Abstract
The plant stomatal lineage manifests features common to many developmental contexts: precursor cells are chosen from an initially equivalent field of cells, undergo asymmetric and self-renewing divisions, communicate among themselves and respond to information from a distance. As we review here, the experimental accessibility of these epidermal lineages, particularly in Arabidopsis, has made stomata a conceptual and technical framework for the study of cell fate, stem cells, and cell polarity in plants.
Keywords: Stomatal development, Cell polarity, Intercellular communication
Introduction
Developmental models distill essential problems faced by cells, tissues and organisms into simplified and experimentally accessible systems. Stomata (the epidermal valves that mediate gas exchange between plants and the atmosphere) and the lineage from which they are derived have emerged as a pre-eminent model for answering questions about cell fate and pattern in plants. Stomata are present in all large land plants and are crucial for allowing the intake of atmospheric carbon dioxide (used for photosynthesis) while minimizing plant water loss. This balance is achieved both by physiological modulation of stomatal pore aperture and through developmental control over the proliferation and distribution of stomatal precursors (Nadeau and Sack, 2002a).
Stomata are distributed on the surfaces of above-ground organs and consist of two epidermally derived sister guard cells (GCs; see Glossary, Box 1) surrounding a pore that leads to an airspace in the mesophyll cell layers below. The making and patterning of stomata requires processes fundamental to developmental biology: cell fate specification, cell-cell communication, asymmetric and stem cell-like divisions, and the creation of cell polarity (Bergmann and Sack, 2007; Pillitteri and Torii, 2012). Because the epidermis is readily accessible for examination, and because a wealth of genetic and molecular tools is available for studying the model plant Arabidopsis thaliana, stomatal development provides an excellent system with which to study these fundamental developmental processes. In this Primer (see Box, Development: the big picture), we review the abundant molecular genetic data and highlight the broadly applicable regulatory themes that have emerged from studies of stomatal development.
Box 1. Glossary
- Entry/amplifying/spacing divisions.
Three distinct asymmetric divisions in the stomatal lineage generate one meristemoid and one SLGC as their daughters. The origin of the meristemoid mother cell (MMC) differs for each division type. Entry divisions initiate lineages and the MMC is derived from a protodermal cell. Amplifying divisions result from successive divisions of meristemoids. Spacing divisions are asymmetric divisions of an SLGC-derived MMC in which the newly formed meristemoid is oriented away from pre-existing stoma or stomatal precursors.
- Guard cell (GC).
Guard cells are specialized epidermal cells formed in functional pairs through a single, symmetric division of a GMC. A pair of guard cells can regulate gas and water exchange by controlling the aperture of the stomatal pore that they surround.
- Guard mother cell (GMC).
The immediate precursor cell to the stomatal guard cells. GMCs divide once, symmetrically, to produce the pair of guard cells that make up a single stoma.
- Hypocotyl.
The embryonically derived stem of a seedling.
- Meristemoid.
An early stomatal precursor cell that has limited self-renewing properties (regenerating itself and producing an SLGC through an asymmetric division). A meristemoid eventually differentiates into a GMC.
- Meristemoid mother cell (MMC).
The earliest stomatal lineage cell and precursor to the meristemoid and SLGC. An MMC also has limited self-renewing properties.
- Stomatal lineage ground cell (SLGC).
The larger daughter cell from an asymmetric division of either an MMC or a meristemoid; it can either differentiate into a pavement cell or become an MMC and found secondary or satellite lineages.
- Split-GFP assay.
This technique, also known as bimolecular fluorescence complementation (BiFC), is employed to study protein-protein interaction in vivo. Two proteins of interest are tagged independently with complementary fragments of a fluorescence protein, e.g. green fluorescent protein (GFP). Individually, the GFP fragments are not functional. However, if the two test proteins interact, the attached GFP fragments can reconstitute a functional fluorescent protein, which can be detected with a fluorescence microscope.
- Pavement cell.
The general epidermal cell type. Pavement cells possess thick cuticles and are lobed and interdigitated in the plane of the epidermis to create a water- and gas-impermeable barrier.
- Polarity switching.
A model for pattern formation in the stomatal lineage whereby cell division orientation and acquisition of stomatal precursor cell fate are guided by the divisional history of the cell. The model requires two components: an identity factor maintained in a single daughter after division, and a polarity factor for which asymmetric localization in a cell (a consequence of previous division orientations) dictates future division planes.
Development: the big picture
This Primer is part of a series entitled ‘Development: the big picture’. This series aims to highlight key developmental systems or processes that have been the subject of intense study because they have broad implications for other developmental, cell and molecular systems, or for disease and therapeutics. Keep an eye out for other articles in this series over the coming months!
An overview of stomatal development in Arabidopsis thaliana
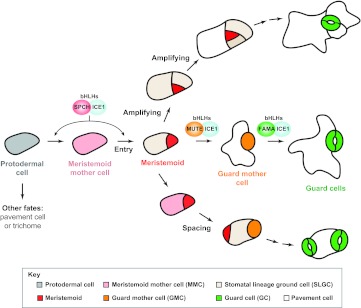
Plant leaves develop in an approximate tip-to-base gradient, with cell division prevalent at the base of the leaf and differentiation occurring near the tip. In Arabidopsis, stomatal production roughly follows this trend, but is ultimately organized by the behaviors of a specialized epidermal cell lineage (Nadeau and Sack, 2002a; Bergmann and Sack, 2007; Pillitteri and Torii, 2012). This stomatal lineage consists of five major cell types (Fig. 1): meristemoid mother cells (MMCs; see Glossary, Box 1), meristemoids (see Glossary, Box 1), stomatal lineage ground cells (SLGCs; see Glossary, Box 1), guard mother cells (GMCs; see Glossary, Box 1) and GCs. The lineage-founding MMCs are derived from a subset of protodermal cells, although the selection process is not well understood and might even be stochastic. MMCs undergo asymmetric divisions to produce small, often triangular-shaped, meristemoids and larger sister SLGCs. When arising from protodermal cells, these divisions are referred to as entry divisions (see Glossary, Box 1). Meristemoids have limited self-renewing properties (Box 2) and can carry out additional asymmetric divisions, termed amplifying divisions (see Glossary, Box 1), which regenerate the meristemoid and create another SLGC. Because these cells are defined by the daughters they produce, this renewing meristemoid is also an MMC. Meristemoids typically divide only a few times before differentiating into a GMC. GMCs, recognizable by their distinctive rounded morphology, undergo a single symmetric division and cell fate transition to form a pair of GCs, the terminal cells in the lineage. The fate of the SLGCs produced in the entry and amplifying divisions is complex; they may differentiate into pavement cells (interdigitated cells that provide the waterproof covering of plant leaves; see Glossary, Box 1), but may also divide asymmetrically to create secondary or satellite meristemoids. Although called the ‘stomatal lineage’, the ability of meristemoids and SLGCs to continue divisions means that this lineage is actually responsible for generating the majority of the epidermal cells in the leaves (Geisler et al., 2000). Developmental flexibility in response to the environment, or recovery from adverse conditions (for example, transient drought stress) is enabled by the behavior of this lineage (Skirycz et al., 2011).
Fig. 1.
Cell fate transitions and divisions during Arabidopsis stomatal development. A protodermal cell (gray) commits to the stomatal lineage when it becomes a meristemoid mother cell (MMC; pink). MMCs undergo an asymmetric division (Entry) and produce a smaller meristemoid (red) and a larger stomatal lineage ground cell (SLGC; beige). Meristemoids may undergo additional self-renewing asymmetric divisions (Amplifying) or differentiate into a guard mother cell (GMC; orange). A GMC divides symmetrically once to form a pair of guard cells (GCs; green). SLGCs have two options: they can differentiate into pavement cells (white), or they can adopt an MMC fate and undergo an asymmetric division (Spacing) to create a new secondary meristemoid that is positioned away from the existing stomatal lineage cell. Three related bHLH transcription factors (bHLHs), SPCH, MUTE and FAMA, form heterodimers with bHLH-LZs ICE1 or SCRM2 (not shown), and are necessary and sufficient for driving the production of meristemoid, GMC and GC fates, respectively. For simplicity, they are shown at only one position in this figure, but they are also required during cell fate transitions in the amplifying and spacing branches.
Box 2. Meristemoids versus stem cells
In plants, the continued generation of above-ground and below-ground organs and increase in girth are mediated by the activities of shoot apical meristems, root apical meristems and the vascular cambium, respectively. By the strictest definitions, these long-lived and pluripotent cell populations would be considered stem cells. But what of meristemoids and MMCs, cells with restricted developmental potential that nonetheless have the ability to recreate themselves as well as producing a sister of a different identity? These cells are more properly compared to adult stem cells of specific lineages, such as muscle satellite cells or subtypes of the hematopoietic systems.
In the literature, two terms used in the stem cell field, niche and transit-amplifying cell, appear in descriptions of stomatal lineage cells, but neither is a perfect fit. In the case of the niche, the term has been used to refer to geometric outcome of orienting cell divisions such that the stoma becomes surrounded by non-stomatal sister cells (Robinson et al., 2011). This usage appropriately describes the nest-like arrangement of cells that isolate a developing stoma from adjacent stomata. It is, however, potentially confusing in that, in contrast to a canonical niche, it is the result, not the source, of stem cell activity. MMCs and meristemoids have also been described as transient-amplifying cells (Nadeau and Sack, 2002a), but this term, traditionally referring to populations born from stem cells that proliferate (usually by symmetric divisions) before differentiating, inadequately captures the division potential of the stomatal system. MMCs and meristemoids may be transient, but their daughters produce several cell types each with fractal-like potential to divide again.
When considering stomatal pattern in the context of the entire developing leaf, it is striking that patterning mechanisms appear to be acting at the local level. Timelapse-enabled monitoring of the descendants of a single MMC reveals that some stomata create their own ‘niches’ by regulating amplifying division orientations to ensure that they are surrounded by non-stomatal cells (Robinson et al., 2011) (Box 2). On the same leaf, descendants of other MMCs may follow different trajectories. Commonly, correct patterning involves new meristemoids (derived from SLGC divisions) placed away from the existing stoma/precursors, and this requires neighbor cell signaling (Geisler et al., 2000) (discussed later). In contrast to well-studied animal epithelial systems, there is no evidence for leaf-wide morphogen gradients that organize stomatal development, and neither stomata nor the division axes of precursor cell divisions exhibit planar polarity.
Cell fate transitions
Two groups of bHLH transcription factors govern stomatal cell fate transitions
The flexible and multistep developmental pathway for stomata might suggest a need for complex cell fate regulatory programs. It was therefore somewhat surprising to find that the system could be explained to a large extent by the behavior of a handful of transcription factors (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008). Three closely related basic helix-loop-helix (bHLH) transcription factors (sharing 90% amino acid similarity in their bHLH domains and 40% similarity overall), SPEECHLESS (SPCH), MUTE and FAMA, are successively required for the transitions between the major cell types in the stomatal lineage (Fig. 1) (MacAlister et al., 2007; Pillitteri et al., 2007; Ohashi-Ito and Bergmann, 2006). SPCH drives MMC formation and the asymmetric entry division of these cells, as well as the subsequent asymmetric amplifying and spacing divisions (see Glossary, Box 1) that expand the lineage (MacAlister et al., 2007; Robinson et al., 2011; Pillitteri et al., 2007). MUTE terminates stem cell behavior by promoting the differentiation of meristemoids into GMCs (MacAlister et al., 2007; Pillitteri et al., 2007), and FAMA promotes the terminal cell division and differentiation of GMCs into GCs (Ohashi-Ito and Bergmann, 2006). Expression patterns and mutant phenotypes correspond to these roles: among stomatal lineage cells, SPCH is expressed only in MMCs and meristemoids, and in spch mutants, no stomatal lineage cells are initiated so the epidermis comprises only pavement cells. Ectopic expression of the bHLH transcription factors also provides valuable insights into the nature of their activities. Constitutive overexpression of MUTE produces an epidermis completely filled with stomata, including some cells that transdifferentiated from pavement cells (MacAlister et al., 2007; Pillitteri et al., 2007). MUTE cannot produce GCs in the absence of FAMA, but it can bypass the need for SPCH, indicating that early asymmetric divisions are not actually required for producing GC fates (MacAlister et al., 2007). Overexpression of FAMA can also produce ectopic GCs, but they are not properly paired, indicating a second role of FAMA in regulating cell division (Ohashi-Ito and Bergmann, 2006). Studies with the MYB transcription factors FOUR LIPS and MYB88 confirm the need for tight cell-cycle regulation during the GMC-to-GC transition (Xie et al., 2010). Overexpression of SPCH, in contrast to the results with MUTE and FAMA, does not produce extra stomata (MacAlister et al., 2007). This unexpectedly mild phenotype revealed two additional levels of regulation: first, the post-translational phosphoregulation of SPCH protein itself, and second, that precursor cell fates can be rerouted midway through the pathway (Lampard et al., 2008). The mechanisms responsible for these additional levels of control are discussed later.
By characterizing a gain-of-function mutant, the epidermis of which was composed entirely of stomata, two related bHLH-leucine zipper (bHLH-LZ) proteins, INDUCER OF CBF EXPRESSION 1 (ICE1) [also known as SCREAM (SCRM)] and SCRM2, were identified as the partners of SPCH, MUTE and FAMA (Kanaoka et al., 2008). Loss of both ICE1 and SCRM2 resembles spch mutants in that the epidermis lacks stomata. Importantly, ICE1 and SCRM2 are also able to interact physically with SPCH, MUTE and FAMA. Together, the genetic and biochemical data suggest that ICE1 and SCRM2 are required for the function of SPCH, MUTE and FAMA, and that they probably work as heterodimers in binding and regulating their downstream targets (Kanaoka et al., 2008). The gain-of-function ICE1 (SCRM-D) allele contains a single arginine to histidine change outside of the bHLH domain, and this change reportedly does not affect its DNA-binding ability (Chinnusamy et al., 2003; Kanaoka et al., 2008). This version of ICE1, however, does exhibit increased SPCH-binding activity in a split-GFP assay (see Glossary, Box 1), suggesting one mechanism for increased activity in the early part of the stomatal lineage.
Parallels between stomatal and muscle development
The successive use of related bHLH transcription factors in stomatal development bears a close resemblance to tissue specification and differentiation processes in animals, most notably Drosophila neurogenesis and mammalian myogenesis (Jan and Jan, 1993; Weintraub et al., 1991). During muscle development, the myogenic regulatory factors (MRFs), four closely related bHLH transcription factors, establish and promote differentiation of the myogenic lineage (Bentzinger et al., 2012; Olson and Klein, 1994). Two of the MRF members, Myf5 and MyoD, are expressed early in the lineage and commit cells to a myoblast fate. Later, myogenin and MRF4 (also known as Myf6) are expressed and drive the terminal differentiation of myotubes and myofibers. These tissue-specific MRFs heterodimerize with ubiquitously expressed bHLH partners to mediate their functions (Lassar et al., 1991). The parallels between this network and stomata are striking, including the partnership between stomatal lineage-specific SPCH, MUTE, FAMA and the more broadly expressed ICE1 and SCRM2. In addition, like the transdifferentiation of non-stomatal lineage cells induced by misexpression of the stomatal bHLH transcription factors, ectopic expression of MRFs can drive non-muscle cultures towards myogenic differentiation (Weintraub et al., 1991). Thus, in many contexts, bHLH transcription factors represent master regulators that are capable of reprogramming cell fate, probably through their placement at the top of an extensive gene regulatory network (Carvajal and Rigby, 2010; Hachez et al., 2011), and potentially through connections to chromatin modifications that maintain stable cell fates (De Falco et al., 2006; Borghi et al., 2010). The similarity in the use of bHLH transcription factors in stomatal and muscle development is a case of convergent evolution, because the last common ancestor of plants and animals was unicellular (Meyerowitz, 2002) and because the plant bHLH transcription factors are a distinct subfamily from that containing the myogenic and neurogenic factors. Given their similar regulatory logic, however, the stomatal bHLH transcription factors provide a powerful outgroup with which to test functions and capabilities of the bHLH transcription factors in general.
Inter- and intracellular signaling
Stomata are distributed across the leaf surface in diverse, species-specific and often beautiful patterns. Because plant cells do not move relative to one another during development, these patterns arise primarily through modulation of cell division numbers, timing and orientation. Arabidopsis stomata are patterned fairly minimally by the ‘one-celled spacing’ rule, which posits that stomata must have at least one pavement cell between them and that this spacing optimizes the efficiency of gas exchange (Geisler et al., 2000). To enforce this rule, existing stomata or stomatal lineage precursors must signal their neighbors to either (1) repress their adoption of the stomatal lineage fate or (2) as in spacing divisions, orient the newly formed secondary meristemoids away from them. As we discuss below, research in the past decade has shown that secreted protein ligands, cell surface receptors and intracellular mitogen-activated protein kinase (MAPK) signal transduction pathways are key elements in the cell-cell communication involved in stomatal patterning (Rowe and Bergmann, 2010; Pillitteri and Torii, 2012).
Membrane-bound receptor-like kinases (RLKs) regulate stomatal patterning
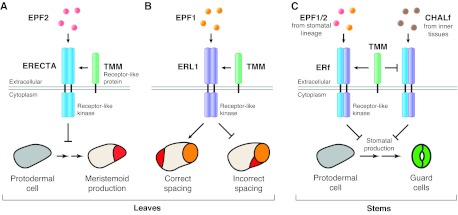
Leucine-rich repeat receptor-like kinases (LRR-RLKs) comprise a major class (>200 members) of plant receptors. Like animal Toll receptors or receptor tyrosine kinases, these LRR-RLKS are single-pass transmembrane proteins with an extracellular ligand-binding domain and an intracellular kinase domain for downstream signaling (Shiu and Bleecker, 2001). ERECTA, ERECTA-LIKE 1 (ERL1) and ERECTA-LIKE 2 (ERL2), together comprising the three-membered LRR-RLK ERECTA family (ERf), were first identified as regulators of plant organ growth, but were later found to control cell proliferation (Shpak et al., 2004) and the proper patterning and differentiation of stomata (Shpak et al., 2005). Detailed genetic analysis of ERf single, double and triple mutants and their dominant-negative versions (lacking kinase domains), together with differential expression patterns within the stomatal lineage, suggests some redundancy between these factors but also distinct roles for each RLK (Shpak et al., 2005; Lee et al., 2012). ERECTA, which is expressed strongly in the protodermal cells but is undetectable thereafter, restricts asymmetric entry division in MMCs (Fig. 2A). Supporting this notion, expression of a kinase-deleted, dominant-negative version of this protein in an erecta mutant background resulted in an increase in asymmetric entry divisions and the production of small cells (Lee et al., 2012). ERL1 and ERL2, which are highly expressed later in meristemoids, GMCs and young GCs, inhibit the differentiation of meristemoids into GMCs. This conclusion stems from the lack of SLGCs in the erl1 erl2 double mutant, which is an indication of a shortened time as a meristemoid (Shpak et al., 2005). ERL1 might also orient asymmetric spacing division, because in mutants expressing a kinase-deleted version of ERL1, stomata are often paired (Fig. 2B) (Lee et al., 2012). ERECTA and ERL1 have been shown to homo- and heterodimerize in vivo. It is not known, however, whether dimerization is important for function or whether it is dependent on ligand binding (Fig. 2).
Fig. 2.
Ligand-receptor interactions regulate stomatal production and patterning. (A) In leaves, the secreted peptide EPF2 (pink) is produced by MMCs and early meristemoids. EPF2 is detected by the receptor-like kinase ERECTA (blue), present in protodermal cells (gray). In partnership with the receptor-like protein TMM (green), the EPF2-ERECTA pair is hypothesized to activate an intracellular signaling cascade that represses production of meristemoids (red). (B) EPF1 (orange, top), which is secreted by late meristemoids, GMCs or GCs, interacts with ERL1 (purple). The EPF1-ERL1 pair, together with TMM, induces signaling that affects the division plane such that the secondary meristemoid (red) forms away from pre-existing stoma or stomatal precursors. Illustrated here is a GMC (orange, bottom), and the results of correct and incorrect spacing of a newly formed secondary meristemoid. The EPF1-ERL1 pair also represses meristemoid differentiation (not shown). (C) In stems, ERf receptors (blue/purple) are subject to inadvertent activation by the EPF-related CHALf peptides (brown), which are normally produced in inner tissues for growth regulation. Stomatal lineage expression of TMM functions as a signaling insulator, repressing CHALf-mediated and promoting EPF1/2-mediated signaling. In A and B, ERECTA and ERL1 are shown as homodimers, but they may also form heterodimers with other members of the ERf.
Stomatal RLKs form specific ligand-receptor pairs with members of the EPIDERMAL PATTERNING FACTOR LIKE (EPFL) family
The EPIDERMAL PATTERNING FACTOR LIKE (EPFL) family is a recently identified group of eleven secreted cysteine-rich peptides (bioactive forms are ~50 aa), and its characterized members have been shown to regulate stomatal development and plant growth (Hara et al., 2007; Hunt and Gray, 2009; Sugano et al., 2010; Abrash et al., 2011; Uchida et al., 2012). Two founding members of the family, EPF1 and EPF2, are stomatal lineage-specific factors and they repress stomatal development in specific stages corresponding to those during which ERL1 and ERECTA, respectively, function (Hara et al., 2007; Hunt and Gray, 2009; Hara et al., 2009). EPF2 is expressed in MMCs and early meristemoids, and inhibits cells from adopting the MMC fate. Similar to the dominant-negative phenotype of kinase-deleted ERECTA, loss-of-function mutants of EPF2 produce excessive numbers of meristemoid-like cells, whereas EPF2 overexpression leads to a loss of stomata (Hunt and Gray, 2009; Hara et al., 2009). EPF1 is expressed in late meristemoids and GMCs. Overexpression of EPF1 arrests the lineage at the meristemoid stage, and epf1 mutants, like plants expressing dominant-negative ERL1, produce clustered stomata, a phenotype associated with defects in orienting spacing divisions (Hara et al., 2007; Lee et al., 2012). Moreover, overexpression phenotypes of EPF1 and EPF2 require functional ERf members (Hara et al., 2007; Hara et al., 2009).
Biochemical evidence for ligand-receptor pairing came recently when EPF1 and EPF2 were shown to interact with both ERECTA and ERL1 in planta and on biosensor chips (Lee et al., 2012). The EPF2-ERECTA pair enables existing MMCs or meristemoids to restrict their neighbors from adopting the stomatal cell fate (Fig. 2A). Thus, this pair plays a role in regulating the number and density of stomata. The EPF1-ERL pair appears to enforce the one-celled spacing rule (Fig. 2B) and also helps control the number of stomata and pavement cells by restricting meristemoid differentiation, which indirectly regulates the amplifying divisions and the production of secondary meristemoids by SLGCs. Establishing the precise spatiotemporal expression patterns of receptors and ligands in the developing leaf is a crucial next step. Not only will this information be needed to understand the stoichiometry of signaling complexes, but it might help solve the longstanding mystery of whether meristemoids, which can express both EPF ligands and ERf receptors, signal in an autocrine fashion to regulate their own behaviors.
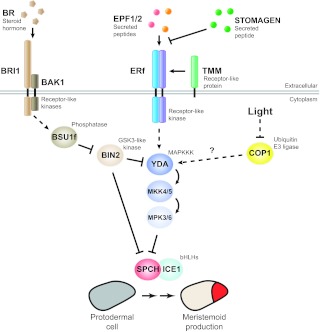
This simple picture of ligand-receptor pairs controlling specific stomatal stages becomes more complicated when a third ligand, STOMAGEN (EPFL9), is considered. STOMAGEN is expressed in the mesophyll tissue below the epidermis and, in contrast to EPF1 and EPF2, STOMAGEN promotes stomatal production (Sugano et al., 2010; Kondo et al., 2010). Nuclear magnetic resonance-based structural studies of the EPFLs indicate that they form a compact knot via their conserved cysteines. The unique regions of the EPFLs are surface-exposed and domain swaps between STOMAGEN and EPF1 indicate that functional specificity does indeed come from this region (Ohki et al., 2011). Whether STOMAGEN and EPF1/2 exert their antagonistic effects by competing for the same receptors or whether STOMAGEN sequesters EPF1 and EPF2 by forming unproductive dimers is not known (Fig. 3). That STOMAGEN is expressed outside of the stomatal lineage is intriguing and suggests that STOMAGEN could help coordinate stomatal production with the photosynthetic tissue below.
Fig. 3.
A mitogen-activated protein kinase (MAPK) pathway transduces and integrates intrinsic and environmental signals during stomatal production. A signal transaction cascade involving the MAPKKK YDA, MKK4/5 and MPK3/6 is employed to repress stomatal production. The EPFL-ERf-TMM module (including the antagonistic STOMAGEN) functioning within the stomatal lineage lies genetically upstream of YDA, activating it through an as yet unknown mechanism. The activated stomatal MAPK module can regulate stomatal development at multiple stages. Shown here is its repression of meristemoid production by MPK3/6 phosphorylation and downregulation of SPCH that is initiated upstream by the EPF2-ERECTA pair. The downstream targets of the MAPK pathway activated by the EPF1-ERL1 pair are not known. An intermediate signaling component in the brassinosteroid (BR) pathway, the GSK3-like kinase BIN2, interfaces with the stomatal development pathway at two levels: by phosphorylating and inhibiting YDA (thus increasing stomata) and by phosphorylating and repressing SPCH (thus decreasing stomata). Light also regulates the number of stomata through the ubiquitin E3 ligase COP1, a central repressor in light signal transduction. Genetic evidence has placed YDA downstream of COP1 but the nature of their molecular link is not known. Arrows indicate positive interactions, T-bars negative interactions. Solid lines indicate confirmed biochemical interactions and dashed lines are indirect or genetic interactions. BAK1, BRI1-ASSOCIATED RECEPTOR KINASE 1; BRI1, BR INSENSITIVE 1; BSU1f, family of BRI1-SUPPRESSOR 1.
A receptor-like protein (RLP) provides signaling specificity to stomatal RLKs
An LRR receptor-like protein (RLP), TOO MANY MOUTHS (TMM), is also required for stomatal patterning. TMM is expressed within the stomatal lineage and appears to provide specificity to the more widely expressed and functioning ERf (Yang and Sack, 1995; Nadeau and Sack, 2002b; Shpak et al., 2005; Lee et al., 2012) (Fig. 2A-C). RLPs lack a C-terminal kinase domain and, thus, are thought to be incapable of transducing signals on their own (Shiu and Bleecker, 2001). Like the ERf, TMM inhibits stomatal lineage proliferation and guides spacing divisions in leaves (Yang and Sack, 1995; Geisler et al., 2000). Furthermore, overexpression phenotypes of EPF1 and EPF2 in leaves depend on TMM (Shpak et al., 2005; Hara et al., 2007; Hara et al., 2009). Because TMM associates with both ERECTA and ERL1 in vivo, the formation of heterodimeric receptor complexes between TMM and the ERf might be required for the initiation of the EPF1/2-induced signal transduction cascade (Lee et al., 2012).
Although TMM promotes ERf-mediated signaling in leaves, it paradoxically functions to antagonize it in stems and hypocotyls (see Glossary, Box 1); tmm mutants produce no stomata in these organs (Yang and Sack, 1995; Bhave et al., 2009). Recent data reconcile these differences, indicating that TMM acts as a stomatal-lineage specificity factor for ERf signaling, promoting reception of EPF1/2 ligands and preventing the inappropriate activation of the ERf by internal EPFLs (Abrash and Bergmann, 2010; Abrash et al., 2011). EPFL4-6 [also known as the CHALLAH family (CHALf) ligands] form a distinct subgroup of EPFLs. CHALf members are highly expressed in internal tissues, especially in those of the hypocotyls and stems, and appear to function as growth (but not normally as stomatal) regulators through the ERf (Abrash et al., 2011; Uchida et al., 2012). Based on these results and on a ‘re-wiring’ experiment in which ligands and receptors were expressed in different spatial domains, it was shown that TMM functions as a stomatal lineage-specific insulator (Fig. 2C) (Abrash et al., 2011). Essentially, TMM promotes signaling of EPF1/2 ligands through ERf while suppressing the inadvertent activation of the stomatal lineage pool of ERf by CHALf ligands. This mechanism allows the use of the same set of receptors simultaneously for different developmental processes, while still maintaining signaling specificity.
A similar role, in which a co-receptor modulates differential responses to related ligands, is seen in the case of animal Cripto-1. Cripto-1, which is involved in embryogenesis and is expressed in many cancers, enhances signaling of some TGFβ superfamily ligands, such as Nodal, but represses others, such as activin (reviewed by Nagaoka et al., 2012). Because Cripto-1 signaling studies were performed in cultured cells, it will be interesting to examine whether Cripto-1 provides similar discrimination when in its native developmental contexts.
Peptide ligand-RLK signaling is common during plant development. Receptors and ligands, however, exist in large families, and the extensive processing of many ligands (for example, the CLE family has >40 members and the active forms are nearly identical dodecamers) (Katsir et al., 2011) has made it difficult to establish specific reagents and circumvent genetic redundancy. The relatively large size and sequence diversity of bioactive EPFLs (Rychel et al., 2010), new biochemical techniques to assay ligand and receptor interactions (Lee et al., 2012), and mounting evidence that EPFL/ERf signaling guides decisions outside of stomatal development (Abrash et al., 2011; Uchida et al., 2012) suggest that the ERf/EPFL system will be an effective and broad platform for mechanistic exploration of plant cell-cell communication.
Integrating intrinsic and environmental signals through the MAPK pathway
Intracellular transduction of the signaling initiated by the EPFL-ERf-TMM module is likely to be through a MAPK pathway (Bergmann et al., 2004; Wang et al., 2007; Lampard et al., 2009). Plant MAPK pathways, like their yeast and animal counterparts, are used to mediate developmental, stress and defense responses, and involve the sequential phosphorylation and activation of a three kinase module: the MAPK kinase kinases (MAPKKKs or MEKKs), the MAPK kinases (MKKs or MEKs) and the MAPKs (Rodriguez et al., 2010). In plants, the repertoire of kinases at each of these levels is greatly expanded relative to animals (e.g. there are >80 MAPKKKs). This expanded family offers opportunities to use this cascade in many processes, but creates significant problems with regards to cross-talk and achieving specificity. At least seven kinases can modulate stomatal development: the MAPKKK YODA (YDA), MKK4, MKK5, MKK7, MKK9, MPK3 and MPK6 (Fig. 3) (Bergmann et al., 2004; Wang et al., 2007; Lampard et al., 2009). Although, thus far, only genetic evidence exists to place the MAPK cascade downstream of ERf signaling, a biochemical connection was established between MAPKs and the fate-promoting bHLH transcription factors when MPK3 and MPK6 were shown to directly phosphorylate and repress SPCH (Lampard et al., 2008).
Besides interpreting intralineage signals for patterning, the stomatal lineage must also respond to systemic signals, such as hormones, and environmental signals, such as light and carbon dioxide, to optimize the number of stomata to the state of the plant and the environment. Recent studies on brassinosteroid (BR) and light highlight the MAPK cascade as an integration point for communicating these signals to stomatal production. BR is a plant steroid hormone that regulates diverse growth and developmental processes (reviewed by Kim and Wang, 2010). BR can also regulate stomatal production through its signaling intermediate, the glycogen synthase kinase 3 (GSK3)-like kinase, BR INSENSITIVE 2 (BIN2; Fig. 3), but not, interestingly, through its major transcriptional effectors (Kim et al., 2012). Inactivation of BIN2 suppresses stomatal production, and quadruple mutants of its direct upstream inhibitors (the BSU1 family of phosphatases) have an epidermis that is composed mostly of stomata, similar to yda mutants. Importantly, BIN2 was found to interact with, phosphorylate and inhibit YDA. Thus, BR negatively regulates stomatal development through the inactivation of BIN2, leading to de-repression of YDA (Kim et al., 2012). The GSK3 and MAPKKK phosphoregulation also represents a novel connection between the two widely used and conserved signaling cascades. BIN2 has also been found to phosphorylate and inactivate SPCH directly, a result that, by contrast, is consistent with BR promoting stomatal development (Gudesblat et al., 2012). This discrepancy between the stomatal promoting and repressing roles of BR could represent a fine-tuning mechanism for targeting SPCH during its successive rounds of activity (in entry versus amplifying or spacing divisions) and must be addressed in future studies.
YDA is also a point of integration between light signaling and stomata (Kang et al., 2009). Light is one of the environmental signals that dramatically affects the number and density of stomata (Casson and Hetherington, 2010). Light signals are perceived by multiple photoreceptors that converge to repress the RING E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) (reviewed by Lau and Deng, 2012; Jiao et al., 2007). Loss-of-function mutants of COP1, as well as mutants of other COP genes, such as COP10, produce stomatal clusters (Kang et al., 2009; Wei et al., 1994; Delgado et al., 2012). The clustering phenotype in the cop1 mutants can be suppressed by the expression of a constitutively active form of YDA, suggesting that COP1 functions upstream of YDA (Kang et al., 2009) (Fig. 3). Rigorous biochemical tests of the hypothesis that COP1 suppresses stomatal production through activating YDA, however, have not yet been reported.
Plants have remarkable sensitivity to environmental cues, and the magnitude of this response has been used to great advantage to dissect a number of universal regulatory mechanisms. For example, COP1 and the COP9 signalosome (CSN), both crucial for regulated protein turnover in both plants and animals, were first identified because they mediate plant environmental responses (Yi and Deng, 2005; Wei et al., 2008). Over both evolutionary and experimentally tractable timescales, stomatal production and pattern shows significant responses to the environment (Hetherington and Woodward, 2003). Because this developmental pathway is composed of simple, discrete, quantifiable steps, there is great potential to assay in situ the multifactorial inputs into development in a real, and changing, world.
Cell polarity
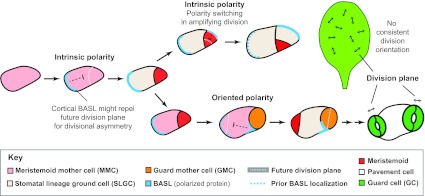
Positive-acting transcription factors and negative-acting signaling systems regulate the cell fate and renewal decisions in the stomatal lineage. After fate decisions are made, however, our understanding of the pathway controlling the execution of asymmetric and oriented cell divisions becomes murky. Moreover, these ‘decision-making’ elements cannot explain how asymmetric MMC divisions occur in a field of equivalent cells without reference to any apparent landmarks, nor do they explain the switching of cell polarity axes evident in self-renewing asymmetric divisions (Robinson et al., 2011) (Fig. 4). Plant genomes are devoid of recognizable homologs of animal or fungal cell polarity genes. This, combined with the mechanical constraints imposed by cell walls, dictates that plants must utilize alternative strategies to create cellular asymmetries.
Fig. 4.
Asymmetric cell division and division polarity of stomata in Arabidopsis. The asymmetric divisions of MMCs with and without influences from stomatal lineage neighbors are referred to as having intrinsic and oriented polarity, respectively. Polarity is seen in cell sizes and division orientations, but also in the localization of the cortical proteins BASL (blue) and POLAR (not shown, but following the same trajectory as BASL). Prior to MMC asymmetric division, BASL localizes at the cortex of the MMC in crescents on the side at which the future SLGC will form. Models predict that such localization could generate cell polarity if the polarized protein acts to repel the nucleus and subsequent division plane. During amplifying divisions (top pathway), self-renewing meristemoids reorient cortical crescents away from the most recently formed walls, leading to a stomatal progenitor surrounded by lineally related non-stomatal sisters (see also Fig. 1, top pathway). White dotted lines indicate the future division plane and blue dotted lines indicate the previous position of BASL. In SLGCs adopting MMC fates (lower pathway), BASL must redistribute within the same cell into a crescent abutting the pre-existing meristemoid or GMC. For simplicity, the nuclear localization of BASL is not indicated. Although divisions of MMCs are precisely oriented relative to immediate neighbors, the divisions of MMCs and GMCs do not appear to be oriented relative to any landmarks or axes of the leaf.
Asymmetric cell division regulators
The predictability of divisions and the accessibility of the epidermis have allowed a number of novel and plant-specific polarity proteins to be identified in the stomatal lineage, through genetic screens (for example, BASL) (Dong et al., 2009) and through transcriptional profiling approaches (for example, POLAR) (Pillitteri et al., 2011). In BASL null mutants, early stomatal lineage divisions often generate daughter cells similar in size, marker expression and identity (Dong et al., 2009). These incorrect early divisions lead to a hyperproliferation of self-renewing cells, a phenotype akin to the tumors resulting from loss of Drosophila neuroblast asymmetry regulators (Neumüller and Knoblich, 2009). BASL RNA and protein are expressed in dividing MMCs, meristemoids and SLGCs, with the protein exhibiting striking dynamic relocalization during repeated asymmetric divisions (Dong et al., 2009). Prior to a typical asymmetric division, BASL is initially nuclear, but as the cell ages and elongates, BASL begins to accumulate in a cortical crescent (Fig. 4). The position of this crescent is such that it is always inherited by the larger daughter. After division, both daughters possess a nuclear pool of BASL, so this is not a segregated factor per se, but only the larger daughter possesses BASL at the cortex. Structure-function studies of BASL indicate that the cortical pool BASL is necessary and sufficient for rescuing activity (Dong et al., 2009), but whether BASL activity is required before division (to define division axes) or after division to reinforce daughter cell differences, or both, is not yet known (Box 3). When expressed in non-stomatal lineage cells, BASL still localizes to the cortex in a polarized fashion, indicating that the ability to localize and maintain polarized domains of the cortex is not a unique property of an unusual cell type, but rather a general (but previously unrecognized) feature of most plant cells. POLAR is localized exclusively at the cortex of stomatal lineage cells and is also inherited preferentially by the larger daughter cell. No mutant phenotype has yet been described for POLAR, but this protein does appear to depend on BASL for its localization (Pillitteri et al., 2011). POLAR mislocalization in basl mutants could be due to a specific interaction with BASL, or because of cell identity defects; here, the incomplete penetrance of the basl phenotype might be revealing, as it has been reported that in the few correctly asymmetric cell divisions, POLAR is correctly localized (Pillitteri et al., 2011).
Box 3. Do asymmetric divisions and segregated fate determinants matter for plant development?
In plants, physically asymmetric cell divisions (ACDs) commonly correlate with the generation of new tissue layers and new cell types. Whether plants actually require the ACDs to produce cells of given identities and whether ACDs segregate fate determinants is still unresolved. Mutant plants in which nearly every cell division plane is misoriented still develop roots, shoots and (sterile) flowers, complete with appropriate cell types, arguing against a strict need for segregated fate determinants (Traas et al., 1995). Differential expression of transcription factors in the ACD daughters can be observed, but the best resolved temporal observations are consistent with differences in post-divisional protein stability that would account for this pattern (Nakajima et al., 2001; Robinson et al., 2011). In the stomatal lineage, expression of FAMA or MUTE can create GCs without asymmetric divisions (Ohashi-Ito and Bergmann, 2006; Pillitteri et al., 2007), and pavement cells (the other product of the lineage) can be made through alternative pathways.
With evidence against segregated determinants guiding plant cell fate, it is provocative that stomatal lineage cells nonetheless have mechanisms for polarizing and segregating cortical factors such as BASL and POLAR. Is there something special about this lineage? We hypothesize (also considering that BASL and POLAR homologs are undetectable outside of a subset of flowering plants), that the stomatal lineage has a unique requirement to create intrinsically asymmetric divisions. Bounded by rigid cell walls, most plant cells have an unchanging spatial relationship to their neighbors and could rely upon signaling (lateral inhibition or proximity to a source) to dictate fate and pattern. Early MMC division orientations, however, bear no obvious alignment to leaf axes or other landmarks. To reliably create cells of different behaviors in this situation requires the generation of intrinsically asymmetric divisions and, thus, additional machinery to do so.
Polarity switching and polarity factors
The phenomenon of polarity switching (see Glossary, Box 1) during rounds of asymmetric amplifying divisions, described developmentally by Sack and colleagues (Geisler et al., 2000; Bergmann and Sack, 2007) and formalized in models by Robinson et al. (Robinson et al., 2011) (Fig. 4), relies on a polarizing factor that can read division history (in other words, it must remember the orientations of previous divisions) and influence future divisions. This ‘polarizer’, over time, must be transient and able to relocate. Time-lapse imaging of BASL and/or POLAR is consistent with these proteins behaving as such factors. POLAR localization at the cell cortex is initially fairly uniform, but then condenses into a discrete crescent just before the mother cell undergoes an asymmetric cell division. When a cell switches polarity, the existing POLAR-GFP crescent delocalizes before reappearing as a crescent on the opposite cell side (Pillitteri et al., 2011). For BASL-GFP, nuclear sequestration appears to precede relocalization of the crescent to the opposite cell side (Dong et al., 2009). In both cases, continued asymmetric division potential correlates with the presence of the protein in a given cell, and differentiation into a pavement cell or stoma follows the disappearance of these reporters.
The polarized localization and loss-of-function phenotype of BASL make its functions potentially comparable to those of intrinsic polarity proteins or fate determinants in animal systems, both of which display polarized localization and are required for asymmetric fate specification in daughters. BASL bears a greater resemblance to polarity generators, such as the partition-defective/atypical protein kinase C (PAR/aPKC) complex, in that divisions lose physical as well as cell fate asymmetry in its absence (Suzuki and Ohno, 2006). POLAR function remains obscure in these divisions, but its coiled-coil domains suggest that it could act as a scaffold. What is clearly needed is a connection between these novel proteins of unknown function and biochemical or cellular processes. An interesting preliminary link comes from the observation of BASL and the RHO OF PLANTS (ROP) family GTPases. ROPs can organize the actin cytoskeleton and are major regulators of polarized outgrowths in the lobed pavement cells neighboring stomata (Fu et al., 2002). Overexpression of BASL can induce ectopic cellular outgrowths, but these outgrowths are suppressed in plants with compromised ROP signaling, suggesting that BASL could generate polarized domains via regulation of ROP localization or activity (Dong et al., 2009). ROP activity also guides localization of other polarized proteins (Nagawa et al., 2012). In plants expressing constitutively active or dominant-negative forms of ROP2, however, BASL crescents form and are correctly positioned (Dong et al., 2009); thus, as yet, there is no evidence for the reciprocal interplays that characterize other ROP-mediated polarization events.
Although there is still far to go, BASL and POLAR provide footholds from which to approach the generation of plant cell polarity. Previous studies of the PIN family of integral membrane auxin transporters (polarly localized in differentiated cells) led to numerous insights into the mechanisms plant cells use to generate subcellular differences (reviewed by Grunewald and Friml, 2010). As non-transmembrane proteins polarized during asymmetric cell divisions, BASL, POLAR and other stomatal lineage proteins provide a complementary set of probes for additional aspects of fundamental polarity-generating processes in plants.
Conclusions and perspectives
Stomatal development provides a framework for studying the fundamental processes of plants at different scales, from molecules, cells and tissues, up to whole plants and ecosystems. In this Primer, we have focused on exemplary modules for cell fate, polarity and pattern derived from studies of the reference plant Arabidopsis. Plants, as an outgroup to animals, have created many different strategies for life, but it is interesting to see some examples of striking conservation in the molecular and developmental solutions both groups have used for many developmental decisions. Looking forward, tools developed for the Arabidopsis stomatal lineage can be leveraged to obtain detailed information about stem cell-like developmental trajectories, especially as modulated by the environment. It is already possible to isolate pure populations of specialized plant cell types to obtain transcriptomic, proteomic and epigenomic information. In the future, it might be possible to generate models of stem cell behavior based on comprehensive profiles of individual stomatal lineage cells as they transit through their self-renewing divisions in an intact, environmentally responsive tissue.
As useful as stomata can be as a mirror for animal development, it is important not to ignore their essential role in photosynthesis and global climate cycles, a role that enabled a world in which humans can even exist to ponder developmental questions. Stomata are essential structures and have been found in plant fossils dating from 400 million years ago. Evidence that the genetic networks for cell fate and patterns described in Arabidopsis are conserved comes from comparative genomic studies (Peterson et al., 2010; Vatén and Bergmann, 2012) and functional studies in extant species of the basal plant lineages (MacAlister and Bergmann, 2011), as well as in plants with highly modified leaf and stomatal structures (Liu et al., 2009). An exciting future direction is to identify key genes that correlate with stomatal innovations across plant groups and to create plants with a wide range of stomatal alterations that would allow eco-physiologists to test models about stomata at the whole plant or whole planet level.
Acknowledgements
We thank members of our laboratory and the vibrant and interdisciplinary plant stomatal research community for data and discussions. We apologize to colleagues whose work could not be included owing to space constraints.
Footnotes
Funding
Work on stomatal development in the authors' laboratory was supported by the National Science Foundation and the National Institutes of Health. O.S.L. was supported by the Croucher Foundation and D.C.B. is a Gordon and Betty Moore Foundation Investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Abrash E. B., Bergmann D. C. (2010). Regional specification of stomatal production by the putative ligand CHALLAH. Development 137, 447-455 [DOI] [PubMed] [Google Scholar]
- Abrash E. B., Davies K. A., Bergmann D. C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23, 2864-2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger C. F., Wang Y. X., Rudnicki M. A. (2012). Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 4, a008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann D. C., Sack F. D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58, 163-181 [DOI] [PubMed] [Google Scholar]
- Bergmann D. C., Lukowitz W., Somerville C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494-1497 [DOI] [PubMed] [Google Scholar]
- Bhave N. S., Veley K. M., Nadeau J. A., Lucas J. R., Bhave S. L., Sack F. D. (2009). TOO MANY MOUTHS promotes cell fate progression in stomatal development of Arabidopsis stems. Planta 229, 357-367 [DOI] [PubMed] [Google Scholar]
- Borghi L., Gutzat R., Fütterer J., Laizet Y., Hennig L., Gruissem W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22, 1792-1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal J. J., Rigby P. W. J. (2010). Regulation of gene expression in vertebrate skeletal muscle. Exp. Cell Res. 316, 3014-3018 [DOI] [PubMed] [Google Scholar]
- Casson S. A., Hetherington A. M. (2010). Environmental regulation of stomatal development. Curr. Opin. Plant Biol. 13, 90-95 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.-H., Hong X., Agarwal M., Zhu J.-K. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco G., Comes F., Simone C. (2006). pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene 25, 5244-5249 [DOI] [PubMed] [Google Scholar]
- Delgado D., Ballesteros I., Torres-Contreras J., Mena M., Fenoll C. (2012). Dynamic analysis of epidermal cell divisions identifies specific roles for COP10 in Arabidopsis stomatal lineage development. Planta 236, 447-461 [DOI] [PubMed] [Google Scholar]
- Dong J., MacAlister C. A., Bergmann D. C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137, 1320-1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Li H., Yang Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14, 777-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., Nadeau J., Sack F. D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12, 2075-2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29, 2700-2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat G. E., Schneider-Pizoñ J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C., et al. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548-554 [DOI] [PubMed] [Google Scholar]
- Hachez C., Ohashi-Ito K., Dong J., Bergmann D. C. (2011). Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol. 155, 1458-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K. U., Bergmann D. C., Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720-1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K. M., Torii K. U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019-1031 [DOI] [PubMed] [Google Scholar]
- Hetherington A. M., Woodward F. I. (2003). The role of stomata in sensing and driving environmental change. Nature 424, 901-908 [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864-869 [DOI] [PubMed] [Google Scholar]
- Jan Y. N., Jan L. Y. (1993). HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell 75, 827-830 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Lau O. S., Deng X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217-230 [DOI] [PubMed] [Google Scholar]
- Kanaoka M. M., Pillitteri L. J., Fujii H., Yoshida Y., Bogenschutz N. L., Takabayashi J., Zhu J.-K., Torii K. U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20, 1775-1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.-Y., Lian H.-L., Wang F.-F., Huang J.-R., Yang H.-Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21, 2624-2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Davies K. A., Bergmann D. C., Laux T. (2011). Peptide signaling in plant development. Curr. Biol. 21, R356-R364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-W., Wang Z.-Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61, 681-704 [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Michniewicz M., Bergmann D. C., Wang Z.-Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kajita R., Miyazaki A., Hokoyama M., Nakamura-Miura T., Mizuno S., Masuda Y., Irie K., Tanaka Y., Takada S., et al. (2010). Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 51, 1-8 [DOI] [PubMed] [Google Scholar]
- Lampard G. R., MacAlister C. A., Bergmann D. C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113-1116 [DOI] [PubMed] [Google Scholar]
- Lampard G. R., Lukowitz W., Ellis B. E., Bergmann D. C. (2009). Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21, 3506-3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Davis R. L., Wright W. E., Kadesch T., Murre C., Voronova A., Baltimore D., Weintraub H. (1991). Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66, 305-315 [DOI] [PubMed] [Google Scholar]
- Lau O. S., Deng X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. doi:10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Lee J. S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M. M., McAbee J. M., Sarikaya M., Tamerler C., Torii K. U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26, 126-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K., Bergmann D. C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136, 2265-2276 [DOI] [PubMed] [Google Scholar]
- MacAlister C. A., Bergmann D. C. (2011). Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol. Dev. 13, 182-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister C. A., Ohashi-Ito K., Bergmann D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537-540 [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M. (2002). Plants compared to animals: the broadest comparative study of development. Science 295, 1482-1485 [DOI] [PubMed] [Google Scholar]
- Nadeau J. A., Sack F. D. (2002a). Stomatal development in Arabidopsis. The Arabidopsis Book 1, e0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. A., Sack F. D. (2002b). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697-1700 [DOI] [PubMed] [Google Scholar]
- Nagaoka T., Karasawa H., Castro N. P., Rangel M. C., Salomon D. S., Bianco C. (2012). An evolving web of signaling networks regulated by Cripto-1. Growth Factors 30, 13-21 [DOI] [PubMed] [Google Scholar]
- Nagawa S., Xu T., Lin D., Dhonukshe P., Zhang X., Friml J., Scheres B., Fu Y., Yang Z. (2012). ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 10, e1001299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P. N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307-311 [DOI] [PubMed] [Google Scholar]
- Neumüller R. A., Knoblich J. A. (2009). Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 23, 2675-2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493-2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki S., Takeuchi M., Mori M. (2011). The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2, 512 [DOI] [PubMed] [Google Scholar]
- Olson E. N., Klein W. H. (1994). bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8, 1-8 [DOI] [PubMed] [Google Scholar]
- Peterson K. M., Rychel A. L., Torii K. U. (2010). Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell 22, 296-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J., Torii K. U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63, 591-614 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Sloan D. B., Bogenschutz N. L., Torii K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501-505 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Peterson K. M., Horst R. J., Torii K. U. (2011). Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23, 3260-3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S., Barbier de Reuille P., Chan J., Bergmann D., Prusinkiewicz P., Coen E. (2011). Generation of spatial patterns through cell polarity switching. Science 333, 1436-1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M. H., Bergmann D. C. (2010). Complex signals for simple cells: the expanding ranks of signals and receptors guiding stomatal development. Curr. Opin. Plant Biol. 13, 548-555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychel A. L., Peterson K. M., Torii K. U. (2010). Plant twitter: ligands under 140 amino acids enforcing stomatal patterning. J. Plant Res. 123, 275-280 [DOI] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001, re22 [DOI] [PubMed] [Google Scholar]
- Shpak E. D., Berthiaume C. T., Hill E. J., Torii K. U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131, 1491-1501 [DOI] [PubMed] [Google Scholar]
- Shpak E. D., McAbee J. M., Pillitteri L. J., Torii K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290-293 [DOI] [PubMed] [Google Scholar]
- Skirycz A., Claeys H., De Bodt S., Oikawa A., Shinoda S., Andriankaja M., Maleux K., Eloy N. B., Coppens F., Yoo S. D., et al. (2011). Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23, 1876-1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Rodriguez M. C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621-649 [DOI] [PubMed] [Google Scholar]
- Sugano S. S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463, 241-244 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Ohno S. (2006). The PAR-aPKC system: lessons in polarity. J. Cell Sci. 119, 979-987 [DOI] [PubMed] [Google Scholar]
- Traas J., Bellini C., Nacry P., Kronenberger J., Bouchez D., Caboche M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676-677 [Google Scholar]
- Uchida N., Lee J. S., Horst R. J., Lai H.-H., Kajita R., Kakimoto T., Tasaka M., Torii K. U. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 109, 6337-6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A., Bergmann D. C. (2012). Mechanisms of stomatal development: an evolutionary view. EvoDevo 3, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J. C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Kwok S. F., von Arnim A. G., Lee A., McNellis T. W., Piekos B., Deng X.-W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6, 629-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Serino G., Deng X. W. (2008). The COP9 signalosome: more than a protease. Trends Biochem. Sci. 33, 592-600 [DOI] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S., et al. (1991). The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251, 761-766 [DOI] [PubMed] [Google Scholar]
- Xie Z., Lee E., Lucas J. R., Morohashi K., Li D., Murray J. A. H., Sack F. D., Grotewold E. (2010). Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22, 2306-2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Sack F. D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7, 2227-2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C., Deng X. W. (2005). COP1-from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618-625 [DOI] [PubMed] [Google Scholar]