT Val/Met polymorphism on COMT enzyme activity, (ii) the known effects of COMT on prefrontal dopamine metabolism, and (iii) the effect of dopamine on prefrontal neuronal function and working memory, the COMT Val/Met allele is the causative genetic locus for the association with prefrontal function. Using a COMT knockout mouse model, others have shown that prefrontal dopamine levels are increased (33) and that performance on a memory task is actually improved relative to the wild-type animal.‖ This remarkable improvement in memory performance supports our model that the Met allele, with its reduced activity, accounts for improved prefrontal function, and not another nearby gene.
Finally, is it plausible that a common allele with such weak effects could increase risk for schizophrenia? In some respects, our results with COMT and schizophrenia are similar to the calpain-10 association with diabetes (63), and the association of the APO e4 allele with Alzheimer's disease, although the APO e4 effect is much greater (64). The calpain-10 allele is found in 75% of the general population and in 80% of diabetics, a weak association that is not easily replicated across populations, and the biologic effect of the polymorphism is unknown. It is assumed that such polygenes interact with other genes and environmental factors to incrementally increase risk. The COMT Val allele is certainly not a necessary or sufficient causative factor for schizophrenia, nor is it likely to increase risk only for schizophrenia. However, its biological effect on prefrontal function and the relevance of prefrontal function for schizophrenia susceptibility implicate a mechanism by which it could increase liability for this disorder. The data presented here provide convergent evidence that the Val allele compromises prefrontal function and thereby impacts directly on the biology of schizophrenia. Despite the apparent disadvantage of the Val allele, the Met allele may increase susceptibility to other disorders, such as estrogenic cancer (23), suggesting that a heterozygote advantage could maintain the high Met and Val allele frequencies observed in a variety of human populations. Finally, it should be noted that the COMT polymorphism affects performance and prefrontal cortical function in both ill and healthy subjects. Thus, the recent Met mutation, which has not been reported in nonhuman primates (12), enhances an important component of normal human cognition, suggesting a possible role in the evolution of human brain function.
Acknowledgments
We thank the following for their assistance: Lew Bigelow, Mary Weirick, Venkatta Mattay, Tonya Gscheidle, Ashley Bone, Tom Weickert, Andreas Myer-Lindenberg, Alan Barnett, and the patients and their families whose participation made this project possible. This project was supported by funding from the National Institute of Mental Health and the Stanley Foundation (to D.R.W.).
Abbreviations
- COMT
catechol-O-methyltransferase
- fMRI
functional MRI
- WCST
Wisconsin Card Sorting Test
- TDT
transmission disequilibrium test
- WRAT
Wide Range Achievement Test
Footnotes
Callicott, J., Egan, M., Mattay, V., Bertolino, A., Jones, K., Goldberg, T. & Weinberger, D. (1998) NeuroImage 7, S895 (abstr.).
Kneavel, M., Gogos, J., Karayiorgou, K. & Luine, V., Society for Neuroscience 30th Annual Meeting, November 5–10, 2000, New Orleans, 571.20 (abstr.).
Mattay, V. S., Tessitore, A., Callicott, J. H., Bertolino, A., Duyn, J., Frank, J. A., Goldberg, T., Chase, T., Hyde, T. & Weinberger, D. R., Society for Neuroscience 30th Annual Meeting, November 5–10, 2000, New Orleans, 746 (abstr.).
References
- 1.Brzustowicz L M, Hodgkinson K A, Chow E W, Honer W G, Bassett A S. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straub R E, MacLean C J, O'Neill F A, Burke J, Murphy B, Duke F, Shinkwin R, Webb B T, Zhang J, Walsh D, et al. Nat Genet. 1995;11:287–293. doi: 10.1038/ng1195-287. [DOI] [PubMed] [Google Scholar]
- 3.Pulver A E, Karayiorgou M, Wolyniec P S, Lasseter V K, Kasch L, Nestadt G, Antonarakis S, Housman D, Kazazian H H, Meyers D, et al. Am J Med Genet. 1994;54:36–43. doi: 10.1002/ajmg.1320540108. [DOI] [PubMed] [Google Scholar]
- 4.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 5.Risch N. Am J Hum Genet. 1990;46:222–228. [PMC free article] [PubMed] [Google Scholar]
- 6.Weinshilboum R M, Otterness D M, Szumlanski C L. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson A, Waters N, Waters S, Carlsson M L. Brain Res Brain Res Rev. 2000;31:342–349. doi: 10.1016/s0165-0173(99)00050-8. [DOI] [PubMed] [Google Scholar]
- 8.Kunugi H, Vallada H P, Sham P C, Hoda F, Arranz M J, Li T, Nanko S, Murray R M, McGuffin P, Owen M, et al. Psychiatr Genet. 1997;7:97–101. doi: 10.1097/00041444-199723000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Li T, Sham P C, Vallada H, Xie T, Tang X, Murray R M, Liu X, Collier D A. Psychiatr Genet. 1996;6:131–133. doi: 10.1097/00041444-199623000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Li T, Ball D, Zhao J, Murray R M, Liu X, Sham P C, Collier D A. Mol Psychiatry. 2000;5:77–84. doi: 10.1038/sj.mp.4000638. [DOI] [PubMed] [Google Scholar]
- 11.Karayiorgou M, Gogos J A, Galke B L, Wolyniec P S, Nestadt G, Antonarakis S E, Kazazian H H, Housman D E, Pulver A E. Biol Psychiatry. 1998;43:425–431. doi: 10.1016/s0006-3223(97)00202-3. [DOI] [PubMed] [Google Scholar]
- 12.Palmatier M A, Kang A M, Kidd K K. Biol Psychiatry. 1999;46:557–567. doi: 10.1016/s0006-3223(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 13.Freedman R, Coon H, Myles-Worsley M, Orr-Urtreger A, Olincy A, Davis A, Polymeropoulos M, Holik J, Hopkins J, Hoff M, et al. Proc Natl Acad Sci USA. 1997;94:587–592. doi: 10.1073/pnas.94.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kremen W S, Seidman L J, Pepple J R, Lyons M J, Tsuang M T, Faraone S V. Schizophr Bull. 1994;20:103–119. doi: 10.1093/schbul/20.1.103. [DOI] [PubMed] [Google Scholar]
- 15.Cannon T D, Huttunen M O, Lonnqvist J, Tuulio-Henriksson A, Pirkola T, Glahn D, Finkelstein J, Hietanen M, Kaprio J, Koskenvuo M. Am J Hum Genet. 2000;67:369–382. doi: 10.1086/303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg T E, Ragland J D, Torrey E F, Gold J M, Bigelow L B, Weinberger D R. Arch Gen Psychiatry. 1990;47:1066–1072. doi: 10.1001/archpsyc.1990.01810230082013. [DOI] [PubMed] [Google Scholar]
- 17.Weinberger D R, Berman K F, Zec R F. Arch Gen Psychiatry. 1986;43:114–124. doi: 10.1001/archpsyc.1986.01800020020004. [DOI] [PubMed] [Google Scholar]
- 18.Carter C S, Perlstein W, Ganguli R, Brar J, Mintun M, Cohen J D. Am J Psychiatry. 1998;155:1285–1287. doi: 10.1176/ajp.155.9.1285. [DOI] [PubMed] [Google Scholar]
- 19.Manoach D S, Press D Z, Thangaraj V, Searl M M, Goff D C, Halpern E, Saper C B, Warach S. Biol Psychiatry. 1999;45:1128–1137. doi: 10.1016/s0006-3223(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 20.Callicott J H, Bertolino A, Mattay V S, Langheim F J P, Duyn J, Coppola R, Goldberg T E, Weinberger D R. Cereb Cortex. 2000;10:1078–1092. doi: 10.1093/cercor/10.11.1078. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg T E, Weinberger D R. Schizophr Bull. 1988;14:179–183. doi: 10.1093/schbul/14.2.179. [DOI] [PubMed] [Google Scholar]
- 22.Park S, Holzman P S, Goldman-Rakic P S. Arch Gen Psychiatry. 1995;52:821–828. doi: 10.1001/archpsyc.1995.03950220031007. [DOI] [PubMed] [Google Scholar]
- 23.Lavigne J A, Helzlsouer K J, Huang H Y, Strickland P T, Bell D A, Selmin O, Watson M A, Hoffman S, Comstock G W, Yager J D. Cancer Res. 1997;57:5493–5497. [PubMed] [Google Scholar]
- 24.Egan, M., Goldberg, T., Gscheidle, T., Bigelow, L., Hyde, T. & Weinberger, D. R. (2001) Biol. Psychiatry, in press. [DOI] [PubMed]
- 25.Lidow M S, Williams G V, Goldman-Rakic P. Trends Pharmacol Sci. 1998;19:136–140. doi: 10.1016/s0165-6147(98)01186-9. [DOI] [PubMed] [Google Scholar]
- 26.Gao W J, Krimer L S, Goldman-Rakic P S. Proc Natl Acad Sci USA. 2001;98:295–300. doi: 10.1073/pnas.011524298. . (First Published December 26, 2000, 10.1073/pnas.011524298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawaguchi T, Goldman-Rakic P S. Science. 1991;251:947–950. doi: 10.1126/science.1825731. [DOI] [PubMed] [Google Scholar]
- 28.Williams G V, Goldman-Rakic P S. Nature (London) 1995;376:572–575. doi: 10.1038/376572a0. [DOI] [PubMed] [Google Scholar]
- 29.Seamans J K, Floresco S B, Phillips A G. J Neurosci. 1998;18:1613–1621. doi: 10.1523/JNEUROSCI.18-04-01613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniel D G, Weinberger D R, Jones D W, Zigun J R, Coppola R, Handel S, Bigelow L B, Goldberg T E, Berman K F, Kleinman J E. J Neurosci. 1991;11:1907–1917. doi: 10.1523/JNEUROSCI.11-07-01907.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattay V S, Berman K F, Ostrem J L, Esposito G, Van Horn J D, Bigelow L B, Weinberger D R. J Neurosci. 1996;16:4816–4822. doi: 10.1523/JNEUROSCI.16-15-04816.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karoum F, Chrapusta S J, Egan M F. J Neurochem. 1994;63:972–979. doi: 10.1046/j.1471-4159.1994.63030972.x. [DOI] [PubMed] [Google Scholar]
- 33.Gogos J A, Morgan M, Luine V, Santha M, Ogawa S, Pfaff D, Karayiorgou M. Proc Natl Acad Sci USA. 1998;95:9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis D A, Sesack S R, Levey A I, Rosenberg D R. Adv Pharmacol. 1998;42:703–706. doi: 10.1016/s1054-3589(08)60845-5. [DOI] [PubMed] [Google Scholar]
- 35.Sesack S R, Hawrylak V A, Matus C, Guido M A, Levey A I. J Neurosci. 1998;18:2697–2708. doi: 10.1523/JNEUROSCI.18-07-02697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I, Taskinen J. Biochemistry. 1995;34:4202–4210. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- 37.Faraone S V, Seidman L J, Kremen W S, Pepple J R, Lyons M J, Tsuang M T. J Abnorm Psychol. 1995;104:286–304. doi: 10.1037//0021-843x.104.2.286. [DOI] [PubMed] [Google Scholar]
- 38.Berman K F, Ostrem J L, Randolph C, Gold J, Goldberg T E, Coppola R, Carson R E, Herscovitch P, Weinberger D R. Neuropsychologia. 1995;33:1027–1046. doi: 10.1016/0028-3932(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 39.Cohen J D, Perlstein W M, Braver T S, Nystrom L E, Noll D C, Jonides J, Smith E E. Nature (London) 1997;386:604–608. doi: 10.1038/386604a0. [DOI] [PubMed] [Google Scholar]
- 40.Mattay V S, Callicott J H, Bertolino A, Heaton I, Frank J A, Coppola R, Berman K F, Goldberg T E, Weinberger D R. NeuroImage. 2000;12:268–275. doi: 10.1006/nimg.2000.0610. [DOI] [PubMed] [Google Scholar]
- 41.Egan M, Goldberg T E, Gscheidle T, Weirick M, Bigelow L, Weinberger D R. Am J Psychiatry. 2000;157:1309–1316. doi: 10.1176/appi.ajp.157.8.1309. [DOI] [PubMed] [Google Scholar]
- 42.First M B, Gibbon M, Spitzer R L, Williams J B W. User's Guide for the SCID-I for DSM-IV Axis I Disorders-Research Version. New York: Biometrics Research; 1996. [Google Scholar]
- 43.Heaton R K, Chelune G J, Talley J L, Kay G G, Curtiss G. Wisconsin Card Sorting Test Manual. Odessa, FL: Psychological Assessment Resources; 1993. [Google Scholar]
- 44.Jastak S, Wilkinson G S. Wide Range Achievement Test. Wilmington, DE: Jastak Associates; 1984. [Google Scholar]
- 45.Yang Y, Glover G H, van Gelderen P, Mattay V S, Santha A K, Sexton R H, Ramsey N F, Moonen C T, Weinberger D R, Frank J A, Duyn J H. Magn Reson Med. 1996;36:620–626. doi: 10.1002/mrm.1910360418. [DOI] [PubMed] [Google Scholar]
- 46.Friston J K, Holmes A P, Worsley J, Poline J B, Frith C D, Frackowiak R S J. Hum Brain Mapp. 1995;2:189–210. [Google Scholar]
- 47.Lachman H M, Papolos D F, Saito T, Yu Y M, Szumlanski C L, Weinshilboum R M. Pharmacogenetics. 1996;6:243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 48.Straub R E, Speer M C, Luo Y, Rojas K, Overhauser J, Ott J, Gilliam T C. Genomics. 1993;15:48–56. doi: 10.1006/geno.1993.1008. [DOI] [PubMed] [Google Scholar]
- 49.Spielman R S, McGinnis R E, Ewens W J. Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 50.Allison D B, Heo M, Kaplan N, Martin E R. Am J Hum Genet. 1999;64:1754–1763. doi: 10.1086/302404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pritchard J K, Rosenberg N A. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daniels J K, Williams N M, Williams J, Jones L A, Cardno A G, Murphy K C, Spurlock G, Riley B, Scambler P, Asherson P, et al. Am J Psychiatry. 1996;153:268–270. doi: 10.1176/ajp.153.2.268. [DOI] [PubMed] [Google Scholar]
- 53.Chen C H, Lee Y R, Wei F C, Koong F J, Hwu H G, Hsiao K J. Biol Psychiatry. 1997;41:985–987. doi: 10.1016/S0006-3223(97)00045-0. [DOI] [PubMed] [Google Scholar]
- 54.Strous R D, Bark N, Woerner M, Lachman H M. Biol Psychiatry. 1997;41:493–495. doi: 10.1016/s0006-3223(96)00474-x. [DOI] [PubMed] [Google Scholar]
- 55.de Chaldee M, Laurent C, Thibaut F, Martinez M, Samolyk D, Petit M, Campion D, Mallet J. Am J Med Genet. 1999;88:452–457. doi: 10.1002/(sici)1096-8628(19991015)88:5<452::aid-ajmg2>3.3.co;2-s. [DOI] [PubMed] [Google Scholar]
- 56.Ohmori O, Shinkai T, Kojima H, Terao T, Suzuki T, Mita T, Abe K. Neurosci Lett. 1998;243:109–112. doi: 10.1016/s0304-3940(98)00100-1. [DOI] [PubMed] [Google Scholar]
- 57.Servan-Schreiber D, Printz H, Cohen J D. Science. 1990;249:892–895. doi: 10.1126/science.2392679. [DOI] [PubMed] [Google Scholar]
- 58.Weinberger D R, Berman K F, Illowsky B P. Arch Gen Psychiatry. 1988;45:609–615. doi: 10.1001/archpsyc.1988.01800310013001. [DOI] [PubMed] [Google Scholar]
- 59.Akil M, Pierri J N, Whitehead R E, Edgar C L, Mohila C, Sampson A R, Lewis D A. Am J Psychiatry. 1999;156:1580–1589. doi: 10.1176/ajp.156.10.1580. [DOI] [PubMed] [Google Scholar]
- 60.Riley B P, McGuffin P. Am J Med Genet. 2000;97:23–44. doi: 10.1002/(sici)1096-8628(200021)97:1<23::aid-ajmg5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 61.Pulver A E, Karayiorgou M, Lasseter V K, Wolyniec P, Kasch L, Antonarakis S, Housman D, Kazazian H H, Meyers D, Nestadt G, et al. Am J Med Genet. 1994;54:44–50. doi: 10.1002/ajmg.1320540109. [DOI] [PubMed] [Google Scholar]
- 62.Gill M, Vallada H, Collier D, Sham P, Holmans P, Murray R, McGuffin P, Nanko S, Owen M, Antonarakis S, et al. Am J Med Genet. 1996;67:40–45. doi: 10.1002/(SICI)1096-8628(19960216)67:1<40::AID-AJMG6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 63.Horikawa Y, Oda N, Cox N J, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner T H, Mashima H, Schwarz P E, et al. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 64.Roses A D. Ann NY Acad Sci. 1998;855:738–743. doi: 10.1111/j.1749-6632.1998.tb10653.x. [DOI] [PubMed] [Google Scholar]