Abstract
Various muscle diseases present with aberrant muscle cell morphologies characterized by smaller myofibers with mispositioned nuclei. The mechanisms that normally control these processes, whether they are linked, and their contribution to muscle weakness in disease, are not known. We examined the role of Dynein and Dynein-interacting proteins during Drosophila muscle development and found that several factors, including Dynein heavy chain, Dynein light chain and Partner of inscuteable, contribute to the regulation of both muscle length and myonuclear positioning. However, Lis1 contributes only to Dynein-dependent muscle length determination, whereas CLIP-190 and Glued contribute only to Dynein-dependent myonuclear positioning. Mechanistically, microtubule density at muscle poles is decreased in CLIP-190 mutants, suggesting that microtubule-cortex interactions facilitate myonuclear positioning. In Lis1 mutants, Dynein hyperaccumulates at the muscle poles with a sharper localization pattern, suggesting that retrograde trafficking contributes to muscle length. Both Lis1 and CLIP-190 act downstream of Dynein accumulation at the cortex, suggesting that they specify Dynein function within a single location. Finally, defects in muscle length or myonuclear positioning correlate with impaired muscle function in vivo, suggesting that both processes are essential for muscle function.
Keywords: Drosophila, Dynein, Nuclear positioning, Muscle size
INTRODUCTION
The size, shape and organization of tissues and individual cells directly impact their physiology. A pressing question in cell and developmental biology is how different morphogenetic processes that are necessary for the functional development of a common tissue are coordinated. Skeletal muscle provides an ideal system with which to study structure-function relationships as it is highly organized with a physiologically testable output.
The cellular unit of all metazoan muscle is the myofiber, a syncytial cell that is formed from the iterative fusion of myoblasts. The correlation between structure and function is highlighted by the organization of myosin and actin within the sarcomere that provides the contractile apparatus (Squire, 1997). However, other aspects of skeletal muscle structure, including myofiber size and organelle positioning, are also crucial for its function.
Myofiber size encompasses a variety of measurements, including length, width, depth and volume, and size is important for muscle output, as smaller myofibers provide less contractile force than larger muscles (Fitts et al., 1991). During muscle formation in Drosophila, muscle cell growth occurs in three distinct phases (Volk, 1999; Schnorrer and Dickson, 2004). As fusion begins, the muscle cell breaks symmetry, adds mass, and elongates to form polarized structures with a long axis and a short axis. Once the long axis is established, the muscle cell extends filopodia at each end in search of an attachment with a tendon cell. Finally, the filopodial protrusions stop, the muscle cell poles become smooth, and stable muscle-tendon attachments are made. In Drosophila embryos, muscle size has often been correlated with the number of fusion events (Bate, 1990), but it is unlikely that fusion alone determines myofiber size. In Drosophila larvae and in mammals, a number of signaling pathways have been demonstrated to control muscle size through growth regulation (Demontis and Perrimon, 2009; Rommel et al., 2001). Although several guidance cues and signaling mechanisms are known to regulate the direction of muscle growth (Schejter and Baylies, 2010; Schweitzer et al., 2010), the factors and mechanisms that directly provide the necessary forces that promote oriented and regulated muscle growth during early myogenesis are not known.
The importance of organelle positioning is highlighted by the localization of the myonuclei. In skeletal muscle, nuclei reside above the sarcomere at the periphery of the muscle fiber and are positioned to maximize their internuclear distance. Moreover, mispositioned myonuclei, in conjunction with smaller myofibers, are a hallmark of many muscle disorders and have been used diagnostically for decades (Pierson et al., 2007; Romero, 2010). The mechanisms by which nuclei are moved throughout the muscle (Metzger et al., 2012) and anchored at the neuromuscular junction (NMJ) (Grady et al., 2005; Lei et al., 2009) are beginning to emerge; however, only a small number of factors necessary for these processes have been identified. Additionally, potential links between muscle size and myonuclear position have not been explored. In other systems, the microtubule cytoskeleton regulates both polarized growth and nuclear position. Cytoplasmic Dynein (Dynein) is a crucial factor in both processes. During nuclear movement in the first cell division of C. elegans and in migrating neurons, cortically anchored Dynein pulls microtubule minus ends and the attached nuclei towards itself (Gönczy, 2002; Tsai et al., 2007). During polarized cell growth, Dynein contributes to the establishment of the polarity axis (Etienne-Manneville and Hall, 2001; Palazzo et al., 2001) that is necessary to localize the factors for cell protrusion (Prigozhina and Waterman-Storer, 2004; Schmoranzer et al., 2003). Given these functions, Dynein may contribute to both myofiber size and myonuclear positioning and might integrate the two processes.
To investigate how myofiber size and myonuclear positioning are regulated, and whether the two processes are co-dependent, we have studied the role of Dynein during muscle morphogenesis in Drosophila. We establish muscle length, which is an aspect of muscle size, and myonuclear positioning as independent aspects of muscle morphogenesis. We further show that the microtubule cytoskeleton contributes to both processes. Specifically, Dynein regulates muscle length and myonuclear positioning with overlapping, but distinct, sets of proteins. The Dynein heavy chain (Dhc64C), Dynein light chain (Dlc90F; Tctex1 in mammals) and Partner of inscuteable [Pins; Rapsynoid (Raps) – FlyBase] are necessary regulators of both muscle length and myonuclear positioning. Subsequently, the two pathways become distinct: muscle length is regulated by a Dynein-Lis1-dependent mechanism, whereas myonuclear positioning is regulated by a Dynein–CLIP-190–Glued-dependent mechanism.
Mechanistically, in mutant embryos that lack Dynein (Dhc64C4-19) or fail to localize Dynein to the muscle pole (raps193), both muscle length and myonuclear positioning are affected, suggesting that Dynein activity at the muscle pole is required for both processes. However, Dynein localizes to the muscle pole in both Lis1 and CLIP-190 mutant embryos, indicating that Dynein-dependent regulation of muscle length and Dynein-dependent regulation of myonuclear positioning are specified downstream of its arrival at the muscle pole. Lis1 mutant embryos exhibit hyperaccumulation of Dynein at the muscle pole with a tighter focus of peak intensity, suggesting that Lis1-dependent activation of Dynein retrograde trafficking is essential for muscles to achieve their proper length. CLIP-190 mutant embryos have fewer microtubules near the myofiber poles despite normal Dynein localization, suggesting that CLIP-190 promotes microtubule-cell cortex interactions that are necessary for Dynein-dependent myonuclear positioning. Together, these data suggest that both CLIP-190 and Lis1 work downstream of Dynein localization to the muscle pole and thus specify Dynein function within this single location. Differential regulation of Dynein activity within a single location is novel and likely to be important during morphogenetic processes for which proper timing is essential. Finally, both muscle length and myonuclear positioning are important for muscle function, as defects in either process correlate with larvae that crawl more slowly than controls.
MATERIALS AND METHODS
Drosophila genetics
Stocks were grown under standard conditions. Stocks used were apME-NLS::dsRed (Richardson et al., 2007), Dhc64C4-19 and Dhc64C6-10 (Gepner et al., 1996), UAS-Dhc64C-RNAi (Vienna Drosophila RNAi Center, v28053), dlc90F05089 (Caggese et al., 2001), lis1K11702 (Lei and Warrior, 2000), lis1G10.14 (Liu et al., 1999), UAS-Lis1-RNAi (Vienna Drosophila RNAi Center, v6216), clip190KG06490 (Bloomington Drosophila Stock Center, 14493), UAS-Clip190-RNAi (Vienna Drosophila RNAi Center, v107176), raps193 (Parmentier et al., 2000), inscP49 (Kraut and Campos-Ortega, 1996) and UAS-Glued-RNAi (Vienna Drosophila RNAi Center, v3785). Mutants were balanced and identified using CTG (CyO, twi-Gal4, UAS-2xeGFP), TTG (TM3, twi-Gal4, UAS-2xeGFP), TM6B, CyO P[w+wgen11lacZ] and TM3 Sb1 Dfd-lacZ.
Immunohistochemistry
Embryos were collected at 25°C and fixed with 4% EM-grade paraformaldehyde (Polysciences, 00380) diluted in PBS (Roche, 11666789001) and an equal volume of heptane to allow permeabilization. In all cases, embryos were devitellinized by vortexing in a 1:1 methanol:heptane solution. Larvae were dissected in ice-cold HL3.1 as previously described (Brent et al., 2009) and fixed with 10% formalin (Sigma, HT501128-4L). Embryos and larvae were mounted in ProLong Gold (Invitrogen) for fluorescent stainings. Antibodies were preabsorbed where noted (PA) and used at the following final dilutions: rabbit anti-dsRed (PA, 1:400, Clontech 632496), rat anti-Tropomyosin (PA, 1:500, Abcam ab50567), mouse anti-GFP (PA, 1:200, Clontech 632381), mouse anti-DHC (1:50, Developmental Studies Hybridoma Bank), mouse anti-Tubulin (1:500, Sigma T9026), chicken anti-β-galactosidase (PA, 1:1000, Novus Biologicals NB100-2045), mouse anti-β-PS-Integrin (1:100, Developmental Studies Hybridoma Bank) and mouse anti-Discs large (1:200, Developmental Studies Hybridoma Bank). We used Alexa Fluor 488-, Alexa Fluor 555- and Alexa Fluor 647-conjugated fluorescent secondary antibodies (1:200), Alexa Fluor 647-conjugated phalloidin (1:100) and Hoechst 33342 (1 μg/ml) for fluorescent stains (all Invitrogen). Fluorescent images were acquired on a Leica SP5 laser-scanning confocal microscope equipped with a 63× 1.4 NA HCX PL Apochromat oil objective and LAS AF 2.2 software. Images were processed using Adobe Photoshop CS4. Maximum intensity projections of confocal z-stacks were rendered using Volocity Visualization software (Improvision).
Nuclear position and muscle length measurements
Embryos were staged based on standard laboratory procedures: overall embryo shape, the intensity of the apRed and Tropomyosin signals, gut morphology, and the morphology of the trachea (Beckett and Baylies, 2007). Measurements were taken from confocal projections of embryos and acquired using the Line function of ImageJ software (NIH). For each genotype, all four lateral transverse (LT) muscles were measured in three hemisegments from each of 20 embryos from four independent experiments. Three measurements were taken within each LT muscle: (1) myotube length and (2, 3) the shortest distance between the myotube poles and the outermost edge of the nearest nucleus (supplementary material Fig. S1). The edges of the nuclei and ends of the myotubes were defined by the clear change from signal to background within the relevant viewing channel. Similar measurements were performed using β-PS-Integrin to identify the ends of the LT muscles. The statistical significance of differences in measurements was assessed using a Student's t-test to compare each experimental genotype with wild-type apRed controls. Statistical analysis was performed with Prism 4.0 (GraphPad).
Because the live stage 17 embryos offer few markers for accurate staging, staging was achieved by timed lays: after a pre-lay period, embryos were collected for 15 minutes and then aged for 16.5 hours. Embryos were then dechorionated and mounted on coverslips in halocarbon oil and images were acquired on a Zeiss Axioplan 2 microscope with a 20× PlanNeoFluor 0.50 NA objective and an Axiocam MRm camera. Nuclear spread measurements were performed using the Line function in ImageJ to measure the distance between the dorsal edge of the dorsal nucleus and the ventral edge of the ventral nucleus. Additionally, these same images of stage 17 embryos were used to count the number of nuclei per hemisegment, which was used as a measure of the number of myoblast fusion events.
Microtubule organization measurements
Confocal z-stacks were sequentially acquired to maximize the signal and resolution at the myofiber poles and to minimize interference from the internal muscles and the epidermis. Images were acquired with a 63× 1.4 NA HCX PL Apochromat oil objective and a 10× optical zoom. z-stacks through a depth of 2 μm with a step size of 0.25 μm were acquired for all analyzed images. z-stacks were then assembled into confocal projections using Volocity, which were then exported to ImageJ for analysis. Analysis was performed on 20 embryos from four independent experiments. To count the number of microtubules in contact with the muscle pole, a 2×2 μm box was drawn from the tip of the muscle toward the center and the number of microtubules in this region were counted.
To determine the average intensity, the Measure Average Intensity Function in ImageJ was performed on the distal 3 μm of the myofiber. The intensity of Tubulin staining was standardized against the intensity of Tropomyosin staining to control for sample variation. For each genotype, all four LT muscles were measured in three hemisegments from each of 20 embryos from four independent experiments. Student's t-test was used to compare each experimental genotype with wild-type apRed controls. Statistical analysis was performed with Prism 4.0.
Dynein localization measurements
Embryos were collected and fixed with 10% formalin diluted 1:1 in heptane for 20 minutes, then rinsed three times in PBS containing 0.6% Triton X-100, then fixed with 4% paraformaldehyde diluted 1:1 in heptane for 20 minutes. Antibody incubations were performed in PBS supplemented with 1% BSA and 0.6% Triton X-100. Image acquisition and processing were as for microtubule quantification. Dynein immunofluorescence intensity was assessed by linescan analysis using ImageJ. Positions of linescans were chosen randomly on the Tropomyosin image and the line then copied to the identical position on the Dynein image. The intensity of Dynein immunofluorescence at each point was measured relative to Tropomyosin intensity at that same point to control for sample variation, and the ratios were multiplied by 100. Three hemisegments from 20 embryos from four independent experiments were analyzed. The greatest pixel intensity from each embryo was used to determine the peak intensity. To determine the width of peak intensity, the pixels that were ≥80% of peak intensity were determined and the distance between the first pixel and the last pixel that fitted these criteria was measured. For each genotype, all four LT muscles were measured in three hemisegments from each of five embryos. Student's t-test was used to compare each experimental genotype with wild-type apRed controls. Statistical analysis was performed with Prism 4.0. The heat map representation of Dynein immunofluorescence was generated using the Cool application of the standard Lookup tools in ImageJ.
Larval behavior
Larval speed was assessed as previously described (Louis et al., 2008; Metzger et al., 2012) with minor modifications. Stage 16 and 17 embryos were selected for the absence of the fluorescent balancer and placed on yeast-coated apple juice agar plates at 22°C overnight. L1 larvae were selected the following day and placed into vials of standard fly food containing Bromophenol Blue (Bio-Rad, 161-0404). L3 larvae were picked from the vial 6 days later and individually tracked as they crawled towards an odor source of ethyl butyrate (3.3%; Sigma, E15701) diluted in paraffin oil (Fluka, 76235). Their movements were recorded with a CCD camera for either five total minutes or until they reached the odor source or the walls of the apparatus. The tracks were processed using Ethovision software (Nodlus). At least 20 larvae were tracked for each genotype. Tracked larvae were dissected, stained and analyzed as follows. Internuclear distance was measured in ImageJ by drawing lines between the center of each nucleus and its nearest neighbor using projection images. Muscle size was measured by determining the area of confocal projections of each muscle. To calculate the internuclear distance as a function of muscle size, the average internuclear distance for a given muscle was divided by the area of that specific muscle. Nine muscles from three larvae were used for quantification.
RESULTS
Dynein regulates muscle length and myonuclear position
Each body wall muscle in the Drosophila embryo and larva consists of a single syncytial myofiber similar to the individual myofibers in mammalian muscle. To assess the role of Dynein during Drosophila muscle morphogenesis, the distribution of nuclei within the lateral transverse (LT) muscles was examined in wild-type embryos and in embryos in which Dynein was zygotically removed with either the Dhc64C4-19 (null) or the Dhc64C6-10 (hypomorphic) allele (Gepner et al., 1996). The myonuclei in the LT muscles were visualized by expression of the apterous mesodermal enhancer fused to a nuclear localization signal and the fluorescent protein DsRed (apRed) (Richardson et al., 2007), which specifically labels the nuclei of the LT muscles. Using this reporter, live stage 17 embryos [16.5-20 hours after egg lay (AEL)] were examined. Dhc64C mutant embryos had the same number of apRed nuclei per hemisegment as controls, indicating that myoblast fusion was unaffected (Fig. 1A,B). Although the nuclei were aligned in columns within each LT muscle in embryos of each genotype, the distance between the most ventral and the most dorsal nuclei was reduced in Dhc64C mutant embryos (Fig. 1A,C). To determine whether the difference in the distribution of nuclei was caused by defective nuclear positioning or effects on muscle size, we examined late stage 16 embryos (16 hours AEL), which are amenable to whole-mount immunostaining.
Fig. 1.
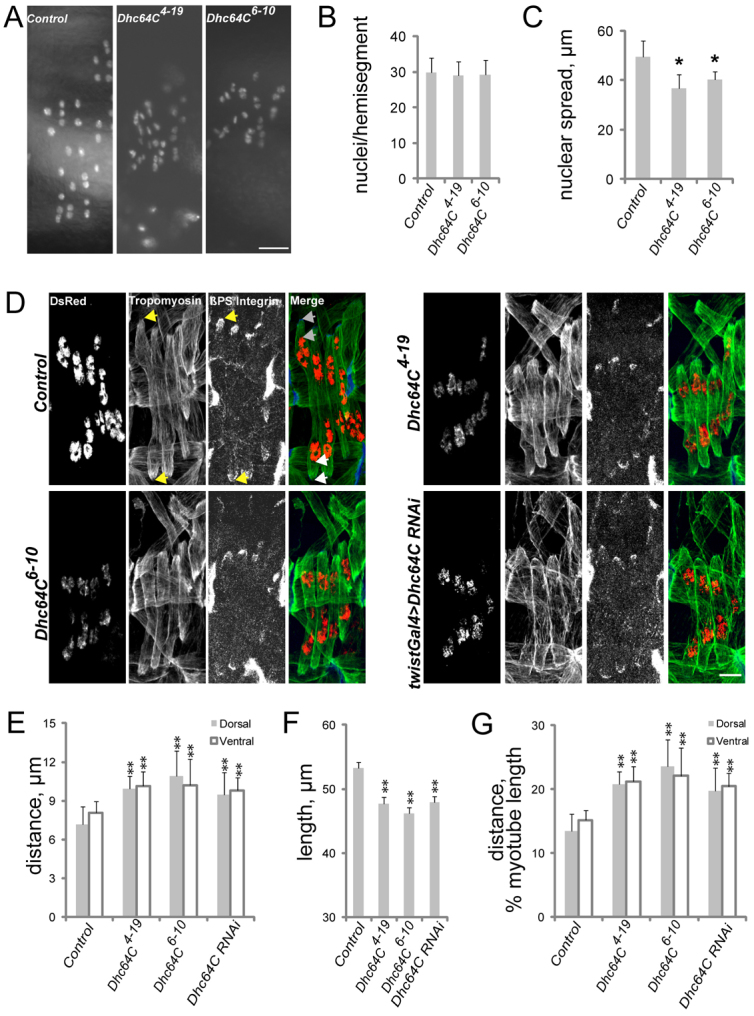
Cytoplasmic Dynein regulates muscle length and myonuclear position. (A) Fluorescence images of apRed nuclei in the lateral transverse (LT) muscles of live stage 17 Drosophila embryos (16.5-20 hours AEL) of the indicated genotypes. (B) The number of nuclei per hemisegment in stage 17 embryos of the indicated genotypes. (C) The distance between the dorsalmost and ventralmost nuclei in stage 17 embryos of the indicated genotypes. (D) Immunofluorescence images of stage 16 (16 hours AEL) embryos of the genotypes noted to the left and antigen listed at the top of the first image. Green, Tropomyosin (muscle); red, DsRed (nuclei); blue, β-PS-Integrin in merge. Arrows in the control panels denote points from which measurements were made for all genotypes. The distance between the points indicated by the yellow arrows in the Tropomyosin and β-PS-Integrin panels was used to determine muscle length. The distance between the pair of gray arrows at the top of the merged image was used to determine the distance between the nuclei and the dorsal pole. The distance between the pair of white arrows at the bottom of the merged image was used to determine the distance between the nuclei and the ventral pole. (E) The shortest distance between the indicated pole of the LT muscles (gray, dorsal; white, ventral) and the nearest cluster of nuclei in stage 16 embryos. (F) LT muscle length in stage 16 embryos. (G) The shortest distance between the indicated pole of the LT muscles (gray, dorsal; white, ventral) and the nearest cluster of nuclei normalized for muscle length in stage 16 embryos of the indicated genotypes. Error bars indicate s.d.; **P<0.01, *P<0.05. Scale bars: 10 μm.
The general muscle pattern as revealed by Tropomyosin staining (Fig. 1D) was normal in Dhc64C mutant embryos. Additionally, β-PS-Integrin staining was normal. Thus, all muscles were present, properly oriented and attached to tendon cells, indicating that specification, guidance and attachment mechanisms are functional. In late stage 16 (16 hours AEL) control embryos, the myonuclei were in two clusters within each LT muscle: one cluster at the dorsal pole and the other at the ventral pole (Fig. 1D; supplementary material Fig. S1) (Metzger et al., 2012). In embryos homozygous for either Dhc64C allele, the nuclei were also in two clusters; however, the clusters were mispositioned 50% further from the muscle poles than in wild-type and heterozygous controls (Fig. 1D,E; supplementary material Fig. S2). Additionally, the length of the LT muscles was reduced by 15% in embryos homozygous for either Dhc64C allele as compared with wild-type and heterozygous controls (Fig. 1D,F; supplementary material Fig. S2). To account for the difference in muscle length, the distance from each cluster of nuclei to the nearest muscle pole was calculated as a percentage of muscle length (Fig. 1G). This measurement was used with respect to all subsequent genotypes. To validate the use of Tropomyosin as a marker for muscle ends, we performed identical measurements using β-PS-Integrin to mark the muscle ends. These analyses produced identical results (Fig. 1D; supplementary material Fig. S2C,D).
To confirm that the effects of Dynein were autonomous to muscle, we used the GAL4/UAS system to deplete Dynein specifically in the mesoderm using twist-GAL4 to express UAS-Dhc64C-RNAi (Dietzl et al., 2007). Tissue-specific depletion of Dhc64C recapitulated the effects of the Dhc64C alleles: the LT muscles were 12% shorter than controls and the nuclei were positioned 40% further from the muscle poles than in controls (Fig. 1D-G). These data indicate that Dynein is specifically required in the muscle to generate muscles of proper length and to properly position myonuclei.
Given that myonuclear position seems to be affected similarly in stage 16 and stage 17 embryos, we examined embryos at earlier stages in their development to determine when the defects in muscle length and myonuclear positioning become evident. Using Tropomyosin to identify the muscles and a combination of trachea morphology and gut morphology to assess embryo age, we determined the length of muscles and the positioning of myonuclei in late stage 14 (11 hours AEL), stage 15 (13 hours AEL) and stage 16 (16 hours AEL) embryos (Fig. 2). At stage 14, the length of the muscles in control and Dhc64C4-19 embryos was similar. However, by stage 15 and through stage 16 the muscles in the Dhc64C4-19 embryos were significantly shorter (17% and 14%, respectively) than those in the control embryos.
Fig. 2.
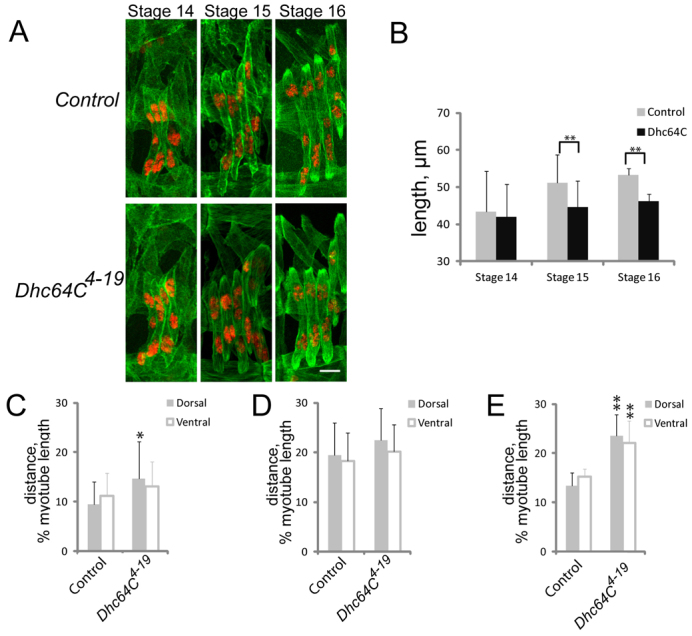
Stage-dependent effects of Dynein on muscle length and myonuclear positioning. (A) Immunofluorescence images of the muscles (green, Tropomyosin) and nuclei (red, DsRed) at stage 14, 15 and 16 in control and Dhc64C4-19 Drosophila embryos. Scale bar: 10 μm. (B) The length of the muscles in control (gray) and Dhc64C4-19 (black) embryos at stage 14, 15 and 16. (C-E) The distance between the nearest nucleus and either the dorsal (gray) or ventral (white) pole of the muscle in stage 14 (C), 15 (D) and 16 (E) embryos. Error bars indicate s.d.; *P<0.05, **P<0.01.
The effects on myonuclear positioning at the different stages were more complex. The nuclei were 30% further from the muscle poles in late stage 14 Dhc64C4-19 embryos than in controls (Fig. 2C); however, only the positioning relative to the dorsal muscle pole was statistically significant. At stage 15, myonuclei were similarly positioned in control and Dhc64C4-19 embryos (Fig. 2D). In stage 16 embryos, the myonuclei were significantly mispositioned (50% further from the poles) at both the dorsal and ventral poles of the muscle in Dhc64C4-19 mutant embryos relative to controls (Fig. 2E).
These data indicate that the defects in muscle length arise prior to muscle-tendon attachment and, together with the muscle-specific depletion of Dhc64C, suggest that the effects are muscle autonomous and not due to defects in tendon specification.
Dynein-interacting proteins contribute to Dynein-dependent regulation of muscle length and myonuclear positioning
To determine whether the roles of Dynein in specifying muscle length and myonuclear positioning are functionally coupled, we tested known Dynein-interacting proteins for their contributions to each process. We focused on factors that are general regulators of Dynein activity or have been found to specifically contribute to Dynein-mediated nuclear movements in other systems. We examined Dynein light chains, which can regulate Dynein activity both positively (Sönnichsen et al., 2005) and negatively (O'Rourke et al., 2007); Glued, which increases Dynein motor processivity and mediates Dynein-cargo interactions (Gill et al., 1991; Vaughan et al., 2002; Waterman-Storer et al., 1997); Pins, which anchors Dynein at the cortex to move nuclei in other systems (Gotta et al., 2003; Hughes et al., 2004); Lis1, which maintains Dynein in an active state and has been implicated in nuclear movements in yeast and neurons (McKenney et al., 2010; Mesngon et al., 2006; Sheeman et al., 2003); and CLIP-190, which works with Lis1 to regulate Dynein activity (Tanenbaum et al., 2008).
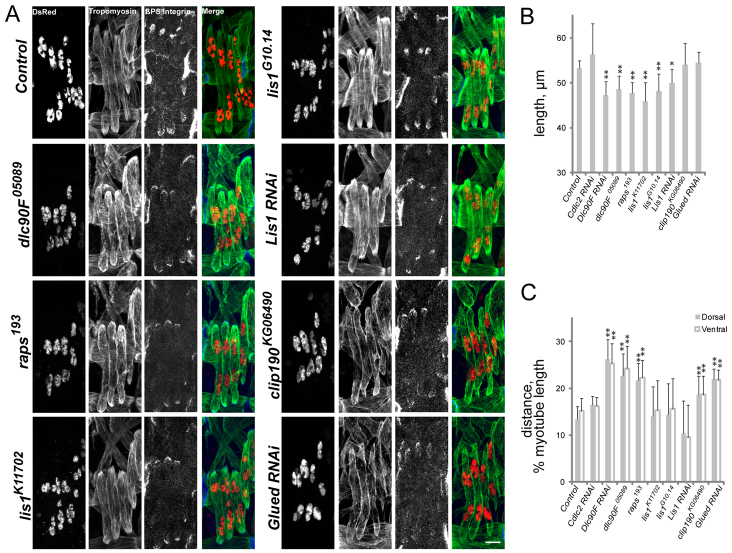
We tested two Dynein light chains, Cdlc2 and Dlc90F, which are the Drosophila orthologs of Dynein light chain 1/2 and Tctex1, respectively (Caggese et al., 2001; Goldstein and Gunawardena, 2000), by RNAi-mediated depletion (Dietzl et al., 2007). Depletion of Cdlc2 specifically in mesoderm and muscle did not affect muscle length or myonuclear positioning, whereas depletion of Dlc90F resulted in shorter muscles with mispositioned nuclei. Additionally, embryos that were homozygous for dlc90F05089 (Caggese et al., 2001) or the pins allele raps193 (Parmentier et al., 2000), produced muscles that were 12% shorter with nuclei that were 40-50% further from the muscle poles than in wild-type and heterozygous control embryos (Fig. 3A-C; supplementary material Fig. S2). These data indicate that Dlc90F and Pins contribute to both muscle length and myonuclear positioning.
Fig. 3.
Dynein-interacting proteins regulate myotube length and/or myonuclear position. (A) Immunofluorescence images of stage 16 Drosophila embryos of the indicated genotypes. Green, Tropomyosin; red, DsRed; blue, β-PS-Integrin in merge. The antigen for grayscale images is listed at the top of the first image. Scale bar: 10 μm. (B) The length of the LT muscles in stage 16 embryos. (C) The shortest distance between the indicated pole of the LT muscles (gray, dorsal; white, ventral) and the nearest cluster of nuclei normalized for muscle length. Error bars indicate s.d.; *P<0.05, **P<0.01.
Embryos homozygous for either of two Lis1 alleles, lis1K11702 (Lei and Warrior, 2000) and lis1G10.14 (Liu et al., 1999), produced muscles that were 15% shorter than those of wild-type and heterozygous controls. However, myonuclear positioning was normal in Lis1 homozygous embryos. Conversely, embryos that were depleted of Glued (Dietzl et al., 2007) in muscle and embryos that were homozygous for clip190KG06490 (Bellen et al., 2004) produced muscles of similar length to controls. However, in these embryos the nuclei were positioned 50% further from the muscle poles than in wild-type and heterozygous controls (Fig. 3A-C; supplementary material Fig. S2). The effects seen on myonuclear positioning in the clip190KG06490 homozygote were phenocopied in embryos that were transheterozygous for clip190KG06490 and each of three different deficiencies that removed CLIP-190: Df(2L)BSC294, Df(2L)Exel7068 and Df(2L)Exel8036 (supplementary material Fig. S2). Additionally, we measured the ventral longitudinal muscle VL3 and found that its length in the embryo was similar in each genetic background (supplementary material Fig. S2). These data suggest that a unique feature of the LT muscles makes them more susceptible to Dynein levels and/or activity with respect to muscle growth in the embryo.
The number and position of nuclei in stage 17 embryos were also examined. Embryos from each genotype had the same number of apRed nuclei per hemisegment, indicating that the defects in muscle length did not result from impaired fusion (Fig. 1B). The clusters of nuclei in each genotype did resolve into columns during stage 17, but the distance between the ventral and dorsal nuclei was reduced by 15% compared with controls. Moreover, the distance between the most distal nuclei was similar in stage 16 and stage 17 embryos for each genotype (supplementary material Fig. S3).
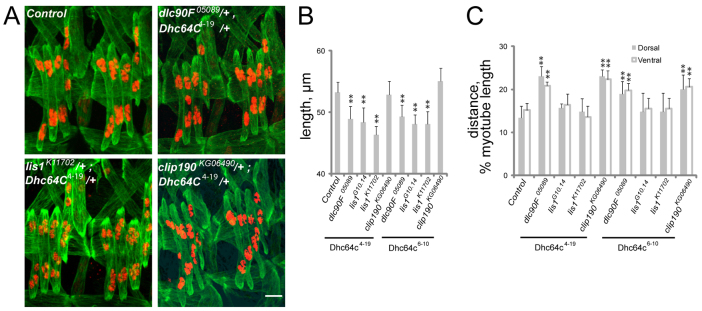
To determine whether the Dynein-interacting proteins work with, or independently of, Dynein, we examined embryos that were doubly heterozygous for Dhc64C mutations and mutations in each of the associated genes. In these, and all subsequent genetic interactions, maternal effects were controlled for by providing each allele from the mother or the father in reciprocal crosses. Both Dhc64C4-19 and Dhc64C6-10 were used in combination with alleles for each putative Dynein-interacting gene. Double heterozygotes of dlc90F05089 and Dhc64C4-19 or of dlc90F05089 and Dhc64C6-10 produced muscles that were 15% shorter than controls and with their nuclei 50% further from the muscle poles (Fig. 4A-C). These data indicate that Dlc90F (Tctex1) regulates Dynein activity for both muscle length and myonuclear positioning. All Lis1−/+; Dhc64C−/+ doubly heterozygous embryos produced muscles that were 15% shorter than those in control embryos, but with properly positioned myonuclei at the muscle poles (Fig. 4A-C). Conversely, CLIP-190−/+; Dhc64C−/+ doubly heterozygous embryos produced muscles that were the same length as those of control embryos but with nuclei 40% further from the muscle poles (Fig. 4A-C; supplementary material Fig. S3). Dhc64C−, +/+, raps193 doubly heterozygous embryos and embryos depleted of Glued in the Dhc64C−/+ background died prior to stage 16 and could not be evaluated. These data indicate that Lis1 and CLIP-190 regulate muscle length and myonuclear position, respectively, via interactions with Dynein, and not by Dynein-independent mechanisms.
Fig. 4.
Interactions between Dynein and associated genes during muscle development. (A) Immunofluorescence images of stage 16 Drosophila embryos that are doubly heterozygous for Dhc64C4-19 and the indicated allele. Green, Tropomyosin; red, DsRed. Scale bar: 10 μm. (B) LT muscle length in stage 16 embryos that are doubly heterozygous for the Dhc64C allele shown beneath the histogram and the listed alleles. (C) The shortest distance between the indicated pole of the LT muscles (gray, dorsal; white, ventral) and the nearest cluster of nuclei normalized for muscle length in stage 16 embryos that are doubly heterozygous for the Dhc64C allele indicated beneath the histogram and the listed alleles. Error bars indicate s.d.; **P<0.01.
The two Dynein-dependent pathways are separable
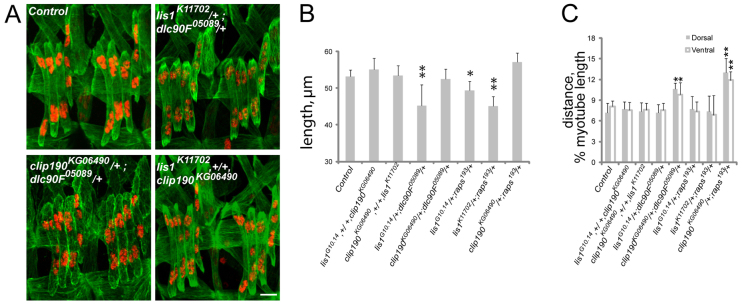
To confirm that Dynein-dependent myonuclear positioning and Dynein-dependent determination of muscle length are regulated by separate mechanisms, we examined the interactions between Dynein regulatory proteins. Both Lis1 and CLIP-190 functionally interacted with dlc90F05089, and these interactions recapitulated their interactions with Dhc64C: lis1K11702/+; dlc90F05089/+ and lis1G10.14/+; dlc90F05089/+ double heterozygotes had muscles that were 20% shorter than those of control embryos, whereas clip190KG06490/+; dlc90F05089/+ double heterozygotes had muscles in which the nuclei were mispositioned, 30% further from the muscle poles compared with controls. Similarly, pins functionally interacted with both Lis1 and CLIP-190: lis1K11702/+; raps193/+ double heterozygotes had muscles that were 20% shorter than those of control embryos and clip190KG06490/+; raps193/+ double heterozygotes had normal sized muscles with mispositioned myonuclei that were 60% further from the muscle poles compared with controls (Fig. 5A-C; supplementary material Fig. S3). The interaction of dlc90F05089 with pins was also examined but resulted in embryonic death prior to stage 16 and could not be evaluated.
Fig. 5.
The pathways controlling muscle length and myonuclear position do not genetically interact. (A) Immunofluorescence images of stage 16 Drosophila embryos that are doubly heterozygous for the indicated alleles. Green, Tropomyosin; red, DsRed. Scale bar: 10 μm. (B) LT muscle length in stage 16 embryos that are doubly heterozygous for the indicated alleles. (C) The shortest distance between the indicated pole of the LT muscles (gray, dorsal; white, ventral) and the nearest cluster of nuclei in stage 16 embryos normalized for muscle length. Error bars indicate s.d.; *P<0.05, **P<0.01.
Lis1 contributes only to muscle length, whereas CLIP-190 contributes solely to myonuclear positioning. We tested whether Lis1 functionally interacts with CLIP-190. Lis1−, +/+, clip190KG06490 double heterozygotes were similar to control embryos in both muscle length and myonuclear positioning. This indicates that Dynein-dependent muscle length and Dynein-dependent myonuclear positioning are indeed mechanistically distinct processes (Fig. 5A-C; supplementary material Fig. S3).
Lis1 affects Dynein localization and CLIP-190 affects microtubule-cortex interactions
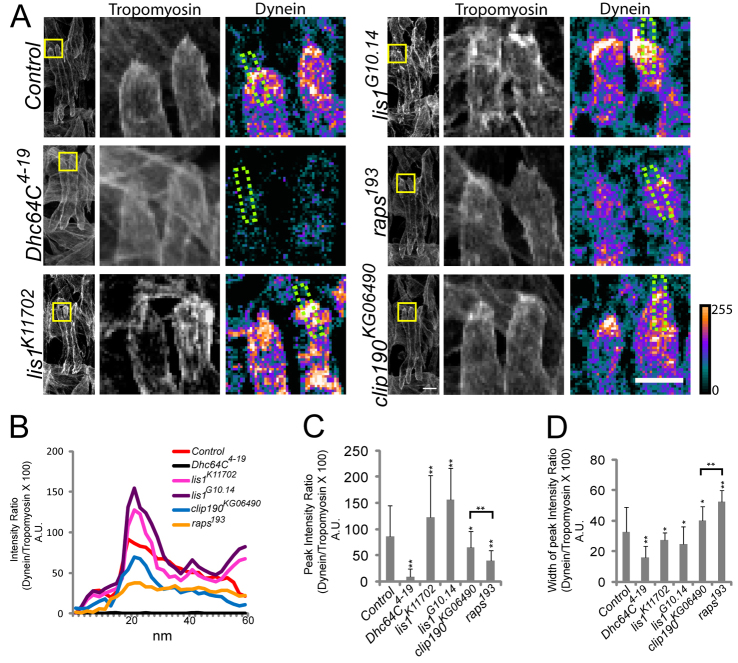
We next examined the effects of Dhc64C, Lis1, CLIP-190 and raps mutants on Dynein localization (Fig. 6A) and microtubule organization (Fig. 7A) to understand how Dynein differentially regulates muscle length and myonuclear positioning. Using confocal projections, linescans of Dynein immunofluorescence intensity (Fig. 6B) were employed to measure Dynein localization. To control for sample variation, the intensity of Dynein immunofluorescence was measured relative to the intensity of Tropomyosin immunofluorescence at the same point. Both the peak of Dynein immunofluorescence (Fig. 6C) and width of peak immunofluorescence intensity (Fig. 6D) were quantified. clip190KG06490 did not significantly affect Dynein localization. Lis1 and raps mutants did affect Dynein localization, although differently. In raps193 mutant embryos, the peak intensity of Dynein immunofluorescence was decreased compared with controls, whereas the width of peak intensity was increased (Fig. 6A-D), suggesting that Pins is required for Dynein localization to the muscle poles. In Lis1 mutant embryos, the peak intensity of Dynein immunofluorescence was increased compared with controls and the width of peak intensity was slightly decreased (Fig. 6A-D). The higher peak intensity combined with the sharper localization suggested that Lis1 does not contribute to the initial localization of Dynein to the muscle pole, but rather to the retrograde trafficking of Dynein away from the muscle pole. Together, these data indicate that proper muscle length requires Dynein localization to the muscle pole, and that raps193 results in shorter muscles due to poor accumulation of Dynein at the myofiber pole. Additionally, tightly focused hyperaccumulation of Dynein, as seen in Lis1 mutant embryos, also affects muscle length, suggesting that trafficking of Dynein away from the myofiber pole is mediated by Lis1 and is necessary to generate muscles of the proper length.
Fig. 6.
Dynein hyperaccumulates in Lis1 mutant embryos. (A) Confocal projections of a single hemisegment from stage 16 Drosophila embryos immunostained for Tropomyosin (green) and Dynein heavy chain (red). The boxed regions are shown at higher magnification to the right, with Tropomyosin shown in grayscale and Dynein shown as a heat map (the scale indicates relative intensity). The regions outlined by the green dotted lines were used for linescan analysis. Scale bars: left-hand panel, 5 μm; right-hand panel, 10 μm. (B) Representative linescan analysis indicating the intensity of Dynein heavy chain immunofluorescence as a function of position across the muscle pole. (C) The peak intensity signal for Dynein heavy chain immunofluorescence in the indicated genotypes. (D) The width of the peak intensity of Dynein heavy chain immunofluorescence. Error bars indicate s.d.; *P<0.05, **P<0.01, compared with control.
Fig. 7.
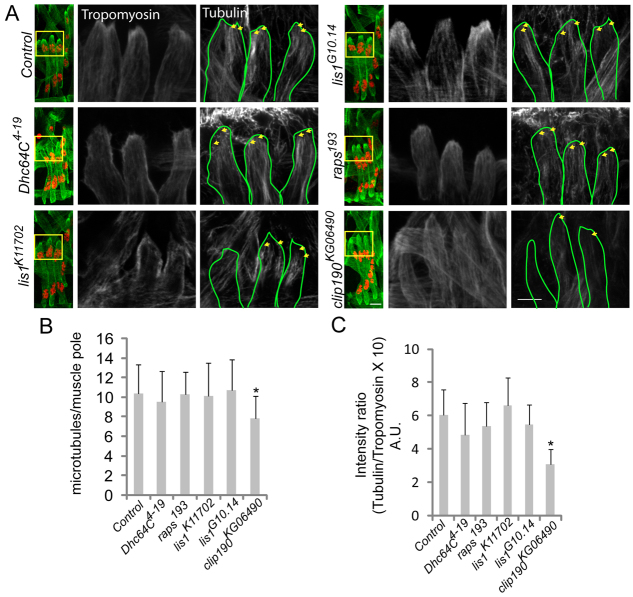
Microtubule organization is disrupted in CLIP-190 mutant embryos. (A) Confocal projections of a single hemisegment from stage 16 Drosophila embryos immunostained for Tropomyosin (green) and Tubulin (grayscale). The boxed regions are shown at higher magnification to the right. Yellow arrows indicate individual and/or bundles of microtubules that reach the muscle pole. Scale bars: 10 μm. (B) The number of microtubules within 3 μm of the myofiber pole in the indicated genotypes. (C) The average intensity of Tubulin immunofluorescence in the 3 μm near the myotube pole in the indicated genotypes. Error bars represent s.d.; *P<0.05, compared with control.
Similarly, confocal projections of embryos immunostained for Tubulin and Tropomyosin were used to assess the presence of microtubules near myofiber poles (Fig. 7A). Two different measures – the number of microtubules in contact with the myofiber pole (Fig. 7B) and the average intensity of tubulin immunofluorescence in the distal 3 μm of the myofiber (Fig. 7C) – indicated that only clip190KG06490 homozygotes had significantly fewer microtubules near the muscle pole than all other genotypes. These data suggest that CLIP-190 is necessary to mediate interactions between microtubules and the myofiber cortex that are then utilized by Dynein to move nuclei towards the muscle pole.
Defects in muscle size and myonuclear position impair muscle function
Larval crawling towards an odorant stimulus was used to assess the importance of Dynein-dependent muscle size regulation and Dynein-dependent myonuclear positioning to muscle physiology. Each genotype tested crawled towards the stimulus, indicating that all genotypes perceived and responded to the stimulus. However, L3 larvae that were homozygous for clip190KG06940 or lis1G10.14 crawled towards the stimulus 33% more slowly than controls (Fig. 8A). Dhc64C4-19, Dhc64C6-10 and lis1K11702 were early larval lethal, thus their locomotive ability could not be tested. However, when Dhc64C was depleted during embryonic and larval development by driving the UAS-Dhc64C-RNAi construct with the muscle-specific driver DMef2-Gal4, these larvae also crawled towards the stimulus 40% more slowly than controls (Fig. 8A,B). Additionally, muscle-specific depletion of CLIP-190 and Lis1 inhibited larval locomotion by 20% (Fig. 8A,B). That the depletion was muscle specific confirms that the effects on crawling are muscle autonomous. Furthermore, the NMJ, as measured by bouton number and distribution, was similar to that of the control in each of the genotypes tested (supplementary material Fig. S4). Finally, lis1G10.14, +/+, clip190KG06490 doubly heterozygous larvae crawled towards the stimulus at the same speed as control larvae (Fig. 8A,B), further indicating the independence of these two genetic pathways.
Fig. 8.
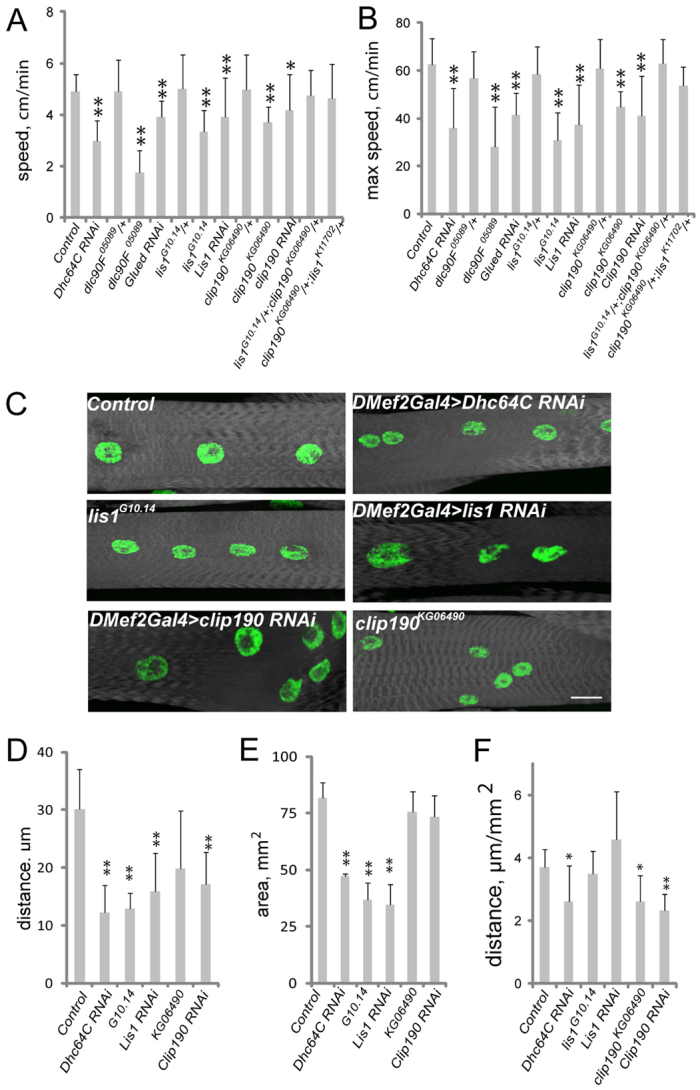
Larval muscle organization and physiology are affected by Dynein and associated proteins. (A,B) The average (A) and maximum (B) speed of Drosophila larvae as they crawl towards a stimulus. (C) Fluorescence images of VL3 muscles from L3 larvae that were used in locomotion assays just prior to dissection. White, phalloidin; green, Hoechst. Scale bar: 20 μm. (D) The average distance between nuclei in larval muscles from the indicated genotypes. (E) The surface area of the muscles in larvae of the indicated genotypes. (F) The distance between nuclei in the larval muscles normalized for muscle size. Error bars indicate s.d. *P<0.05, **P<0.01.
The larvae were then dissected and their musculature was examined. The internuclear distance was reduced by 50-60% in lis1G10.14 and clip190KG06490 larvae and in muscle depleted of Dhc64C, Lis1 or CLIP-190 (Fig. 8C,D). However, the muscles in lis1G10.14, Lis1-depleted and Dhc64C-depleted larvae were 35% smaller than in control and clip190KG06490 larvae (Fig. 8E). Because the muscle size was variable between genotypes, internuclear distance was normalized as a function of muscle size. Using this measure, both muscle size and internuclear distance were diminished in Dhc64C-depleted larvae compared with controls. However, internuclear distance was specifically reduced in clip190KG06490 and CLIP-190-depleted larvae, whereas muscle size was specifically reduced in lis1G10.14 (Fig. 8D-F). These data demonstrate a correlation between muscle output and both muscle size and myonuclear position, suggesting that the defects seen in the embryo are bona fide effects and not merely developmental delays.
DISCUSSION
We have used the Drosophila musculature to investigate the mechanisms that control muscle size and intracellular organization. We find that muscle length is regulated independently of the number of fusion events and demonstrate that perturbations that affect embryonic muscle length correlate with decreased larval muscle size and poor muscle function. Additionally, we find that intracellular organization, specifically myonuclear positioning, is essential for muscle function, consistent with our recent observations (Metzger et al., 2012). Moreover, we have shown that the length and intracellular organization of the myofiber are mechanistically independent. Although a number of factors link both processes, we have identified factors that contribute solely to muscle length or myonuclear position and demonstrate that we can independently manipulate each feature.
Dynein regulates both muscle length and myonuclear positioning. Some Dynein-interacting proteins, such as Dlc90F and Pins, are necessary for both processes. That these factors regulate both muscle length and myonuclear positioning suggests that specific aspects of muscle growth and the positioning of myonuclei can indeed be coordinated. However, Lis1 affects muscle length specifically, whereas CLIP-190 and Glued specifically affect myonuclear position. Within the contexts of muscle length and myonuclear position CLIP-190 and Lis1 do not genetically interact, illustrating that, although linked, the two processes are mechanistically distinct.
Our identification of CLIP-190 and Lis1 as regulators of Dynein is not novel. CLIP-190 and Lis1 are known to interact with Dynein both physically and functionally, and they usually cooperate toward a single goal (Coquelle et al., 2002). Here, we provide the first example of CLIP-190 and Lis1 serving completely independent, Dynein-dependent functions.
Mechanistically, our data suggest that Dynein function, with respect to the regulation of muscle length and myonuclear positioning, is specified by Lis1 and CLIP-190 downstream of its localization. That Dynein localization to the myofiber pole is important is illustrated by pins (raps193) mutants in which Dynein does not accumulate at the myofiber pole, and we observe defects in both muscle length and myonuclear positioning. This suggests that Pins recruits and stabilizes Dynein at the cortex as it does during mitosis (Gotta et al., 2003; Hughes et al., 2004). Lis1 also affects Dynein localization, but in Lis1 mutants Dynein is localized to the muscle pole and is tightly restricted to the pole compared with controls. We interpret this to mean that Lis1 is necessary for retrograde trafficking of Dynein away from the myofiber pole. We hypothesize that, in the absence of Dynein retrograde trafficking, cellular components accumulate at the extending muscle end, thus inhibiting the trafficking of factors necessary for further directed growth. Thus, when Dynein is incapable of moving cargo away from the myofiber pole, muscles are shorter than in control embryos. Indeed, a similar correlation between retrograde trafficking and directed growth was recently reported during mechanosensory bristle growth and axonal transport (Otani et al., 2011; Yi et al., 2011). Interestingly, it was recently reported that decreased retrograde transport of Dynein results in longer processes in both fibroblasts and neurons in culture (Rishal et al., 2012).
This contradictory finding raises several interesting questions. Is there an opposing pathway in muscle or is another aspect of muscle size increased? For example, does the length of the muscle impact its volume? Complications due to muscle contraction in the live embryo make such analysis difficult, however. Likewise, working in vivo on a dynamically changing tissue located 30-150 μm below the surface of the developing embryo presents challenges for imaging the rapid cellular processes that underlie motor activity, microtubule organization, organelle positioning and cell size. Nevertheless, the link between different aspects of cell size and their relationships to each other and to motor activity are being explored.
It is not clear why LT muscle growth/length is more sensitive to Dynein activity than the VL muscles in the embryo. A simple explanation is that the maternally loaded Dynein persists longer in the VL muscles than in the LT muscles. Additionally there might be a physical explanation. Although a cluster of potential tendon cells for the VL muscles exist, they are clustered at the segment border. Therefore, the size of the hemisegments, and thus the distance from segment border to segment border, determines muscle length. Conversely, the cluster of potential tendon cells for the LT muscles might be more broadly dispersed and the location of the muscle pole at the time of tendon specification determines the length of the muscle. Under this hypothesis, inefficient extension of the LT muscles would result in the muscles being shorter during tendon specification/maturation and therefore shorter throughout embryonic development. Alternatively, differences in guidance/signaling systems could explain why the LT muscles, but not the VL muscles, are shorter when Dynein activity is compromised. Different signaling mechanisms are employed by these different muscle types (Schnorrer and Dickson, 2004; Volk, 1999; Schweitzer et al., 2010; Schejter and Baylies, 2010). For example, Derailed (Drl) plays a crucial role in the ability of LT muscles to recognize their target (Callahan et al., 1996). Perhaps, altered trafficking of Drl or another factor in the signaling pathway causes slight, but significant, changes in LT muscle length.
It is interesting that the LT muscles are smaller in Dhc64C, Dlc90F, pins and Lis1 mutant embryos, but that other muscles are unaffected, whereas in larvae all muscles appear to be smaller. During the larval stages, muscles remain stably attached to tendon cells and grow through insulin signaling/Foxo- and dMyc-dependent pathways (Demontis and Perrimon, 2009). We interpret our observations to mean that Dynein and its regulatory proteins Dlc90F, Pins and Lis1 contribute to insulin receptor-mediated muscle growth as previously described (Huang et al., 2001; Varadi et al., 2003). Indeed, it would not be surprising to find that Dynein-dependent cellular trafficking is essential to that signaling pathway.
CLIP-190 does not dramatically affect Dynein localization, but it does affect microtubule organization, which, in turn, affects nuclear positioning. CLIP-190 mutant embryos have fewer microtubules at the myofiber pole, suggesting that, similar to its functions in other systems, the role of CLIP-190 is to stabilize microtubule-cortex interactions (Neukirchen and Bradke, 2011; Watanabe et al., 2004), which Dynein then uses to move nuclei towards the myofiber pole. The specification of Dynein function, downstream of its localization, is novel. Although phospho-regulation has been shown to alter Dynein function during mitosis (Whyte et al., 2008) and the possibility for competitive regulation of Dynein has been suggested (McKenney et al., 2011), this is, to our knowledge, the first example in which Dynein at a single location has its activity modified through interactions with unique binding partners. The ability to specify Dynein function without dramatically altering its localization is likely to be an important factor during development when temporal constraints are high.
With regards to physiology, the small, but significant, changes in myonuclear positioning and muscle size seen in the embryo continue throughout larval development and are associated with impaired muscle function. Additionally, that the same defects and impairments are found in larvae that were depleted of Dynein, Lis1 or CLIP-190 specifically in the muscle shows that the effects are, at least in part, muscle specific.
Many muscle myopathies are characterized by smaller myofibers and mispositioned nuclei. However, it is unclear whether these pathologies are linked and which of these defects are paramount in causing the muscle weakness associated with these myopathies. We have shown that in Drosophila these two processes are linked via the requirement for Dynein activity at the muscle pole. We have further shown that these two processes are mechanistically distinct, yet that both are necessary for muscle function. Together, these data suggest that therapeutics aimed at improving the functional capacity of diseased muscles must counteract effects on both muscle size and myonuclear positioning. This highlights Drosophila as an ideal model system with which to identify the genes and mechanisms required for distinct aspects of muscle morphogenesis and to shed light on key features of muscle disease.
Supplementary Material
Acknowledgements
We thank the Bloomington Stock Center and the Vienna Stock Center for fly stocks and the Developmental Hybridoma Bank for antibodies.
Footnotes
Funding
Our work is supported by Muscular Dystrophy Association (MDA) and National Institutes of Health (NIH) [GM078318 to M.K.B.]. V.K.S. is supported by an NIH Training Grant in Developmental Biology [T32HD060600]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.079178/-/DC1
References
- Bate M. (1990). The embryonic development of larval muscles in Drosophila. Development 110, 791-804 [DOI] [PubMed] [Google Scholar]
- Beckett K., Baylies M. K. (2007). 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Dev. Biol. 309, 113-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., Tsang G., Evans-Holm M., Hiesinger P. R., Schulze K. L., Rubin G. M., et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent J. R., Werner K. M., McCabe B. D. (2009). Drosophila larval NMJ dissection. J. Vis. Exp. 24, 1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggese C., Moschetti R., Ragone G., Barsanti P., Caizzi R. (2001). dtctex-1, the Drosophila melanogaster homolog of a putative murine t-complex distorter encoding a dynein light chain, is required for production of functional sperm. Mol. Genet. Genomics 265, 436-444 [DOI] [PubMed] [Google Scholar]
- Callahan C. A., Bonkovsky J. L., Scully A. L., Thomas J. B. (1996). derailed is required for muscle attachment site selection in Drosophila. Development 122, 2761-2777 [DOI] [PubMed] [Google Scholar]
- Coquelle F. M., Caspi M., Cordelières F. P., Dompierre J. P., Dujardin D. L., Koifman C., Martin P., Hoogenraad C. C., Akhmanova A., Galjart N., et al. (2002). LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 22, 3089-3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F., Perrimon N. (2009). Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 36, 983-993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. (2001). Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106, 489-498 [DOI] [PubMed] [Google Scholar]
- Fitts R. H., McDonald K. S., Schluter J. M. (1991). The determinants of skeletal muscle force and power: their adaptability with changes in activity pattern. J. Biomech. 24 Suppl. 1, 111-122 [DOI] [PubMed] [Google Scholar]
- Gepner J., Li M., Ludmann S., Kortas C., Boylan K., Iyadurai S. J., McGrail M., Hays T. S. (1996). Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics 142, 865-878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. R., Schroer T. A., Szilak I., Steuer E. R., Sheetz M. P., Cleveland D. W. (1991). Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol. 115, 1639-1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L. S., Gunawardena S. (2000). Flying through the drosophila cytoskeletal genome. J. Cell Biol. 150, 63F-68F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. (2002). Mechanisms of spindle positioning: focus on flies and worms. Trends Cell Biol. 12, 332-339 [DOI] [PubMed] [Google Scholar]
- Gotta M., Dong Y., Peterson Y. K., Lanier S. M., Ahringer J. (2003). Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 13, 1029-1037 [DOI] [PubMed] [Google Scholar]
- Grady R. M., Starr D. A., Ackerman G. L., Sanes J. R., Han M. (2005). Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc. Natl. Acad. Sci. USA 102, 4359-4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Imamura T., Olefsky J. M. (2001). Insulin can regulate GLUT4 internalization by signaling to Rab5 and the motor protein dynein. Proc. Natl. Acad. Sci. USA 98, 13084-13089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. R., Bullock S. L., Ish-Horowicz D. (2004). Inscuteable mRNA localization is dynein-dependent and regulates apicobasal polarity and spindle length in Drosophila neuroblasts. Curr. Biol. 14, 1950-1956 [DOI] [PubMed] [Google Scholar]
- Kraut R., Campos-Ortega J. A. (1996). inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev. Biol. 174, 65-81 [DOI] [PubMed] [Google Scholar]
- Lei K., Zhang X., Ding X., Guo X., Chen M., Zhu B., Xu T., Zhuang Y., Xu R., Han M. (2009). SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. USA 106, 10207-10212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Warrior R. (2000). The Drosophila Lissencephaly1 (DLis1) gene is required for nuclear migration. Dev. Biol. 226, 57-72 [DOI] [PubMed] [Google Scholar]
- Liu Z., Xie T., Steward R. (1999). Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development 126, 4477-4488 [DOI] [PubMed] [Google Scholar]
- Louis M., Piccinotti S., Vosshall L. B. (2008). High-resolution measurement of odor-driven behavior in Drosophila larvae. J. Vis. Exp. 3, 638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R. J., Vershinin M., Kunwar A., Vallee R. B., Gross S. P. (2010). LIS1 and NudE induce a persistent dynein force-producing state. Cell 141, 304-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R. J., Weil S. J., Scherer J., Vallee R. B. (2011). Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J. Biol. Chem. 286, 39615-39622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesngon M. T., Tarricone C., Hebbar S., Guillotte A. M., Schmitt E. W., Lanier L., Musacchio A., King S. J., Smith D. S. (2006). Regulation of cytoplasmic dynein ATPase by Lis1. J. Neurosci. 26, 2132-2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E. S., Richardson B. E., Gomes E. R., Baylies M. K. (2012). MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484, 120-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukirchen D., Bradke F. (2011). Cytoplasmic linker proteins regulate neuronal polarization through microtubule and growth cone dynamics. J. Neurosci. 31, 1528-1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S. M., Dorfman M. D., Carter J. C., Bowerman B. (2007). Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 3, e128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T., Oshima K., Onishi S., Takeda M., Shinmyozu K., Yonemura S., Hayashi S. (2011). IKKε regulates cell elongation through recycling endosome shuttling. Dev. Cell 20, 219-232 [DOI] [PubMed] [Google Scholar]
- Palazzo A. F., Joseph H. L., Chen Y. J., Dujardin D. L., Alberts A. S., Pfister K. K., Vallee R. B., Gundersen G. G. (2001). Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr. Biol. 11, 1536-1541 [DOI] [PubMed] [Google Scholar]
- Parmentier M. L., Woods D., Greig S., Phan P. G., Radovic A., Bryant P., O'Kane C. J. (2000). Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 20, RC84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson C. R., Agrawal P. B., Blasko J., Beggs A. H. (2007). Myofiber size correlates with MTM1 mutation type and outcome in X-linked myotubular myopathy. Neuromuscul. Disord. 17, 562-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina N. L., Waterman-Storer C. M. (2004). Protein kinase D-mediated anterograde membrane trafficking is required for fibroblast motility. Curr. Biol. 14, 88-98 [DOI] [PubMed] [Google Scholar]
- Richardson B. E., Beckett K., Nowak S. J., Baylies M. K. (2007). SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development 134, 4357-4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishal I., Kam N., Perry R. B., Shinder V., Fisher E. M., Schiavo G., Fainzilber M. (2012). A motor driven mechanism for cell length sensing. Cell Rep. 1, 608-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero N. B. (2010). Centronuclear myopathies: a widening concept. Neuromuscul. Disord. 20, 223-228 [DOI] [PubMed] [Google Scholar]
- Rommel C., Bodine S. C., Clarke B. A., Rossman R., Nunez L., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001). Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3, 1009-1013 [DOI] [PubMed] [Google Scholar]
- Schejter E. D., Baylies M. K. (2010). Born to run: creating the muscle fiber. Curr. Opin. Cell Biol. 22, 566-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J., Kreitzer G., Simon S. M. (2003). Migrating fibroblasts perform polarized, microtubule-dependent exocytosis towards the leading edge. J. Cell Sci. 116, 4513-4519 [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Dickson B. J. (2004). Muscle building; mechanisms of myotube guidance and attachment site selection. Dev. Cell 7, 9-20 [DOI] [PubMed] [Google Scholar]
- Schweitzer R., Zelzer E., Volk T. (2010). Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development 137, 2807-2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M. A., Pellman D. (2003). Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13, 364-372 [DOI] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A. M., Artelt J., Bettencourt P., Cassin E., et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462-469 [DOI] [PubMed] [Google Scholar]
- Squire J. M. (1997). Architecture and function in the muscle sarcomere. Curr. Opin. Struct. Biol. 7, 247-257 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M. E., Macurek L., Galjart N., Medema R. H. (2008). Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 27, 3235-3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. W., Bremner K. H., Vallee R. B. (2007). Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10, 970-979 [DOI] [PubMed] [Google Scholar]
- Varadi A., Tsuboi T., Johnson-Cadwell L. I., Allan V. J., Rutter G. A. (2003). Kinesin I and cytoplasmic dynein orchestrate glucose-stimulated insulin-containing vesicle movements in clonal MIN6 beta-cells. Biochem. Biophys. Res. Commun. 311, 272-282 [DOI] [PubMed] [Google Scholar]
- Vaughan P. S., Miura P., Henderson M., Byrne B., Vaughan K. T. (2002). A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J. Cell Biol. 158, 305-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T. (1999). Singling out Drosophila tendon cells: a dialogue between two distinct cell types. Trends Genet. 15, 448-453 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Takefuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. (2004). Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell 7, 871-883 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C. M., Karki S. B., Kuznetsov S. A., Tabb J. S., Weiss D. G., Langford G. M., Holzbaur E. L. (1997). The interaction between cytoplasmic dynein and dynactin is required for fast axonal transport. Proc. Natl. Acad. Sci. USA 94, 12180-12185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J., Bader J. R., Tauhata S. B., Raycroft M., Hornick J., Pfister K. K., Lane W. S., Chan G. K., Hinchcliffe E. H., Vaughan P. S., et al. (2008). Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J. Cell Biol. 183, 819-834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J. Y., Ori-McKenney K. M., McKenney R. J., Vershinin M., Gross S. P., Vallee R. B. (2011). High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J. Cell Biol. 195, 193-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.