Summary
The coordination of signalling pathways within the cell is vital for normal human development and post-natal tissue homeostasis. Gene expression and function is therefore tightly controlled at a number of levels. We investigated the role that post-translational modifications play during human hepatocyte differentiation. In particular, we examined the role of the small ubiquitin-like modifier (SUMO) proteins in this process. We used a human embryonic stem cell (hESC)-based model of hepatocyte differentiation to follow changes in protein SUMOylation. Moreover, to confirm the results derived from our cell-based system, we performed in vitro conjugation assays to characterise SUMO modification of a key liver-enriched transcription factor, HNF4α. Our analyses indicate that SUMOylation plays an important role during hepatocellular differentiation and this is mediated, in part, through regulation of the stability of HNF4α in a ubiquitin-dependent manner. Our study provides a better understanding of SUMOylation during human hepatocyte differentiation and maturation. Moreover, we believe the results will stimulate interest in the differentiation and phenotypic regulation of other somatic cell types.
Key words: SUMO, HNF4α, Hepatocyte, hESCs, Liver, Cell-based modelling, p450, Transthyretin, Ubiquitin
Introduction
The coordination of signalling pathways within the cell is vital for human embryonic development and post-natal tissue homeostasis (Vaillancourt and Lafond, 2009). This requires the regulation of gene expression at multiple levels (Chen and Rajewsky, 2007), including protein post-translational modification (PTM). Protein PTMs involve the addition of a chemical group, following protein translation, regulating protein behaviour in the cell. PTMs are usually reversible (Hannoun et al., 2010a) and our studies centred on the small ubiquitin-like modifier (SUMO).
SUMOylation is a widely studied modification that elicits a wide range of effects within the cell (Johnson, 2004). SUMO proteins are highly conserved in a large number of species and have been shown to be important in many eukaryotic cell processes (Hannoun et al., 2010a). Three homologues exist in mammals, SUMO1, SUMO2 and SUMO3. SUMO2 and SUMO3 share 95% homology with each other, but only share 50% homology with SUMO1 (Johnson, 2004) resulting in different biological activities. SUMO2 and SUMO3 possess the ability to form polySUMO chains via the lysine residue at the N-terminus consensus motif (Müller et al., 2001). SUMO1 lacks this consensus site and is therefore unable to form polySUMO chains (Kroetz, 2005) and often acts as a polySUMO chain terminator (Ulrich, 2009).
In order to study SUMOylation in a homologous system, we used an efficient and high-fidelity human embryonic stem cell (hESC) hepatocyte differentiation model (Hay et al., 2008). hESCs possess two important qualities: self-renewal and pluripotency (Thomson et al., 1998; Reubinoff et al., 2000). These properties allow large numbers of undifferentiated cells to be scaled up, prior to differentiation, generating a source of primary somatic cell derivatives from stable and known genotype. SUMO modification has already been shown to play an important role in both stem cell self-renewal and in somatic cells (Hannoun et al., 2010a). However, SUMOylation has not yet been studied in a stem-cell-derived developmental system. Our studies centred on the role of SUMOylation in the derivation of human hepatocytes from hESCs.
Given the importance of SUMOylation in cell biology, improving our understanding of this process is vital. Such studies will undoubtedly lead to an improved understanding of human development and cell physiology. Additionally, the development of high-fidelity cell-based resources from renewable sources, such as stem cells, has a significant role to play in many aspects of modern medicine (Hannoun et al., 2010b).
Results
SUMOylation during human embryonic stem cell self renewal and hepatocyte differentiation
In order to gain insight into the roles that post translational modifications play during hESC self-renewal and differentiation, protein SUMOylation was investigated. Human ESCs were differentiated into hepatocytes using a robust procedure (Hay et al., 2008) and cell extracts were harvested at various time points throughout the differentiation process. Cell lysates were separated by SDS-PAGE and probed for SUMO1, SUMO2, SENP7, hepatocyte nuclear factor 4α (HNF4α) and albumin, with β-actin used as a loading control (Fig. 1). The results demonstrated that high levels of SUMO1-, but not SUMO2-modified proteins were present in hESC populations undergoing self-renewal (Fig. 1A,B). The number of SUMO1-modified proteins decreased substantially as the cells differentiated towards the primitive streak (day 1) (Hay et al., 2008; Medine et al., 2011). By contrast, SUMO2 modification was increased at this time point. In both cases, SUMO1- and SUMO2-modified proteins were detected during definitive endoderm specification (days 2 and 3), whereas high levels of SUMO1-, but not SUMO2-modified proteins, were detected during hepatic specification (day 5). During hepatocyte maturation we observed a decrease in the levels of SUMO1- and SUMO2-modified proteins (days 7–17) (Fig. 1A,B). This observation was reinforced by increased expression of the SUMO-deconjugating enzyme SENP7 at the onset of hepatic maturation (Fig. 1C).
Fig. 1.
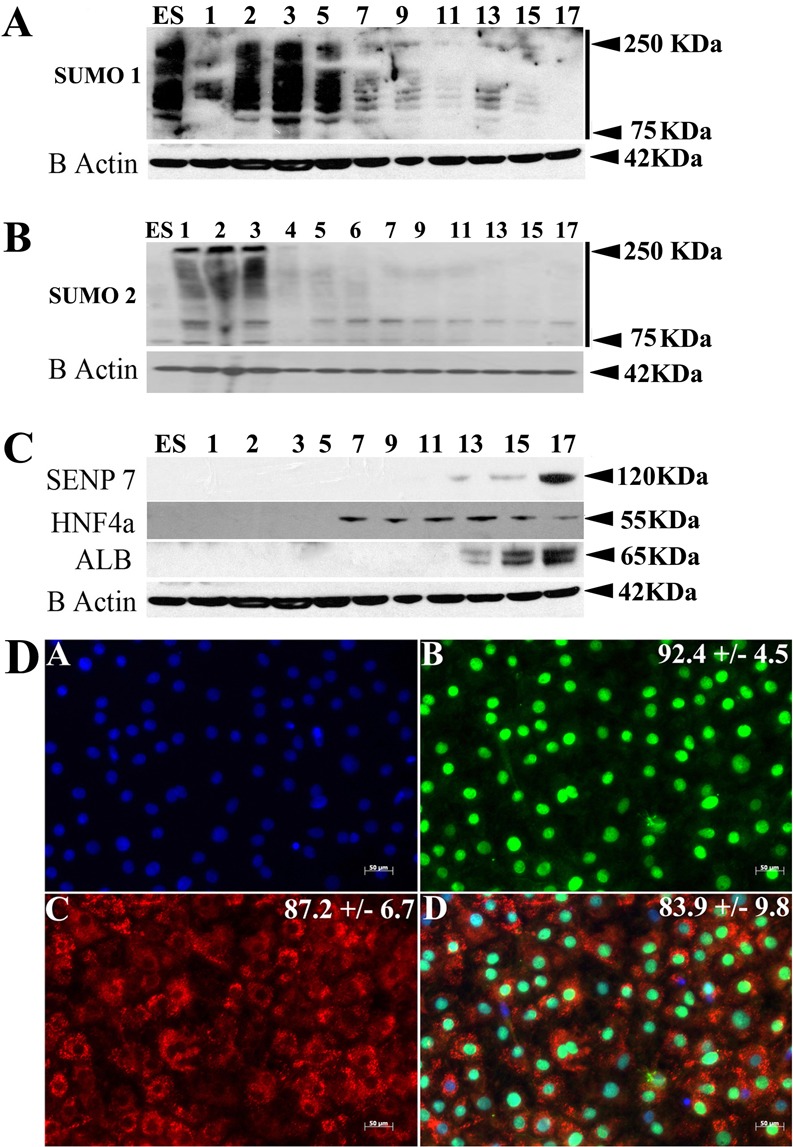
SUMOylation during human embryonic stem cell self-renewal and hepatocyte differentiation. (A,B) Cell extracts were collected at various time points throughout the differentiation procedure (days 0–17) and separated by SDS-PAGE. Western blots were probed for SUMO1, SUMO2, SENP7, hepatocyte nuclear factor 4 α (HNF4α), albumin (ALB) and β-actin. A decrease in SUMO-modified proteins is observed as the cells commit to hepatic endoderm. (C) During stem cell differentiation to hepatocytes we observed an increase in levels of the SUMO-deconjugating enzyme SENP7 and the serum protein albumin. We detected HNF4α expression from day 7, which decreased at days 15 and 17. (D) Day 17 hepatocytes were fixed and stained for DNA (A), HNF4α (B), albumin (C) and overlaid (D). The percentage of positive cells (shown top right on each panel) was calculated from five random fields of view and are quoted ± 1 s.d.
At the point of hepatic commitment (day 7), and in line with human liver development, we observed production of HNF4α. As cellular differentiation progressed, hESC-derived hepatocytes matured and expressed increasing levels of albumin. This was paralleled by decreasing levels of HNF4α (Fig. 1C) and merited further investigation. The reduction of HNF4α protein stability was also accompanied by a decrease in HNF4α message and was observed for all six isoforms of HNF4α during hepatic differentiation (supplementary material Fig. S1). As HNF4α was expressed ubiquitously in hESC-derived hepatocytes (92.4±4.5%) and in most albumin positive cells (83.9±9.8%), the reduction in HNF4α expression was most likely exhibited by all cells in our populations.
Identification and mapping of HNF4α SUMO modification sites in vitro
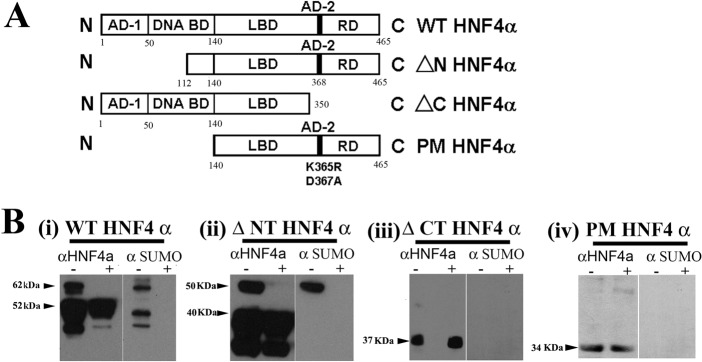
To assess whether SUMO modification was likely to play a role in HNF4α biology we examined the HNF4α amino acid sequence for potential SUMO modification consensus sites. We identified two potential SUMO modification sites. One site was present in the amino terminus and one in the carboxy terminus. In order to determine whether these sites were used during HNF4α SUMOylation we performed deletion analysis. N- and C-terminus deletion mutants of HNF4α were cloned, expressed and purified (Fig. 2A). Subsequently, we assessed the ability of these proteins to undergo SUMOylation using an established in vitro SUMOylation assay (Tatham et al., 2008; Shen et al., 2009). The data presented in Fig. 2B confirmed that HNF4α was modified by SUMO (Fig. 2Bi) and that the SUMOylation site used in this process was not found on the N-terminus (Fig. 2Bii) but on the C-terminus (Fig. 2Biii). These experiments were controlled by the inclusion of a SUMO specific protease control (denoted +) which cleaved SUMO modified HNF4α (Fig. 2B). In order to confirm the absolute requirement for the C-terminal consensus site in this process, we cloned truncated versions of HNF4α, which contained the wild-type or point-mutated SUMO consensus motif Ψ-K-x-D/E at residues 364–367. In the point mutant, the lysine residue at position 365 was mutated to an arginine residue and the aspartic acid residue 367 was mutated to an alanine residue using site-directed mutagenesis. The protein was then expressed, purified and used in our in vitro SUMO assay as before. We did not detect SUMO modification of the point mutated HNF4α indicating that HNF4α SUMOylation proceeds through the SUMO consensus site at the C-terminus in vitro.
Fig. 2.
Mapping SUMO modification of HNF4α in vitro. (A) HNF4α WT and the various deletion mutants were generated in order to identify the region of SUMO modification in HNF4α. HNF4α contains a functional domain structure, which includes the activation domains (AD-1 and AD-2), the DNA binding domain (DNA BD), the ligand binding domain (LBD) and the repression domain (RD). (B) The in vitro SUMO assay contains the specified form of HNF4α and the SUMO conjugation machinery (E1-SAE1/2 and E2-Ubc9). The use of the deletion constructs and the point mutant (Lys365 to Arg365 and Asp367 to Ala367) demonstrates that the C-terminus in HNF4α is absolutely required for SUMOylation in vitro. The control reaction contained the SUMO-deconjugating enzyme SENP1. SUMO conjugation resulted in a ∼10 kDa shift in protein mass, whereas SUMO deconjugation was measured by the disappearance of the band in the presence of SENP1.
SUMO modification of HNF4α during hepatocellular differentiation
In order to establish whether HNF4α was SUMO modified during hepatocyte differentiation, we collected and analysed cell lysates by immunoprecipitation. HNF4α was specifically purified from cell extracts using HNF4α antibody covalently to crosslinked Protein-G–Sepharose beads and compared to an IgG isotype crosslinked control throughout (Fig. 3). Immunoprecipitated complexes were separated by SDS-PAGE, Western blotted and probed for SUMO1 and SUMO2 using specific antisera. HNF4α was modified with SUMO2, but not SUMO1 during hESC hepatocellular differentiation. Interestingly, as the levels of HNF4α decreased during the differentiation process (Fig. 1B), there was an increase in the level of SUMO2-modified HNF4α, indicating that SUMO2 may regulate HNF4α stability (Fig. 3).
Fig. 3.
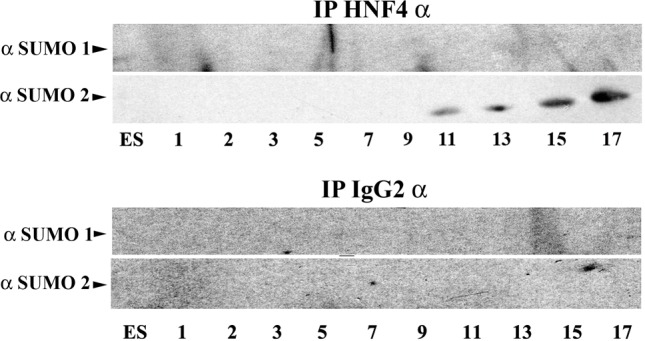
SUMO modification of HNF4α during hepatocellular differentiation. hESCs were differentiated to hepatocytes and samples were harvested at the time points indicated. HNF4α was pulled down using HNF4α antibody covalently crosslinked to Protein-G–Sepharose. An IgG isotype was used as a control throughout. Following incubation and extensive washing, the beads were eluted, separated by SDS-PAGE, western blotted and probed for SUMO1 and SUMO2. We did not detect SUMO1 modification of HNF4α during the differentiation processes. By contrast, we observed an increase in HNF4α SUMO2 modification as differentiation progressed. Our negative control IgG demonstrated that our assay was operating specifically throughout.
PolySUMOylation marks HNF4α for RNF4-mediated ubiquitination in vitro
Recent developments in the field have shown that polySUMOylation can serve as a template for RNF4-mediated ubiquitination and protein degradation (Tatham et al., 2008). RNF4 is an ubiquitin E3 ligase with specific SUMO interaction motifs (SIM) that facilitate its interaction and ubiquitin transfer to polySUMOylated proteins (Sun et al., 2007). In order to study this in vitro, HNF4α was used in its native state or conjugated with SUMO1 or SUMO2 (Fig. 4A). Subsequently, HNF4α was incubated in the presence or absence of RNF4, and ubiquitin. Three hours post incubation HNF4α was immunoprecipitated, separated by SDS-PAGE, probed for ubiquitin and analysed by densitometry (Fig. 4B). RNF4-mediated HNF4α ubiquitination was more than sixfold greater for polySUMOylated (SUMO2) and twofold greater for monoSUMOylated (SUMO1) over unmodified HNF4α controls (Fig. 4C).
Fig. 4.
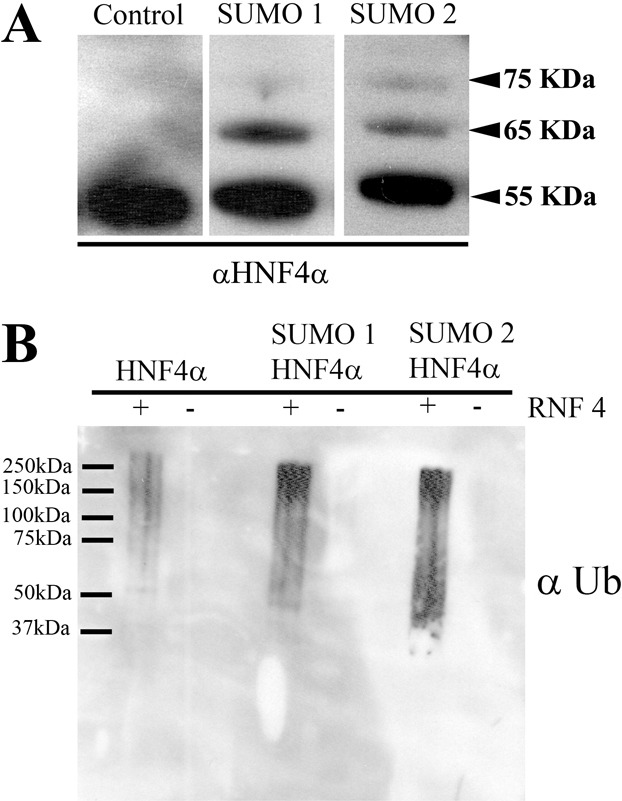
PolySUMOylation of HNF4α enhances RNF4-mediated ubiquitination in vitro. (A) In order to study HNF4α ubiquitin modification in vitro we employed an in vitro ubiquitination assay. WT HNF4α was mono- and polySUMOylated using SUMO1 and SUMO2 but absent from the control lane. (B) SUMOylated HNF4α protein was incubated with ubiquitination machinery (E1 and E2) in the presence and absence of RNF4 (E3). Following assay completion, HNF4α was immunoprecipitated and ubiquitin conjugation analysed using western blotting. Ubiquitination only occurred in the presence of RNF4 and was increased when HNF4α was polySUMOylated. (C) Densitometry analysis was performed using ImageJ software. The level of ubiquitination was increased more than sixfold over the control when HNF4α was polySUMOylated and twofold when HNF4α was monoSUMOylated.
HNF4α stability is regulated by ubiquitination during hepatocellular maturation
To further investigate the effect of polySUMOylation and ubiquitination on HNF4α biology, we followed the expression of RNF4 during hESC differentiation. We observed an upregulation of RNF4 from days 5–17 (Fig. 5A) indicating that RNF4 was expressed in our model at the time that HNF4α was degraded. To test the hypothesis that protein ubiquitination was responsible for regulating HNF4α stability, we incubated day 17 hESC-derived hepatocytes in the presence of proteasome 26S inhibitor (P26S), MG132 (Fiedler et al., 1998). Cell extracts were harvested from two independent differentiation experiments and analysed for HNF4α stability using western blotting and actin-controlled densitometry (Fig. 5B,C). In the presence of MG132, HNF4α levels were increased about fourfold, demonstrating that ubiquitin-mediated proteolysis is an important process that regulates the stability of HNF4α in the cell (Fig. 5B,C). Moreover, we detected a doublet at ∼75 kDa, indicating the likely preservation of SUMO2 and ubiquitin-modified HNF4α in the absence of P26S activity.
Fig. 5.
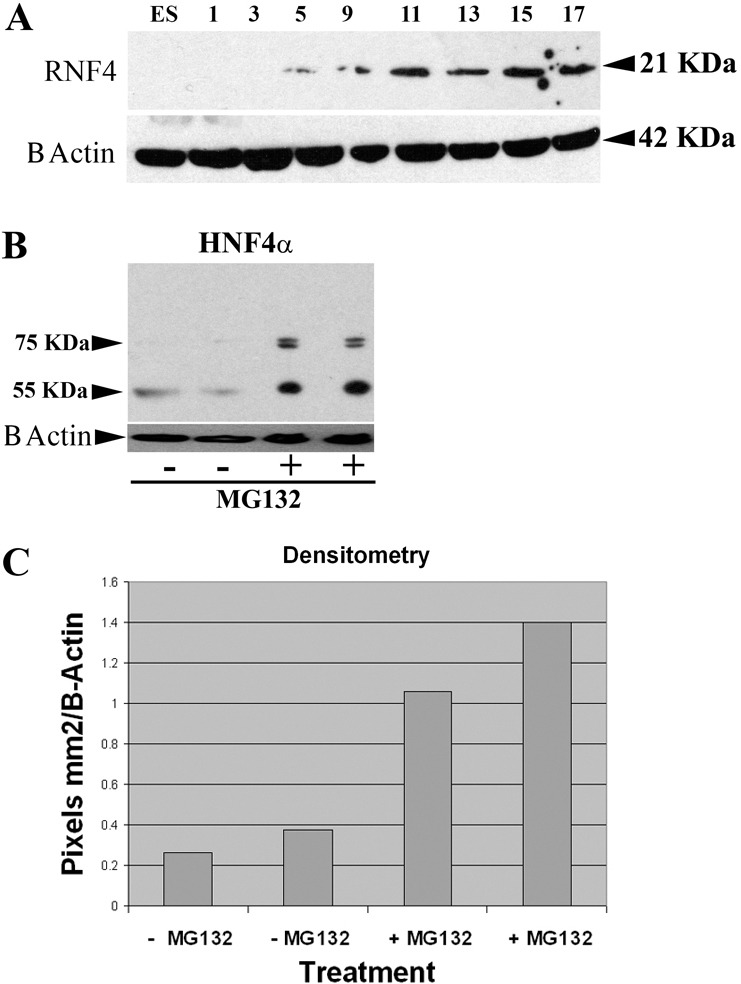
HNF4α is a target for ubiquitin-mediated degradation. (A) Because RNF4 plays an important role in modulating HNF4α stability in vitro, we monitored its expression during human embryonic stem cell differentiation. RNF4 expression was observed and increased during hepatic differentiation and maturation. (B,C) To assess whether ubiquitin-mediated proteolysis played a central role in the stability of HNF4α, we inhibited the 26S proteasome using MG132. hESC-derived hepatocytes were incubated with or without MG132 for 4 hours and cell extracts were collected, separated, western blotted and probed for HNF4α. HNF4α levels increased ∼fourfold in the presence of MG132, as determined by actin-controlled densitometry.
Loss of HNF4α results in reduced hepatocellular function and increased oxidative stress
In order to study the effect of HNF4α proteolysis on hepatocyte biology, we studied liver exocrine and endocrine function. We assayed for drug bio-transformation (Cyp3A) function and serum protein production (TTR) from days 15 to 25 during cellular differentiation. Cyp3A function was detected at day 15 and peaked at day 17 and then decreased until day 25 (Fig. 6A). A similar pattern was observed for TTR with peak function detected at day 15 and decreasing thereafter (Fig. 6B). These results demonstrate that a drop-off in Cyp3A and TTR function was in line with the loss of HNF4α in our system. In addition to the decrease of hepatocellular function, we also detected an increase in cellular oxidative stress as cells lost HNF4α expression and de-differentiated in culture. This was in line with previous studies (Marcil et al., 2010) (Fig. 6C).
Fig. 6.
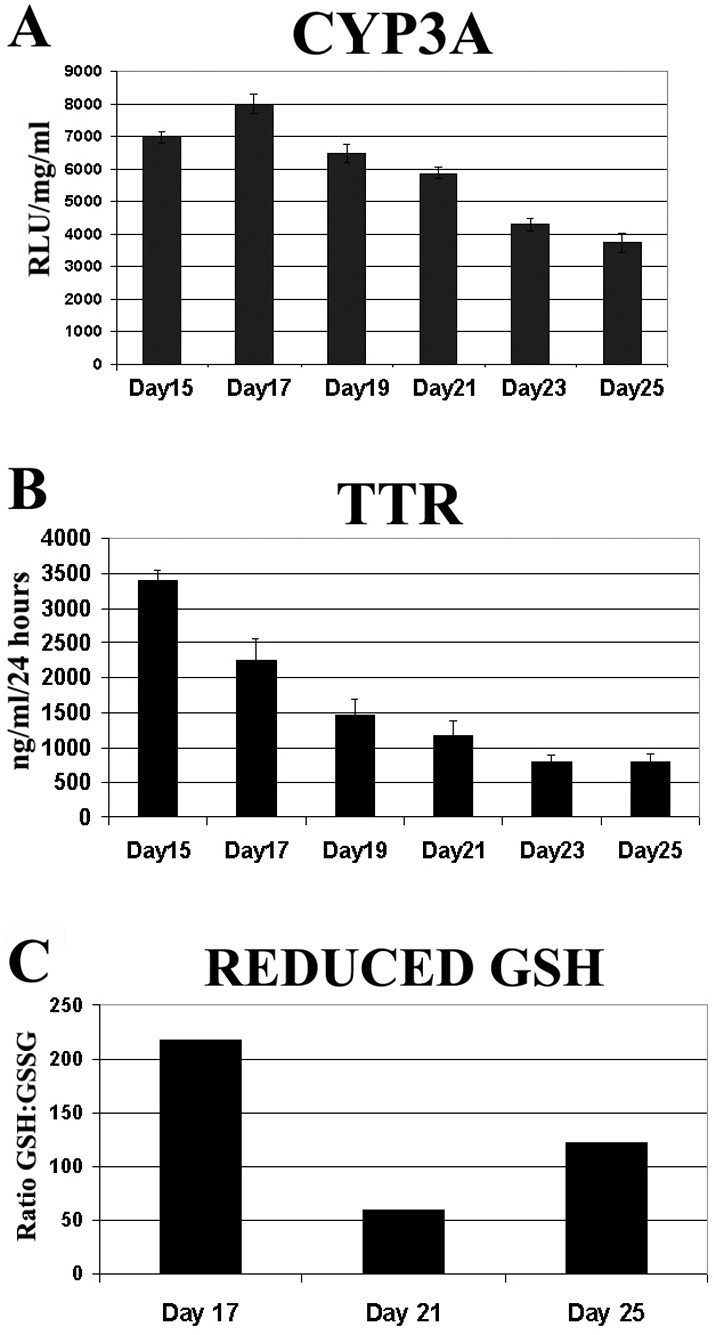
Loss of HNF4α leads to decreased hepatocellular function and oxidative state. (A) Cyp3A metabolic activity of hESC-derived hepatocytes was measured on days 15–25 using CYP3A pGlo substrate. 5 hours post treatment, CYP3A activity was measured on a luminometer. Units of activity are expressed as relative light units (RLU)/mg protein/ml (n = 6). Error bars represent 1 s.d. (B) hESC-derived hepatocytes (days 15–25) were cultured in six-well plates with 1 ml L-15 for 24 hours before culture supernatants were collected and TTR secretion was measured by ELISA. Error bars represent 1 s.d. (C) The redox state of hESC-derived hepatocytes was measured between days 17–25. hESC-derived hepatocytes were incubated with L-15 supplemented with GSH/GSSG pGlo substrate and activity was measured on a luminometer.
Discussion
In order to coordinate human development and maintain cell phenotype in the neonate, it is necessary to tightly regulate gene expression, translation and protein biology. Our studies focussed on protein post-translational modifications. Using a bona fide human model, we demonstrate that protein modification by SUMO is important during hESC self-renewal and hepatic differentiation (Fig. 1). During differentiation, we observed the differential expression and stability of a key hepatocyte transcription factor, HNF4α (Fig. 1, supplementary material Fig. S1). We reasoned that SUMO modification may be regulating HNF4α stability and function. We identified two potential SUMOylation sites; one located in the N-terminus and one at the C-terminus of the protein. Through deletion and point mutation analysis we determined that the critical region for HNF4α modification was at the consensus site found in the C-terminus (Fig. 2). While HNF4α SUMOylation took place in vitro, immune precipitation experiments confirmed that HNF4α was SUMO modified during cellular differentiation and resulted in decreased HNF4α stability (Fig. 3). This process was regulated by ubiquitin-mediated proteolysis (Figs 4, 5) and preceded hepatocyte de-differentiation (Fig. 6).
HNF4α is a member of the nuclear receptor family (NR) of transcription factors. HNF4α possesses a modular structure containing two activation domains (AD-1 and AD-2), a DNA binding domain, a ligand binding (LBD), a dimerization domain and repression domain (Fig. 2) (Bogan et al., 2000). The AD-2 transactivation domain (Dhe-Paganon et al., 2002) was of particular interest in our studies (residues 360–368) because this contained the SUMO consensus motif. The AD-2 domain lies adjacent to another important domain of HNF4α, the ligand binding domain (LBD). The LBD plays a pivotal role in transcriptional activity and has two distinct conformations, open and closed (Bogan et al., 2000). The difference between these forms is in the position of helix α12 (the AD-2 domain). In the open conformation, α12 is extended co-linearly with helices α10 and α11, rendering the ligand-binding pocket accessible. The open structure does not allow the binding of co-activator molecules, therefore it is considered to be the inactive form. In the closed conformation, the α12 helix folds back onto the LBD, sealing the ligand-binding pocket (Bogan et al., 2000). In this form, HNF4α can interact with co-activators, and is considered to be in the active state (Duda et al., 2004). Therefore, it is conceivable that SUMOylation of HNF4α's AD2 may directly affect the conformation of LBD and increase HNF4α transcriptional activity through co-factor binding. Consistent with this, we demonstrate increasing albumin production as HNF4α is increasingly modified by SUMO-2 (Figs 1,3). In addition to the LBD, the RD in the C-terminus also plays a role in HNF4α transcriptional activity and is sufficient by itself to repress the activity of the AD-2 domain (Iyemere et al., 1998). Therefore SUMOylation at AD-2 may also play an important role in displacing the RD and enhancing HNF4α transcriptional activity.
In conclusion, our investigation has provided evidence that SUMOylation of HNF4α regulates protein stability and potentially transcriptional activity. Dysregulation of this process results in the loss of HNF4α and hepatic function, demonstrating its importance in maintaining the hepatocellular phenotype. We believe our studies are novel and will stimulate interest in the differentiation and phenotypic regulation of other cell types.
Materials and Methods
hESC culture
H1 hESCs were cultured as previously described (Fletcher et al., 2008; Hay et al., 2011). H1 hESCs were differentiated into HE as described (Hay et al., 2008) and characterised using standard criteria (Hannoun et al., 2010c).
hESC-derived hepatocyte characterisation
Stem-cell-derived hepatocyte Cyp3A function and stem-cell-derived hepatocyte redox state was determined as per the manufacturer's instructions (Promega; http://www.promega.com/resources/protocols/technical-bulletins/101/p450-glo-assays-protocol/ and http://www.promega.com/products/cell-health-assays/glutathione-measurement/gsh_gssg_glo-assay/). Stem-cell-derived hepatocyte TTR secretion was determined as previously described (Hay et al., 2008). HNF4α and albumin immunostaining were performed as previously described (Medine et al., 2012).
Solexa
RNA was isolated from differentiating hESCs at days 0, 3, 10 and 17. Fetal and adult hepatocyte RNA was also used. The cDNA obtained from the purified RNA was initially cleaved to a uniform length between 200 and 500 bases and then tagged using unique adapters and sequenced. The data was then analysed and normalised to allow comparison of gene expression between the samples. Two types for normalisation were used. First, the raw count of reads for each gene was divided by the total number of reads aligned and multiplied by one million, thus accounting for the depth of sequencing. Second, the results were divided by the length of the gene multiplied by one thousand to obtain a read per kilo bases per million reads value (RPKM) value.
Cellular protein extraction
Cells grown in a six-well plate were lysed in 150 µl of SUMO lysis buffer [2% SDS, 50 mM Tris-HCl, pH 8, 1 mM ethylenediaminetetraacetic acid (EDTA) and 10 mM iodoacetamide (all from Sigma-Aldrich)] for 5 minutes at room temperature. The cell extracts were sonicated and stored at −80°C for later use.
Western blotting
Protein concentrations were measured using the standard BCA assay (Pierce). 40 μg of protein was then used for western blotting. The Nu-Page SURE Lock system from Invitrogen was employed as per the manufacturer's instructions. The membranes were blocked for 1 hour at room temperature with PBS-Tween containing 10% non-fat milk. They were then probed with primary antibody at 4°C overnight under constant rotation. The secondary antibody was then incubated for 1 hour at room temperature, and detected using enhanced chemiluminescence (ECL, Pierce). Details of antibodies used and sources are provided in supplementary material Table S1.
Crosslinking antibodies
HNF4α (Santa Cruz) was crosslinked to Protein G beads (Sigma) as previously described (Harlow, 1999).
Immunoprecipitation
The cell extract was diluted in incubation buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM EDTA, 2 mM DTT and 1% NP40). The cell extract was incubated with the 1.5 µg of the HNF4α antibody (Santa Cruz Biotechnology) and a rabbit IgG (Santa Cruz Biotechnology) was used as a control, for 3 hours at 4°C. Protein G beads (Sigma-Aldrich) were prepared in NP40 buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM EDTA and 1% NP40, Sigma-Aldrich). 20 µl of beads were added to each sample and incubated overnight at 4°C under constant rotation. The samples were spun down at 3500 r.p.m. for 2 minutes, washed three times with the incubation buffer and eluted with 2× Disruption Buffer at 100°C for 2 minutes. The samples were then centrifuged at 13,000 r.p.m. for 5 minutes and analysed by western blot with antibodies against SUMO1 and SUMO2.
Protein expression and purification
HNF4α wild-type and an N-terminal deletion mutant were kindly provided to us by Primorigen Inc. The C-terminal deletions and the point mutant versions of the HNF4α were generated by amplifying the respective fragment using primer specific PCR The fragments were cloned into the pET15b Vector (Novagen), using specific BamHI and NdeI restriction sites. All mutants contained a His tag to confirm the expression and aid in the purification process. The constructs were then transformed into BL21 E. coli (Stratagene) and selected clones were grown at 37°C until they reached an optical density of 0.5. At this point the cultures were induced with 0.5 mM IPTG (Invitrogen) for 1 hour. After induction the cells were spun down, lysed and purified as previously described (Hoffmann and Roeder, 1991). Point Mutagenesis was carried out as directed by the QuikChange II manual (Stratagene), the protein was expressed and purified as described above.
In vitro SUMOylation assay
3 µg of each HNF4α variant was conjugated with the SUMO1 and SUMO2 proteins as previously described (Tatham et al., 2008). The SUMO deconjugation was carried out as previously described (Shen et al., 2009).
In vitro ubiquitination assay
1 µg of HNF4α was either mono or polySUMOylated as described above. The ubiquitination machinery (8 µM of ubiquitin, 40 nM Uba1, 0.7 µM UbcH5a, 0.5 µM RNF4) was added to the reaction to make a total volume of 50 µl under the following conditions: 50 mM Tris-HCl, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, 2 mM ATP and 0.1% NP40. The reaction was incubated at 37°C for 3–4 hours. The HNF4α was immunoprecipitated out of the reaction using the crosslinked beads specific to HNF4α and the eluate was analysed using western blotting.
Supplementary Material
Footnotes
Funding
W.Z. was supported by a scholarship from the Chinese government. Z.H. was supported by an MRC PhD studentship. D.H. was supported by a Research Councils UK (RCUK) fellowship. S.G. was supported by an award from Genomia. C.M. was supported by an award from Edinburgh Bio-Quarter. R.H. and E.J. were supported by CRUK. J.I. was supported by an MRC Programme Grant. These studies also benefited from a generous donation from Emmet and Toni Stephenson, Doctors.com. Deposited in PMC for immediate release.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.102889/-/DC1
References
- Bogan A. A., Dallas–Yang Q., Ruse M. D., Jr, Maeda Y., Jiang G., Nepomuceno L., Scanlan T. S., Cohen F. E., Sladek F. M. (2000). Analysis of protein dimerization and ligand binding of orphan receptor HNF4alpha. J. Mol. Biol. 302, 831–851 10.1006/jmbi.2000.4099 [DOI] [PubMed] [Google Scholar]
- Chen K., Rajewsky N. (2007). The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 8, 93–103 10.1038/nrg1990 [DOI] [PubMed] [Google Scholar]
- Dhe–Paganon S., Duda K., Iwamoto M., Chi Y. I., Shoelson S. E. (2002). Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J. Biol. Chem. 277, 37973–37976 10.1074/jbc.C200420200 [DOI] [PubMed] [Google Scholar]
- Duda K., Chi Y. I., Shoelson S. E. (2004). Structural basis for HNF-4alpha activation by ligand and coactivator binding. J. Biol. Chem. 279, 23311–23316 10.1074/jbc.M400864200 [DOI] [PubMed] [Google Scholar]
- Fiedler M. A., Wernke–Dollries K., Stark J. M. (1998). Inhibition of TNF-alpha-induced NF-kappaB activation and IL-8 release in A549 cells with the proteasome inhibitor MG-132. Am. J. Respir. Cell Mol. Biol. 19, 259–268 [DOI] [PubMed] [Google Scholar]
- Fletcher J., Cui W., Samuel K., Black J. R., Hannoun Z., Currie I. S., Terrace J. D., Payne C., Filippi C., Newsome P., et al. (2008). The inhibitory role of stromal cell mesenchyme on human embryonic stem cell hepatocyte differentiation is overcome by Wnt3a treatment. Cloning Stem Cells 10, 331–340 10.1089/clo.2007.0094 [DOI] [PubMed] [Google Scholar]
- Hannoun Z., Greenhough S., Jaffray E., Hay R. T., Hay D. C. (2010a). Post-translational modification by SUMO. Toxicology 278, 288–293 10.1016/j.tox.2010.07.013 [DOI] [PubMed] [Google Scholar]
- Hannoun Z., Filippi C., Sullivan G., Hay D. C., Iredale J. P. (2010b). Hepatic endoderm differentiation from human embryonic stem cells. Curr. Stem Cell Res. Ther. 5, 233–244 10.2174/157488810791824403 [DOI] [PubMed] [Google Scholar]
- Hannoun Z., Fletcher J., Greenhough S., Medine C., Samuel K., Sharma R., Pryde A., Black J. R., Ross J. A., Wilmut I., et al. (2010c). The comparison between conditioned media and serum-free media in human embryonic stem cell culture and differentiation. Cell. Reprogram. 12, 133–140 10.1089/cell.2009.0099 [DOI] [PubMed] [Google Scholar]
- Hay D. C., Fletcher J., Payne C., Terrace J. D., Gallagher R. C., Snoeys J., Black J. R., Wojtacha D., Samuel K., Hannoun Z., et al. (2008). Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc. Natl. Acad. Sci. USA 105, 12301–12306 10.1073/pnas.0806522105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D. C., Pernagallo S., Diaz–Mochon J. J., Medine C. N., Greenhough S., Hannoun Z., Schrader J., Black J. R., Fletcher J., Dalgetty D., et al. (2011). Unbiased screening of polymer libraries to define novel substrates for functional hepatocytes with inducible drug metabolism. Stem Cell Res. (Amst.) 6, 92–102 10.1016/j.scr.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Roeder R. G. (1991). Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 19, 6337–6338 10.1093/nar/19.22.6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyemere V. P., Davies N. H., Brownlee G. G. (1998). The activation function 2 domain of hepatic nuclear factor 4 is regulated by a short C-terminal proline-rich repressor domain. Nucleic Acids Res. 26, 2098–2104 10.1093/nar/26.9.2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Kroetz M. B. (2005). SUMO: a ubiquitin-like protein modifier. Yale J. Biol. Med. 78, 197–201 [PMC free article] [PubMed] [Google Scholar]
- Marcil V., Seidman E., Sinnett D., Boudreau F., Gendron F. P., Beaulieu J. F., Ménard D., Precourt L. P., Amre D., Levy E. (2010). Modification in oxidative stress, inflammation, and lipoprotein assembly in response to hepatocyte nuclear factor 4alpha knockdown in intestinal epithelial cells. J. Biol. Chem. 285, 40448–40460 10.1074/jbc.M110.155358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medine C N., Lucendo–Villarin B., Zhou W., West C C., Hay D C. (2010). Robust generation of metabolically active hepatocytes from pluripotent stem cells. J. Vis. Exp. 56, e2969 10.3791/2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medine C. N., Hannoun Z., Greenhough S., Payne C. M., Fletcher J., Hay D. C. (2012) Deriving Metabolically Active Hepatic Endoderm from Pluripotent Stem Cells. Human Embryonic and Induced Pluripotent Stem Cells. Part 4 (ed. Ye K, Jin S.), pp. 369–386 New York: Springer Science and Business Media [Google Scholar]
- Müller S., Hoege C., Pyrowolakis G., Jentsch S. (2001). SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2, 202–213 10.1038/35056591 [DOI] [PubMed] [Google Scholar]
- Reubinoff B. E., Pera M. F., Fong C. Y., Trounson A., Bongso A. (2000). Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 18, 399–404 10.1038/74447 [DOI] [PubMed] [Google Scholar]
- Shen L. N., Geoffroy M. C., Jaffray E. G., Hay R. T. (2009). Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem. J. 421, 223–230 10.1042/BJ20090246 [DOI] [PubMed] [Google Scholar]
- Sun H., Leverson J. D., Hunter T. (2007). Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 10.1038/sj.emboj.7601839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008). RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 10.1038/ncb1716 [DOI] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz–Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Ulrich H. D. (2009). The SUMO system: an overview. Methods Mol. Biol. 497, 3–16 10.1007/978-1-59745-566-4_1 [DOI] [PubMed] [Google Scholar]
- Vaillancourt C., Lafond J. (2009). Human embryogenesis: overview. Methods Mol. Biol. 550, 3–7 10.1007/978-1-60327-009-0_1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.