Summary
Following irradiation, numerous DNA-damage-responsive proteins rapidly redistribute into microscopically visible subnuclear aggregates, termed ionising-radiation-induced foci (IRIF). How the enrichment of proteins on damaged chromatin actually relates to DNA repair remains unclear. Here, we use super-resolution microscopy to examine the spatial distribution of BRCA1 and 53BP1 proteins within single IRIF at subdiffraction-limit resolution, yielding an unprecedented increase in detail that was not previously apparent by conventional microscopy. Consistent with a role for 53BP1 in promoting DNA double-strand break repair by non-homologous end joining, 53BP1 enrichment in IRIF is most prominent in the G0/G1 cell cycle phases, where it is enriched in dense globular structures. By contrast, as cells transition through S phase, the recruitment of BRCA1 into the core of IRIF is associated with an exclusion of 53BP1 to the focal periphery, leading to an overall reduction of 53BP1 occupancy at DNA damage sites. Our data suggest that the BRCA1-associated IRIF core corresponds to chromatin regions associated with repair by homologous recombination, and the enrichment of BRCA1 in IRIF represents a temporal switch in the DNA repair program. We propose that BRCA1 antagonises 53BP1-dependent DNA repair in S phase by inhibiting its interaction with chromatin proximal to damage sites. Furthermore, the genomic instability exhibited by BRCA1-deficient cells might result from a failure to efficiently exclude 53BP1 from such regions during S phase.
Key words: DNA double-strand breaks, BRCA1, 53BP1, γH2AX, DNA damage, IRIF, Structured Illumination, Super-resolution microscopy
Introduction
Following the detection of a DNA double-strand break (DSB), the histone variant H2AX is targeted for phosphorylation in chromatin flanking the break site. This phosphorylated isoform, known as γH2AX, serves as a molecular beacon, signalling the presence of damage and marking nucleosomes in up to megabases of DNA surrounding the DSB (Rogakou et al., 1998). γH2AX is paramount in linking this modified chromatin to the DNA damage resolution machinery, directing the recruitment of multiple DNA repair proteins (Fernandez-Capetillo et al., 2004). This recruitment results in the formation of repair centres, microscopically visible nuclear aggregates known as ‘foci’. These contain factors from both major DSB repair pathways: non-homologous end joining (NHEJ), a repair process that re-ligates DSB ends independent of DNA sequence; and Homologous Recombination (HR), which involves extensive end-processing before a homologous DNA sequence is used as a template for error-free repair. Choosing the appropriate DSB repair pathway is crucial, as inappropriately processed DSBs have the potential to trigger mutations and chromosomal translocations that may result in infertility, immunodeficiency, neurodegenerative disease and cancer (Jackson and Bartek, 2009).
Studies using ultraviolet (UV) laserbeams to induce DSBs along subnuclear tracts have revealed that different proteins accumulate in distinct subcompartments at DSB sites (Bekker-Jensen et al., 2006). Yet, as proteins with non-complementary DNA repair roles appear to accumulate within common sub-compartments, it is unclear how this spatial organisation influences choice of appropriate DSB repair pathway. Insight into the function of DSB foci has been hampered by the diffraction limit imposed by light microscopy. This limit, defined by the wavelength of visible light, impedes our ability to resolve structures to less than a theoretical limit of 200 to 350 nm. However, several new super-resolution technologies have been developed that can bypass the diffraction limit (reviewed by Schermelleh et al., 2010). Here, we report using one such technology, three-dimensional structured illumination microscopy (3D-SIM) (Schermelleh et al., 2008), to better dissect the relationships between DSB-responsive factors in damaged chromatin. This has enabled us to resolve the spatial distribution of DSB-responsive proteins within a single DSB focus with an unprecedented increase in nano-scale detail. Our data reveal unexpected insights into the temporal and spatial control of BRCA1 and 53BP1 at DSBs, which suggests that they may regulate DSB repair pathway choice via antagonistic chromatin contacts at DSB sites.
Results and Discussion
Sustained enrichment of the DSB-responsive proteins 53BP1 and BRCA1 in ionising-radiation-induced foci (IRIF) relies on γH2AX, MDC1 binding γH2AX and a ubiquitylation cascade catalysed through recruitment of the RNF8/RNF168 E3-ubiquitin ligases (Lukas et al., 2011). Because of the common upstream requirements for 53BP1 and BRCA1 recruitment to IRIF, it has been speculated that these proteins accumulate in common sub-nuclear compartments spanning DNA damage sites (Bekker-Jensen et al., 2006). However, others have reported very little spatial overlap between BRCA1 and 53BP1, with the majority of their respective IRIF being non-associated (Mok and Henderson, 2010). The perceived enrichment of these two proteins in common chromatin territories spanning damage sites is perplexing considering the opposing roles of 53BP1 and BRCA1 in promoting DSB repair by NHEJ and HR, respectively. To date, how the accumulation of these proteins in IRIF influences DSB repair outcome remains unclear.
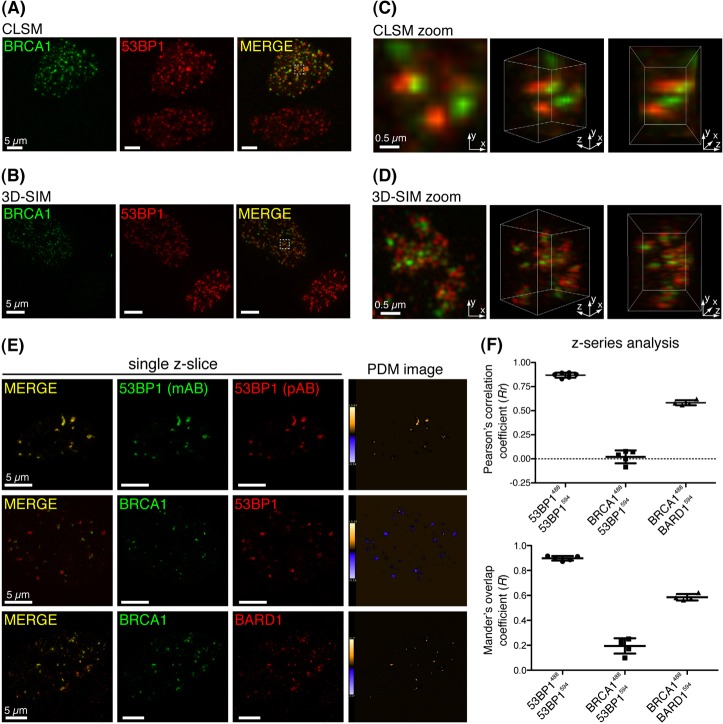
We speculated that the increased resolution offered by 3D-SIM might provide insights into the opposing roles of BRCA1 and 53BP1 within IRIF. To this end, 53BP1 and BRCA1 localisation was examined in γ-irradiated human hTert-RPE1 cells by 3D-SIM, and compared with data acquired in parallel by confocal laser scanning microscopy (CLSM). Two notable patterns of BRCA1 and 53BP1 localisation were evident in Z-series of asynchronous cell-cultures resolved by both techniques: some cells were positive for 53BP1 IRIF but exhibited little or no BRCA1 IRIF; while others comprised IRIF containing both proteins (Fig. 1A,B, lower and upper cells, respectively). In line with a previous report (Mok and Henderson, 2010), CLSM-acquired and 3D-rendered Z-series identified little spatial overlap between 53BP1 and BRCA1 IRIF in cells positive for both proteins (Fig. 1C, projection in left panel and 3D-rendered images in middle and right panels). However, in these cells 3D-SIM revealed a striking co-enrichment pattern of BRCA1 and 53BP1 within IRIF that was not apparent by CLSM (upper cell in Fig. 1B; supplementary material Movie 1). The increased resolution enabled spatial discrimination of BRCA1 and 53BP1 within individual foci, revealing these proteins resided in mutually exclusive yet adjacent sub-focal volumes, positioned at the core and periphery of IRIF, respectively (Fig. 1D). Importantly, colocalisation was observed in cells immunostained with reciprocal 53BP1 antibodies, and when immunostaining of BRCA1 and its interaction partner BARD1 was analysed across multiple 3D-SIM Z-series (Fig. 1E,F). However, this analysis also revealed an inverse correlation between BRCA1 and 53BP1 immunostaining patterns (Fig. 1E,F), indicating that although closely apposed within IRIF, they largely occupy non-overlapping volumes.
Fig. 1.
Subdiffraction-limit imaging of BRCA1 and 53BP1 in IRIF. (A) RPE1 cells recovered for 30 minutes following irradiation (2 Gy), were subjected to detergent pre-extraction and fixation before immunostaining for BRCA1 and 53BP1. Panels represent projection images constructed from multiple CLSM Z-series images. (B) Projection images constructed from 3D-SIM acquired Z-series of cells prepared exactly as in A. (C,D) Projection (left) and 3D-rendered images (other panels) were constructed from Z-series images of the indicated cell volumes (checked box) in A and B, respectively. (E) Pixel-by-pixel analysis of colocalisation of indicated fluorescently labelled proteins was performed across whole-cell 3D-SIM Z-series by intensity correlation analysis (ICA). Representative Z-slices from each dataset are presented, and the product of the differences from the mean intensity of each pixel (PDM) is shown: positive PDM pixels (an indication of co-variance) are orange, and negative PDM pixels (an indication of inverse correlation) are blue. (F) Statistical analysis of colocalisation. ICA analysis was performed across multiple Z-series (8–32 Z-slices/cell) and standard measures of image colocalisation for each labelled antibody pair are plotted. Each dot represents a single cell. Mean and s.d. are shown.
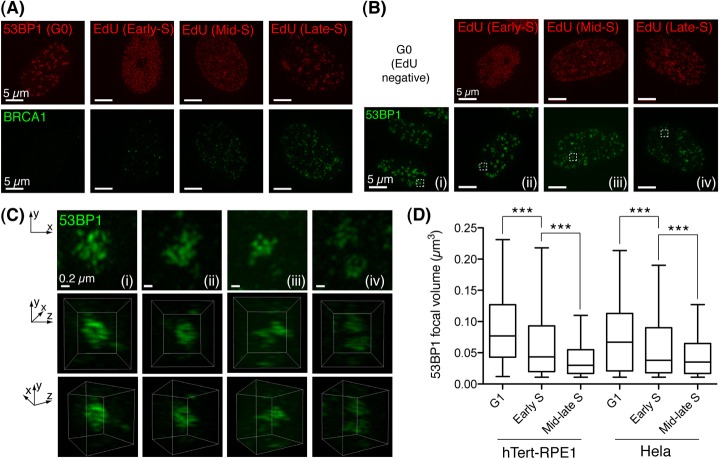
We speculated that the varying patterns of 53BP1-BRCA1 enrichment in different cells might coincide with cell cycle-dependent alterations in the DSB repair program. To address this, RPE1 cells were enriched in G0/G1 by serum starvation. In parallel, we pulse-labelled asynchronous cell cultures with the nucleotide analogue 5′-ethynyl-2′-deoyuridine (EdU) immediately before irradiation, the fluorescent labelling of EdU in fixed cells enabling discrimination of the various stages of S phase (Dimitrova and Berezney, 2002). In this analysis, stationary phase cells were positive for 53BP1 IRIF yet displayed little or no BRCA1 co-enrichment (Fig. 2A, left panel). By contrast, BRCA1 IRIF became apparent in cells exhibiting early S-phase replicon patterns, with increasing numbers of such foci in mid to late S phase (Fig. 2A).
Fig. 2.
Temporal changes to BRCA1 and 53BP1 IRIF distributions. (A) RPE1 cells were G0/G1 arrested by serum starvation or asynchronous cell cultures were pulse-labelled with EdU (5 minutes, 20 µM) immediately before irradiation (2 Gy). 30 minutes later, fixed cells stained for EdU in click reactions, were counterstained for BRCA1. Representative projection images constructed from 3D-SIM-acquired Z-series datasets are shown (n≥10 per stage). (B) Cells treated exactly as in A immunostained for 53BP1. (C) Projections and 3D reconstructions of 53BP1 distribution in IRIF (top and bottom two panels, respectively) were assembled from 3D-SIM datasets of cell volumes indicated in B. (D) G1 to S-phase progression is associated with decreased 53BP1 IRIF occupancy. Volumes of isosurface rendered 53BP1 foci from multiple irradiated (harvested 1 hour following 2Gy) RPE1 (n = 32; >300 foci scored per group) and HeLa cells (n = 36; >700 foci scored per group). S-phase position was discriminated as above. EdU-negative cells with a DAPI content <S or G2 were discriminated as G1. Whiskers indicate the 5th to 95th percentile. ***P<0.0001, Mann–Whitney test.
53BP1 staining was most intense in stationary-phase cells, where it formed large, dense nuclear aggregates (Fig. 2B). However, we observed a progressive reduction in 53BP1 IRIF intensity between early and late S-phase cells (Fig. 2B,C). Temporal changes in 53BP1 sub-focal distribution were also apparent: stationary phase cells exhibiting dense 53BP1 IRIF core staining (Fig. 2C, left panels); while the reduction in 53BP1 IRIF staining intensity during S-phase progression corresponded to its redistribution to more peripheral positions within IRIF (Fig. 2C, right panels). To quantify such alterations, 53BP1 stained volumes were isosurface rendered in 3D-SIM Z-series (supplementary material Movie 2). This enabled measurement of individual 53BP1 IRIF volumes, and statistical analysis of focal populations in cells of defined cell cycle positions. Strikingly, significant reductions to the volume occupied by 53BP1 in IRIF between G1, early and mid to late S-phase stages were observed in both RPE1 and HeLa cells (Fig. 3F). Hence, the cell-cycle-dependent redistribution of 53BP1 in IRIF is coupled to a reduction in the overall volume it occupies.
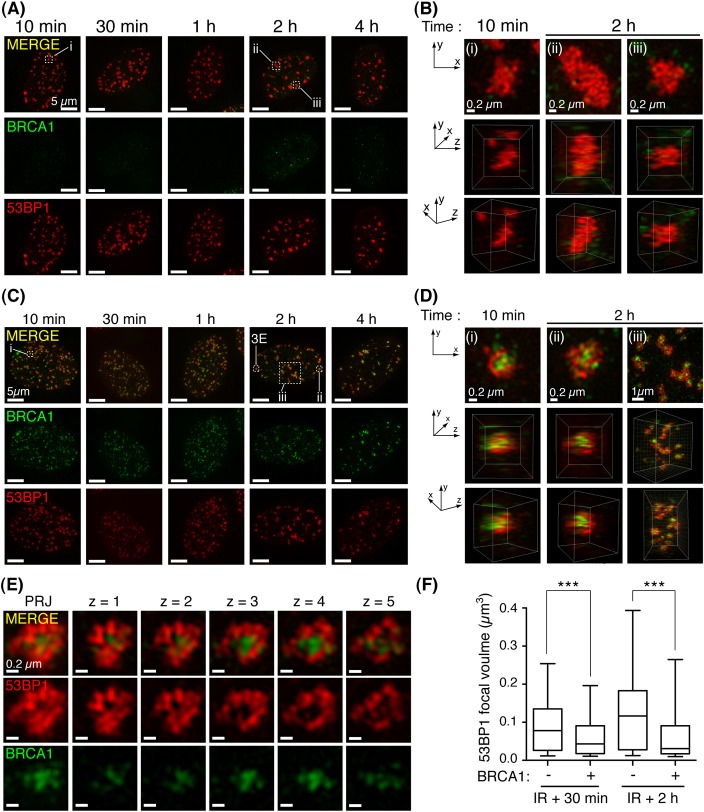
Fig. 3.
The spatial distribution of 53BP1 and BRCA1 at DSB sites is established rapidly and persists following irradiation. (A) Irradiated cells (2 Gy), detergent pre-extracted and fixed at indicated times following irradiation, immunostained for BRCA1 and 53BP1. Representative projection images of 3D-SIM datasets (n≥8 per group). (B) Projection and 3D reconstructions of 3D-SIM datasets of cell volumes indicated in A. (C) Cells positive for co-enrichment of BRCA1 and 53BP1 in IRIF were processed as in A. (D) Projection and 3D-reconstruction images constructed from 3D-SIM datasets of cell volumes indicated in C. (E) Projection (PRJ) and single Z-slices show 53BP1 exclusion from BRCA1-associated IRIF core. (F) BRCA1 enrichment is associated with decreased 53BP1 IRIF occupancy. Volumes of isosurface-rendered 53BP1 foci from multiple irradiated cells (2 Gy) positive or negative for BRCA1 co-enrichment were analysed per time point; n = 3 per group, scoring >300 foci and >100 foci at 30 minute and 2 hour time points, respectively. Whiskers indicate the 5th to 95th percentile. ***P<0.0001, Mann–Whitney test.
It has recently been proposed that BRCA1 IRIF occurring at early times following irradiation are associated with BRCA1 function in promoting HR, while interactions with the RAP80/Abraxas complex that mediate its later-forming IRIF may restrict HR (Hu et al., 2011). As BRCA1 HR function is associated with its suppression of 53BP1-dependent DSB repair (Bothmer et al., 2011; Bunting et al., 2010), we examined for changes in the relative distributions of these proteins at DSB sites at multiple early time-points following irradiation. Thus, we fixed RPE1 cells between 10 minutes and 4 hours following irradiation and then processed the samples for BRCA1 and 53BP1 immunofluorescence by 3D-SIM. As shown in Fig. 3A, dense G1-pattern 53BP1 IRIF were established as early as 10 minutes following irradiation and persisted at later time-points. 3D rendering of single foci identified these IRIF to vary in volume and dimensions, ranging from globular (Fig. 3B, panels i and iii), to extended protein aggregates (Fig. 3B, panel ii; supplementary material Movie 3). Similarly, cells displaying the S-phase pattern of BRCA1-53BP1 IRIF co-enrichment were apparent at all times examined following irradiation (Fig. 3C), excluding the possibility of a gradual progression from one state to another. Strikingly, 3D-rendered datasets revealed BRCA1 and 53BP1 were enriched at the IRIF core and periphery, respectively (Fig. 3D, panels i–iii; supplementary material Movie 4), and Z-series confirmed 53BP1 was excluded from the BRCA1-associated IRIF core (Fig. 3E). By volumetric measurement of isosurface-rendered 53BP1 foci in multiple cells at 30 minutes and 2 hours following γ-irradiation, we found BRCA1 co-enrichment in IRIF correlated with a reduction in the focal volume occupied by 53BP1 (Fig. 3F).
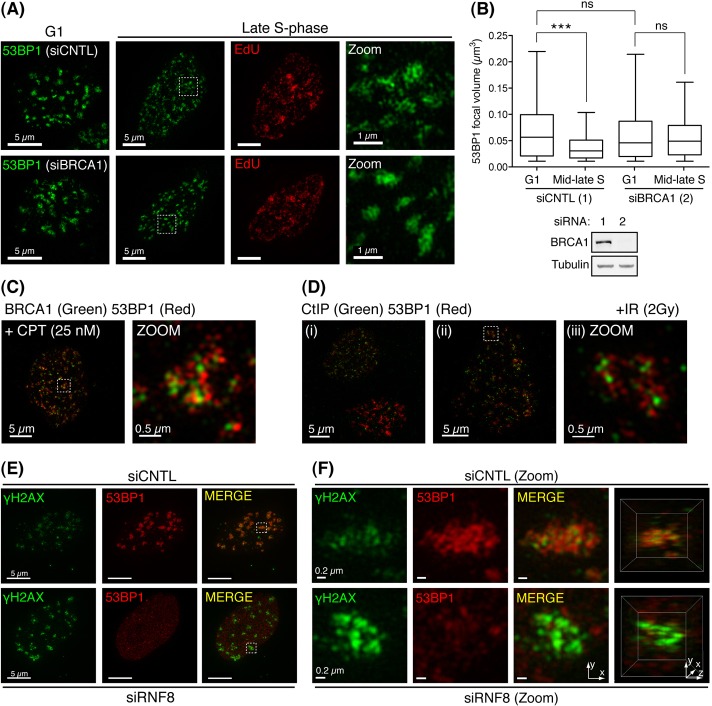
Our data indicate that the reduction in 53BP1 occupancy in IRIF and its exclusion from the core of IRIF coincides with BRCA1's enrichment at such sites. To examine whether these reductions in 53BP1 occupancy in S-phase IRIF are dependent on BRCA1, HeLa cells treated with control or BRCA1-targeting siRNA, were EdU pulse labelled, γ-irradiated, and processed as in Fig. 2. Importantly, BRCA1 siRNA-mediated depletion yielded no significant alterations in cell-cycle distribution compared with control cells (data not shown). Moreover, no significant deviation in 53BP1-stained IRIF volumes was observed between G1 populations of control and BRCA1-depleted cells (Fig. 4A, left panels; Fig. 4B). By contrast, while control-treated cells showed the expected 53BP1 redistribution and reduced occupancy within IRIF in mid to late S phase, these changes were compromised in BRCA1-depeleted cells (Fig. 4A, right panels; Fig. 4B). Thus, BRCA1 influences 53BP1 occupancy in IRIF, perhaps by antagonising its interaction with DSB-associated chromatin in S phase.
Fig. 4.
BRCA1-dependent 53BP1 exclusion from IRIF coincides with S-phase DSB repair and 53BP1-associated chromatin displays reduced γH2AX. 48 hours following control (CNTL) or BRCA1 siRNA treatment, HeLa cells were processed exactly as in Fig. 2D. (A) Representative images of two independent experiments in which IRIF (>40 per cell; n≥19) were scored per experimental group. (B) Quantification of experiment shown in A (top). Whiskers indicate the 5th to 95th percentile. ns, not significant (P = 0.8639). ***P<0.0001, Mann–Whitney test. Western blot (bottom) showing BRCA1 protein depletion in CNTL and BRCA1 siRNA-treated cells (lanes 1 and 2, respectively). (C) Cells recovered for 30 minutes following acute CPT treatment (25 nM for 10 minutes) were detergent pre-extracted and fixed before BRCA1 and 53BP1 immunostaining. Representative projection images are presented (n = 10). (D) Irradiated cells (2 Gy) processed as in C were immunostained for CtIP and 53BP1. (E) 72 hours following siRNA treatment, cells were irradiated (2 Gy), recovered (30 minutes), then processed as in B with 53BP1 and γH2AX antisera. Representative projection images are presented (n≥10 in two independent experiments). (F) Projection and 3D-reconstruction images constructed from two-channel 3D-SIM datasets of cell volumes indicated in E.
To further explore whether BRCA1 enrichment and its associated exclusion of 53BP1 from the IRIF core correlates with inhibition of 53BP1-dependent repair at DSB sites during S phase, we analysed cells recovering from acute treatment with camptothecin (CPT), a topoisomerase I inhibitor that induces DSBs in S phase when replication forks encounter trapped Top1-DNA cleavage complexes (Pommier et al., 2003). In BRCA1-deficient cells, CPT-induced chromosomal aberrations are 53BP1 dependent; indicating BRCA1 normally counteracts 53BP1-mediated repair activities in this context (Bunting et al., 2010). Consistent with this notion, CPT-induced foci closely resembled S-phase IRIF by 3D-SIM, with BRCA1 and 53BP1 adopting equivalent focal positions (Fig. 4C). The repair of DSBs by HR in S phase also relies on CtIP-mediated DNA-end resection (Sartori et al., 2007). Consistent with a model in which the focus core corresponds to sites of HR in S-phase cells, 3D-SIM analysis of CtIP localisation in irradiated asynchronous RPE1 cell cultures revealed CtIP to closely resemble BRCA1 staining patterns, occupying comparable positions in the IRIF core, and surrounded by peripheral 53BP1 staining (Fig. 4D, panels i–iii). Moreover, in accord with CtIP functioning primarily in HR, cells exhibiting large dense G1-like 53BP1 IRIF exhibited minimal associated CtIP enrichment (Fig. 4D, panel i).
The sustained enrichment of 53BP1 and BRCA1 in IRIF requires common γH2AX-dependent molecular determinants, yet surprisingly, we observed minimal overlap between 53BP1 and γH2AX IRIF-staining by 3D-SIM (Fig. 4E, top panels and zoomed images in Fig. 4F). Although associated within IRIF, most prominent γH2AX signals corresponded to regions peripheral of 53BP1 signal (Fig. 4F, top panels). In light of these observations, we examined γH2AX IRIF in cells treated with RNF8 siRNA to abolish γH2AX-dependent 53BP1 and BRCA1 recruitment. As expected, RNF8 depletion abolished 53BP1 IRIF formation (Fig. 4E). However, it also induced a marked increase in γH2AX signal (Fig. 4E, lower panels), and its shift from peripheral to core IRIF positions (Fig. 4F, bottom panels). These data suggest that although primed by γH2AX, RNF8-dependent ubiquitylation might promote chromatin alterations that subsequently restrict γH2AXmaintenance at DSB sites. As these events also direct 53BP1 and BRCA1 recruitment, these factors may influence such changes.
Both 53BP1-chromatin interactions and γH2AX phosphorylation are required for inhibition of DSB resection, suggesting that this is a prime function for γH2AX-dependent 53BP1 IRIF (Bothmer et al., 2011; Bothmer et al., 2010). Such DSB end-protection may favour NHEJ by preserving DNA end integrity and therefore accurate re-ligation. However, it also impairs DSB resection in BRCA1-deficient cells, blocking HR, resulting in chromosomal instability (Bunting et al., 2010). It is plausible that a common activity restricts both γH2AX phosphorylation and DSB resection within the 53BP1-associated chromatin domain. Perhaps the dense globular appearance of 53BP1 IRIF in G1 cells hints at the formation of a condensed or aggregated chromatin state proximal to DSB sites. Establishing such a state might inhibit the resection machinery by simply occluding its DNA substrate. It might also be consistent with recent evidence that the molecular events that drive 53BP1 enrichment at damage sites, also mediate transcriptional silencing in DSB-flanking chromatin (Shanbhag et al., 2010). Our findings raise the possibility that BRCA1 might counteract 53BP1-dependent DNA end-protection in a temporally controlled manner: inhibiting 53BP1 interaction with chromatin proximal to break sites during S phase when this activity threatens genome stability.
There has been considerable debate about how and why 53BP1 and BRCA1, with opposing roles in DNA repair, are recruited to damage sites by apparently common mechanisms. Our data reveal that these proteins actually occupy associated yet adjacent territories at DSB sites. Delineating the relative boundaries between these proteins at nucleotide resolution, perhaps by further improvements in microscopy and/or by biochemical studies, will be useful in corroborating such a model and thereby explaining how the molecular choreography of 53BP1, BRCA1 and other proteins takes place and how such events have evolved to maintain genome integrity.
Materials and Methods
CLSM and 3D-SIM microscopy
An Olympus FLV1000 inverted microscope equipped with a 100× 1.4 NA oil objective was used for CSLM. Fluorochromes were excited with a 405, 488 and 594 nm line lasers. Settings: 512×512 pixel frame size; 1 Airy Unit pinhole diameter; projection and 3D images were of z-sections taken every 125 nm across the cell. 3D-SIM was carried out with an OMX microscope (Applied Precision, WA), equipped with 405, 488, and 594 nm line lasers, and a 100× 1.4 NA oil objective. Settings were as above and data was reconstructed using API SoftWorx software. Following acquisition, images were imported into IMARIS (Bitplane) and Adobe Photoshop, for 3D analysis and processing. IRIF volume analysis was by isosurface rendering in IMARIS with a background subtraction algorithm and settings: experimentally defined threshold; largest sphere diameter of 0.1 µm; noise filter eliminating structures <0.01 µm3. Statistical analysis of data was performed in Prism (Graphpad Software). Colocalisation analysis was performed between 8-bit image datasets in ImageJ using the ‘Intensity Correlation Analysis’ plug-in.
Immunofluorescence
Cells cultured on #1.5 cover-glasses were washed and pre-extracted with ice-cold CSK buffer [5 minutes; 10 mM PIPES pH 6.8, 300 mM sucrose, 50 mM NaCl, 3 mM EDTA, 0.5% Triton X-100, Protease Inhibitors (Complete; Roche)]. Cells were subsequently fixed in 2% paraformaldehyde for 15 minutes. Afterwards cells were blocked (30 minutes; 3% BSA, 0.1% Triton X-100 in PBS), before sequential incubations with primary and secondary fluorophore-conjugated antibodies in block, separated by extensive PBS-Triton washes. Cover glasses cleaned in water were mounted in DAPI-supplemented Prolong Gold (Invitrogen). Anti-rabbit and anti-mouse Alexa Fluor 488 and 594 nm secondary antibodies (Molecular Probes, Invitrogen) were used, and EdU labelled using a Click-IT EdU Alexa Fluor 594 imaging kit (Invitrogen). Primary antibodies were titrated to minimise bleaching during 3D-SIM: mouse anti-BRCA1 (clone D9; Santa Cruz, CA); rabbit anti-53BP1 (pAB, NB100-305, Novus Biologicals, Cambridge, UK); mouse anti-53BP1 (mAB, BP13, Millipore); mouse anti-γH2AX (05-636, Millipore); rabbit anti-BARD1 (A300-263a; Bethyl); mouse anti-CtIP (a gift from Richard Baer, Columbia University, New York, NY).
Cell culture and transfection
RPE1 cells were cultured in DMEM + 10% FBS, G0/G1 arrested by serum starvation for 48 hours (DMEM without FBS), and G0/G1 enrichment verified at >90% by flow cytometry DNA content measurement. RPE1 and HeLa cells were siRNA transfected using Lipofectamine RNAi MAX (Invitrogen) according to manufacturer's instructions. siRNAs were a 1∶1 mixture of RNF8-A: 5′-UGGACA-AUUAUGGACAACA-3′, and RNF8-C 5′-CCAAGAACAAAGAAUUAG-3′ (Galanty et al., 2009) for RNF8-depletion (MWG Biotech AG); a smartpool against BRCA1 (5′-CAGCUACCCUUCCAUCAUAUU-3′; 5′-GGGAUACCAUGCAACAUAAUU-3′; 5′-GAAGGAGCUUUCAUCAUUCUU-3′; 5′-CUAGAAAUCUGUUGCUAUGUU-3′; MU-003461-01, Dharmacon), and 5′-CGUACGCGGAAUACUUCGATT-3′ was used in control (luciferase) depletions.
Supplementary Material
Acknowledgments
We thank Cath Green, Vitor Trovisco, Andreas Bruckbauer, Jo Morris and the Jackson and Boulton lab members for support and discussion. We also thank André Nussenzweig for reagents and discussions, Nicola Lawrence for microscopy support and Richard Baer for CtIP antibody.
Footnotes
Funding
J.R.C. is funded by a Sir Henry Wellcome Postdoctoral Fellowship, with infrastructural support from Cancer Research UK. The lab of S.J.B. is funded by Cancer Research UK and by an ERC Advanced Investigator Grant (RecMitMei). Research in the S.P.J. lab is supported by grants from Cancer Research UK; a European Advanced Investigator Grant (DDREAM); the European Community's 7th Framework Programme grant agreement HEALTH-F2-2012-259893 (DDResponse); and core infrastructure funding from Cancer Research UK and the Wellcome Trust. S.P.J. is salaried by the University of Cambridge, supplemented by Cancer Research UK. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.105353/-/DC1
References
- Bekker–Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006). Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 10.1083/jcb.200510130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A., Robbiani D. F., Feldhahn N., Gazumyan A., Nussenzweig A., Nussenzweig M. C. (2010). 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207, 855–865 10.1084/jem.20100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A., Robbiani D. F., Di Virgilio M., Bunting S. F., Klein I. A., Feldhahn N., Barlow J., Chen H. T., Bosque D., Callen E., et al. (2011). Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 10.1016/j.molcel.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez–Capetillo O., Cao L., et al. (2010). 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova D. S., Berezney R. (2002). The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115, 4037–4051 10.1242/jcs.00087 [DOI] [PubMed] [Google Scholar]
- Fernandez–Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. (2004). H2AX: the histone guardian of the genome. DNA Repair (Amst.) 3, 959–967 10.1016/j.dnarep.2004.03.024 [DOI] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009). Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 10.1038/nature08657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Scully R., Sobhian B., Xie A., Shestakova E., Livingston D. M. (2011). RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25, 685–700 10.1101/gad.2011011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., Bartek J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J., Lukas C., Bartek J. (2011). More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13, 1161–1169 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- Mok M. T., Henderson B. R. (2010). A comparison of BRCA1 nuclear localization with 14 DNA damage response proteins and domains: identification of specific differences between BRCA1 and 53BP1 at DNA damage-induced foci. Cell. Signal. 22, 47–56 10.1016/j.cellsig.2009.09.007 [DOI] [PubMed] [Google Scholar]
- Pommier Y., Redon C., Rao V. A., Seiler J. A., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., et al. (2003). Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 532, 173–203 10.1016/j.mrfmmm.2003.08.016 [DOI] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. (2007). Human CtIP promotes DNA end resection. Nature 450, 509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P. M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M. C., Agard D. A., Gustafsson M. G., et al. (2008). Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Heintzmann R., Leonhardt H. (2010). A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190, 165–175 10.1083/jcb.201002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag N. M., Rafalska–Metcalf I. U., Balane–Bolivar C., Janicki S. M., Greenberg R. A. (2010). ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.