Abstract
Context and Aims:
Rapid, accurate peripheral blood differentials are essential to maintain standards of patient care. CellaVision DM96 (CellaVision AB, Lund, Sweden) (CV) is an automated digital morphology and informatics system used to locate, pre-classify, store and transmit images of platelets, red and white blood cells to a trained technologist who confirms or edits CV cell classification. We assessed our experience with CV by evaluating sensitivity, specificity, positive predictive value and negative predictive value for CV in three different patient populations.
Materials and Methods:
We analyzed classification accuracy of CV for white blood cells, erythroblasts, platelets and artefacts over six months for three different university hospitals using CV.
Results:
CV classified 211,218 events for the adult cancer center; 51,699 events for the adult general hospital; and 8,009 events for the children's hospital with accuracy of CV being 93%, 87.3% and 95.4% respectively. Sensitivity and positive predictive value were <80% for immature granulocytes (band neutrophil, promyelocyte, myelocyte and metamyelocytes) (differences usually within one stage of maturation). Cell types comprising a lower frequency of the total events, including blasts, showed lower accuracy at some sites.
Conclusions:
The reduced immature granulocyte classification accuracy may be due in part to the subjectivity in classification of these cells, length of experience with the system and individual expertise of the technologist. Cells with low sensitivity and positive predictive value comprised a minority of the cells and should not significantly affect the technologist re-classification time. CV serves as a clinically useful instrument in performance of peripheral blood differentials.
Keywords: Accuracy, CellaVision, image analysis, peripheral blood
BACKGROUND
Our center is a major university hospital system comprised of multiple hospital sites. The adult Cancer Center serves as the primary diagnostic and treatment facility for the majority of adult hematology/oncology inpatients and outpatients. The major adult general hospital houses most surgical, trauma, transplant and medical subspecialties of the system. The children's hospital provides a broad range of medical, oncologic and surgical services to children in the area. The need for rapid and accurate peripheral blood differentials is essential to maintain standards of patient care and safety in these settings.
CellaVision DM96 (CellaVision AB, Lund, Sweden; distributed by Beckman Coulter, Inc., Brea, CA, USA and Sysmex Corporation, Kobe, Japan) (hereafter termed CV) is an automated digital morphology and informatics system used to locate, pre-classify, store, and transmit platelet, red blood cell, and white blood cell images to a trained technologist who confirms or edits CV cell classification. Detailed descriptions of the CV image analysis system have been previously given;[1–3] briefly, CV initially scans a Romanowsky-stained peripheral blood smear at 1000× magnification in the “zone of morphology” as defined by the relative density of the cells. It then takes digital images of each cell representing a possible white blood cell (WBC). Artificial neural network-based software is used to analyze the cells by comparing the acquired digital images to those in a reference library provided by the manufacturer. Cells are then pre-classified into 18 categories, including leukocytes (segmented neutrophils, band neutrophils, eosinophils, basophils, lymphocytes, monocytes, blasts, promyelocytes, myelocytes, metamyelocytes, variant lymphocytes, plasma cells and unidentified) and non-leukocytes (smudge cells, artifacts, giant platelets, nucleated red blood cells, platelet clumps). After analysis by CV, a technologist reviews these images and either agrees with the CV pre-classification, leaving the cells in the pre-classified categories, or disagrees with the instrument's determination and moves the cells into different categories. Once the technologist has reviewed all of the images and the differential is approved, it is subsequently released to the laboratory information system. Previous studies have shown that CV has a reproducibility of less than 2.5 standard deviations for all cell classes,[3] and that the overall time for the differential remains the same or decreases with the use of CV. The rate appears to vary by technologist, with more experienced technologists performing manual differential rates similar to those of CV differential rates; for less experienced technologists the CV differential rate is less than the manual differential rate.[1,4]
Timing studies previously performed at our institution demonstrated that a manual differential averaged 5.8 minutes, while the CV differential averaged 3.1 minutes. This translated to a reduction of 2.7 minutes per slide.[5] Theoretically, CV efficiency would increase as the need to edit classification (i.e. technologist time) decreases. The goal of the present study was to define the accuracy of CV peripheral blood differential as compared to the technologist review of the CV differential at three unique hospitals in our institution's medical system.
MATERIALS AND METHODS
Per laboratory standard operating procedures, peripheral blood specimens “flagged” by the automated cell counter for microscopic review were routed to CV for analysis. Separate manual differentials were not performed on these specimens. Peripheral blood smears were performed using premium glass slides with a clipped corner, which were Wright-Giemsa stained on the Beckman-Coulter LH750 slide maker stainer (Beckman Coulter, Inc., Brea, CA, USA) according to standard protocols at three unique hospitals in our institution's medical system. Each slide was then analyzed at 1000x under oil by CV, which acquired images of and classified each of the following cell types: promyelocyte, myelocyte, metamyelocyte, band neutrophil, segmented neutrophil, eosinophil, basophil, monocyte, lymphocyte, variant lymphocyte, plasma cell, blast cell, smudge cell, erythroblast (nRBC), giant platelet, and artefact. For each slide, CV analyzed 110 events and at least 100 leukocytes were ultimately counted; this allowed for non-leukocyte events to be removed if necessary.
A medical technologist or medical laboratory technician (the majority of whom have over 20 years of experience at our institution) then visually reviewed the CV image differential and reclassified cells as necessary; their interpretation served as the gold standard, or true values. When indicated, less-experience technologists operating CV were supervised by their senior counterparts. The final results were subsequently released to the laboratory information system. As these technologist reviews of CV images were part of the clinical service's standard operating procedure, no intra-person or inter-person validation was performed for the differentials.
Using CV's Database Query Tool, Version 4.2 (CellaVision AB, Lund, Sweden), we analyzed the classification accuracy of CV at each site for white blood cells, erythroblasts, platelets, and artefacts over a six month period. We assessed our experience with CV by evaluating the sensitivity and specificity of CV initial classification as well as the positive and negative predictive values of a cell type identified by CV. All analyses and statistical calculations were performed utilizing GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA).
RESULTS
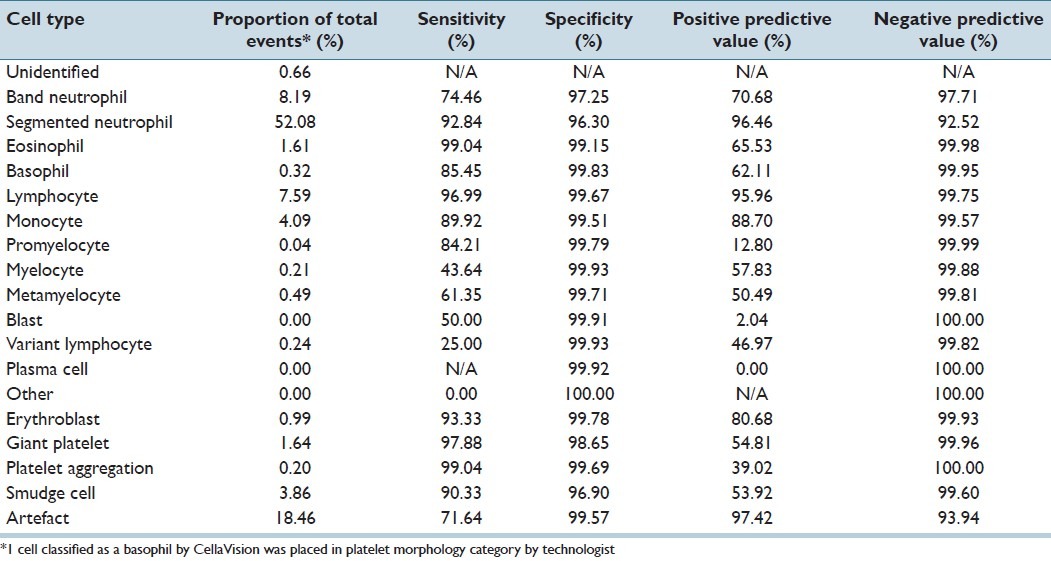
CV classified 211,218 events for the adult cancer center; 51,699 events for the adult general hospital; and 8,009 events for the children's hospital. For all events, the accuracy of CV was 93% for adult cancer center; 87.3% for adult general hospital; and 95.4% for the children's hospital. Tables 1–3 demonstrate the event collection and classification data and Tables 4–6 demonstrate all calculations and analyzes for all of the event collection and classification data for the adult cancer center, adult general hospital and children's hospitals respectively.
Table 1.
Adult cancer center event collection and classification data
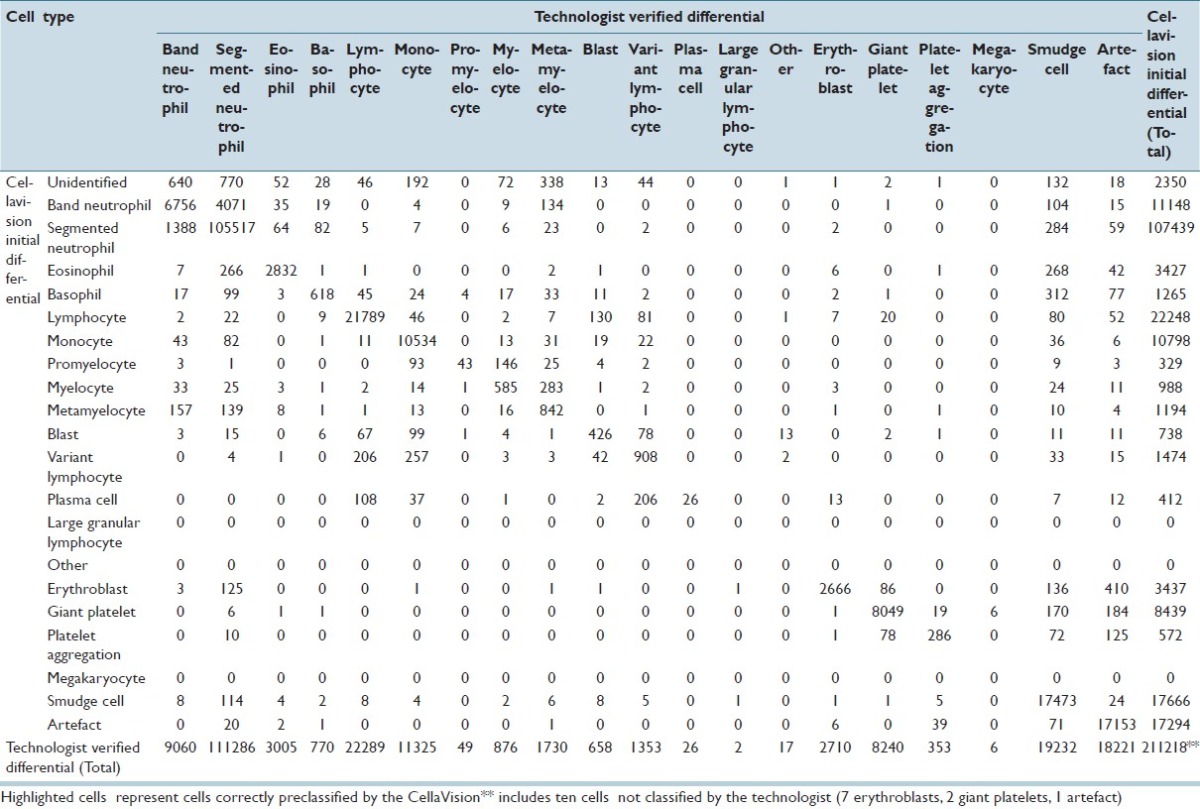
Table 3.
Children's hospital event collection and classification data
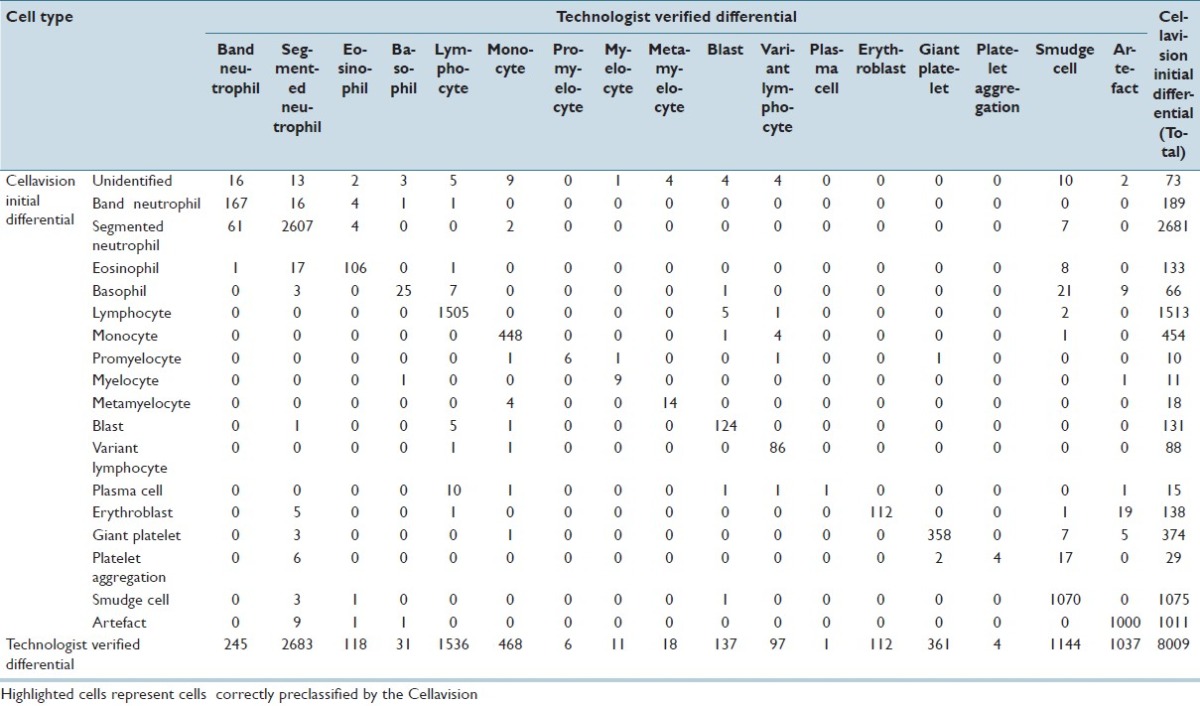
Table 4.
Adult cancer center calculations and analysis
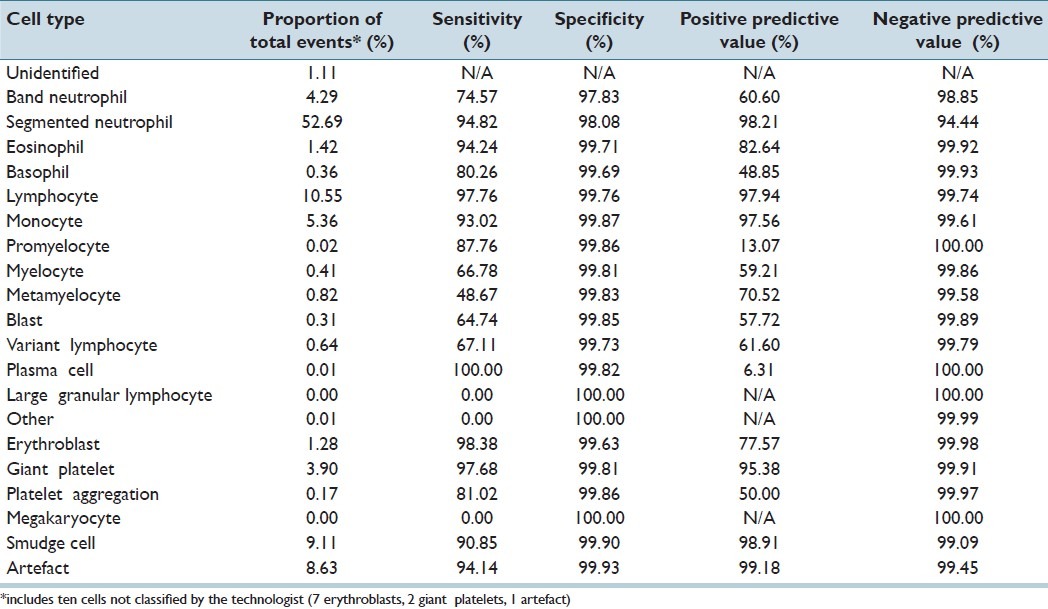
Table 6.
Children's hospital calculations and analysis
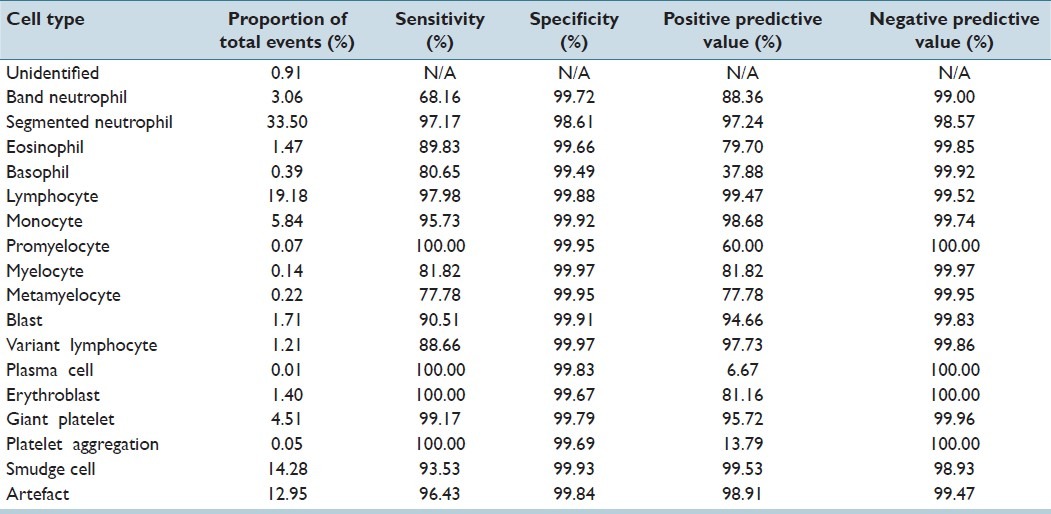
Table 2.
Adult general hospital event collection and classification data
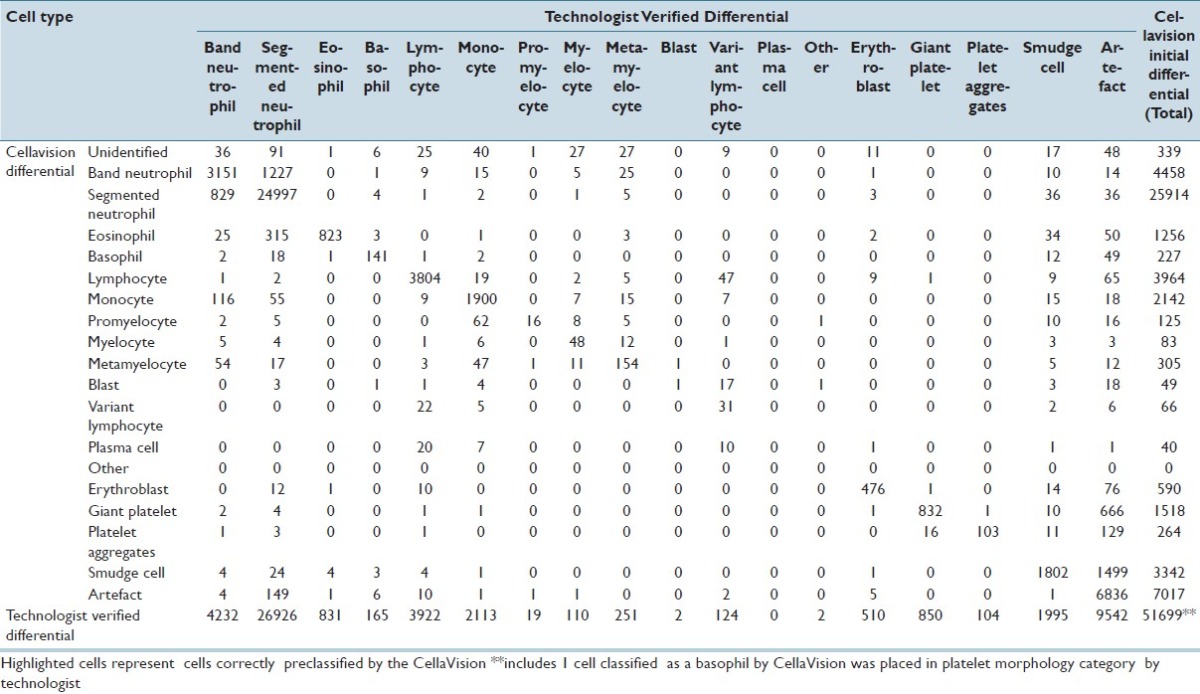
Table 5.
Adult general hospital calculations and analysis
At the adult cancer center, positive predictive values and sensitivities for CV were both >80% for segmented neutrophils, eosinophils, lymphocytes, monocytes, giant platelets, smudge cells, and artefacts. Positive predictive values and sensitivities were both <80% for immature granulocytes (band neutrophils, myelocytes, and metamyelocytes), variant lymphocytes, and blasts. The positive predictive values of CV for promyelocytes and plasma cells were low (13.1% and 6.3%); however, the sensitivities of CV identifying these two cell types were quite high (87.8% and 100%). Negative predictive values and specificities for all cell types were all >94%.
At the adult general hospital, positive predictive values and sensitivities for CV were >80% for segmented neutrophils, lymphocytes, monocytes, and erythroblasts. Similar to the adult cancer center's, positive predictive values and sensitivities were both <80% for immature granulocytes (band neutrophils, myelocytes, metamyelocytes), variant lymphocytes, and blasts. The positive predictive values of CV for blasts and promyelocytes were low (2.0% and 12.8%, respectively). The sensitivity of CV for promyelocytes was 84.2%, however the 50.0% sensitivity for blasts was much lower. Negative predictive values and specificities for all cell types were all >92%.
At the children's hospital, positive predictive values and sensitivities for CV were >80% for segmented neutrophils, lymphocytes, monocytes, myelocytes, blasts, variant lymphocytes, erythroblasts, giant platelets, smudge cells, and artefacts. Unlike the other two sites, both positive predictive value and sensitivity was only <80% for metamyelocytes (and even then only 77.8% and 77.8%). The positive predictive values of CV for plasma cells and platelet aggregation were 6.7% and 13.8%. Sensitivities of CV for these two cell types were 100%. Negative predictive values and specificities for all cell types were all >98%.
DISCUSSION
In this analysis of CV utilization at three different hospitals with distinct patient populations in a large academic hospital system that has been using CV image differentials for 5 years, the overall accuracy of CV in classifying white blood cells is excellent. It has been previously noted that the overall percentage of cells correctly identified by CV was between 82% and 92%,[1,3,6,7] and that specificity was >97% for all cell types;[7] these findings are consistent with the results at all three of our sites. Previous studies have also shown that the pre-classification agreement of CV for identifying segmented neutrophils, lymphocytes, and monocytes was between 81.4% and 99% due to their presence in large amounts.[1,6–8] This degree of accuracy was seen at our institution, as the positive predictive values and sensitivities of these three cell types in our analysis were all >89%. However, other cell types may be misclassified, perhaps due to either the low percentage of these cells in patient samples or to inter-individual morphologic variation. Cell types identified by CV with both low positive predictive values and sensitivities in our analysis comprised a minority of the cell types and should not significantly affect the technologist reclassification time. In analyzing the data, CV had difficulty in reliably identifying three specific cell types: immature granulocytes, plasma cells, and blasts.
Previous analysis of granulocyte precursors with CV have demonstrated less than optimal correlation with direct microscopy.[2,7,9] As this was also true in our analysis, reduced immature granulocyte classification accuracy may be due in part to the subjectivity in classification of these cell types, length of experience with the CV system, and individual expertise of the technologist. However, differences identified by the technologist were generally within one stage of maturation.
For plasma cells, a low event cell type that had low positive predictive values by CV at all three sites, the definition in CV likely needs refinement. However, at the two sites that actually had plasma cells remaining in that category after reclassification by the technologist, those remaining were all placed there by CV (100% sensitivity). This is significantly greater than previously reported 25% sensitivity of CV for identifying cell categories that included plasma cells.[1]
For blast identification in adult patients, the majority of events moved into this category by the technologist were from lymphocytes and lymphocyte variants, suggesting that the technologist identified lymphoblasts missed by CV. The events moved out went to lymphocytes, lymphocyte variants, monocytes, and artefacts, suggesting that CV overcalled some lymphoid forms as blasts as well as overcalled some monocytes and promonocytes as blasts. These are important but sometimes subtle distinctions that may be difficult to make even for an experienced hematopathologist. Interestingly, blast identification in the pediatric population had excellent positive predictive value and sensitivity (94.7% and 90.5%). This was an improvement over the findings of a previous CV study of pediatric WBC differentials that underestimated the percentage of malignant cells.[10] While the specific reason for the more accurate identification of blasts in pediatric patients is not known, it may be due to the narrower range of atypical hematopoietic cells that can resemble blasts in children.
It should be noted that CV can simultaneously display all cells of the same class on the viewing panel. The ability to compare cells head-to-head in real time can be valuable in assisting the technologist in the task of correctly identifying a specific cell.
Other methods of cell analysis and characterization such as flow cytometric studies, either alone or in conjunction with either hematology analyzers or high speed image capture, may further assist in the correct identification of difficult-to-classify cells.[11,12] However, algorithms of sequential analysis with CV followed by other analytic techniques have yet to be described.
The primary limitation of this study is the verification of CV differentials by different technologists. Cell types with poor positive predictive values and sensitivities warrant additional study as smaller group size or range, combined with limited technologist experience with CV technology as well as varying proficiencies, could have contributed to the lower identification accuracy by CV. However, in spite of having different technologists with varying levels of expertise and experiences with CV, overall accuracy of CV at all three labs remained high, thus demonstrating the positive assimilation of CV into the three different types of hematology laboratories. Correlation of CV results with specific disease diagnosis could not be performed in the current study due to de-identification of all data in the CV database prior to analysis. However, it would be interesting in future investigations to determine whether specific clinical conditions affect the ability of CV to accurately identify specific cell types.
Overall, CV serves as a clinically useful instrument in the performance of white blood cell differentials at our institution. Other potential benefits of CV include image storage of abnormal cells for review by pathologists or clinicians either on- or off-site, incorporation of images into patient electronic medical records and reports, and utilization as a teaching tool for technologists, pathology trainees, and clinicians. The potential for reduction of labor costs is an additional important consideration in the evaluation of this technology, especially in high volume laboratories which perform numerous manual differentials. As CV continues its role successfully in hematology laboratories and expands its reach beyond the pathology laboratory information systems, continued assessments will be required to ensure its accuracy and efficacy.
ACKNOWLEDGEMENTS
The authors would like to thank Betty Austin, MT (ASCP) at UPMC Presbyterian Hospital, Susan Waugaman, MT (ASCP) at UPMC-Shadyside Hospital, Marianne Riazzi, MLT (ASCP), MT at Children's Hospital of Pittsburgh of UPMC, and Emily Thompson, MT (ASCP), MSE at CellaVision.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2012/3/1/29/100154
REFERENCES
- 1.Kratz A, Bengtsson HI, Casey JE, Keefe JM, Beatrice GH, Grzybek DY, et al. Performance evaluation of the CellaVision DM96 system: WBC differentials by automated digital image analysis supported by an artificial neural network. Am J Clin Pathol. 2005;124:770–81. doi: 10.1309/XMB9-K0J4-1LHL-ATAY. [DOI] [PubMed] [Google Scholar]
- 2.Briggs C, Longair I, Slavik M, Thwaite K, Mills R, Thavaraja V, et al. Can automated blood film analysis replace the manual differential? An evaluation of the CellaVision DM96 automated image analysis system. Int J Lab Hematol. 2009;31:48–60. doi: 10.1111/j.1751-553X.2007.01002.x. [DOI] [PubMed] [Google Scholar]
- 3.Ceelie H, Dinkelaar RB, van Gelder W. Examination of peripheral blood films using automated microscopy; evaluation of Diffmaster Octavia and Cellavision DM96. J Clin Pathol. 2007;60:72–9. doi: 10.1136/jcp.2005.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornet E, Perol JP, Troussard X. Performance evaluation and relevance of the CellaVision DM96 system in routine analysis and in patients with malignant hematological diseases. Int J Lab Hematol. 2008;30:536–42. doi: 10.1111/j.1751-553X.2007.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contis L, Williams C. In: International Society for Laboratory Hematology. Amsterdam: 2006. Evaluation of an automated digital cell morphology and informatics system for use in a large hematology oncology center. [Google Scholar]
- 6.Seeto M, Ledesma A, Duff J, Houliston P, Lupscha M, Kabral A. In: International Society for Laboratory Hematology. Sydney: 2008. Evaluation of the CellaVision DM96 system: Familiarisation study in a large laboratory setting. [Google Scholar]
- 7.Francois M, Francoise T, Gothot A. In: International Society for Laboratory Hematology. Brighton: 2010. A performance evaluation of the CellaVision Dm96 for automated routine leucocyte differential in an academic hospital. [Google Scholar]
- 8.Brusselmans C. In: International Society for Laboratory Hematology. Las Vegas: 2009. Cellular analysis by digital microscopy. [Google Scholar]
- 9.Merino A, Brugues R, Garcia R, Kinder M, Bedini JL, Escolar G. In: International Society for Laboratory Hematology. New Orleans: 2011. Comparative study of peripheral blood morphology by conventional microscopy and Cellavision DM96 in hematological and non hematological diseases. [Google Scholar]
- 10.Billard M, Lainey E, Armoogum P, Alberti C, Fenneteau O, Da Costa L. Evaluation of the CellaVision DM automated microscope in pediatrics. Int J Lab Hematol. 2010;32:530–8. doi: 10.1111/j.1751-553X.2009.01219.x. [DOI] [PubMed] [Google Scholar]
- 11.Mirabelli P, Scalia G, Pascariello C, D’Alessio F, Mariotti E, Di Noto R, et al. ImageStream promyelocytic leukemia protein immunolocalization: In search of promyelocytic leukemia cells. Cytometry A. 2012;81:232–7. doi: 10.1002/cyto.a.22013. [DOI] [PubMed] [Google Scholar]
- 12.Kleine TO, Nebe CT, Lower C, Geilenkeuser WJ, Dorn-Beineke A. Cell analysis in cerebrospinal fluid (CSF) using Sysmex(R) hematology analyzers XT-4000i and XE-5000: Evaluation with CSF controls of the Joint German Society for Clinical Chemistry and Laboratory Medicine (DGKL) Cytometry A. 2012;81:255–64. doi: 10.1002/cyto.a.22014. [DOI] [PubMed] [Google Scholar]


