Abstract
Iron (Fe) is an essential element for living organisms. However, under aerobic conditions, its use is complicated because of its high insolubility and its potential toxicity through reactivity with reduced forms of oxygen. In plants, Fe overload can lead to intracellular concentrations beyond the storage and detoxification capacities of cells. Such a displacement toward a pro-oxidant state can activate antioxidant defenses, including Fe-mediated expression of ascorbate peroxidase genes. In this work, we demonstrate that Fe overload specifically induces the AtAPX1 gene encoding a cytosolic ascorbate peroxidase in Arabidopsis leaves. The strong constitutive expression of the AtAPX1 gene in roots is unaffected by Fe and depends on the first 5′-untranslated region intron. Presence of an AtAPX1 expressed sequence tag in the Arabidopsis database, longer in its 5′ region than what could be predicted from the published AtAPX1transcription initiation site, leads to define a new transcription initiation region for this gene. A minimal promoter sequence enabling Fe-induced expression of the AtAPX1 gene is defined by following expression of various AtAPX1::β-glucuronidase constructs in transformed Arabidopsis plantlets. This 118-bp minimal promoter sequence contains an Fe-dependent regulatory sequence-like cis-element known to be necessary for maize (Zea mays) and Arabidopsis ferritin gene derepression in response to Fe overload. Site-directed mutagenesis of this element within the AtAPX1 promoter sequence does not abolish the Fe-dependent activation of a reporter gene, indicating that it is likely not involved in the Fe-regulated expression of the AtAPX1 gene.
Oxygen and iron (Fe) are both essential and noxious for all aerobic organisms. Reactive oxygen species (ROS), which are by-products of the oxygen consumption, are responsible for degradation of proteins, lipids, and nucleic acids and are thought to play a major role in aging and cell death. The most ROS is hydroxyl radical, which is produced from hydrogen peroxide (H2O2) in the presence of metals such as Fe, copper (Cu), and manganese through the Haber-Weiss reaction (for review, see Briat, 2002). Therefore, metals potentiate the toxicity of ROS. To avoid the accumulation of these toxic compounds to an unacceptable level, animals and plants possess several detoxifying enzymatic systems. Superoxide dismutases and catalases are found in animals, plants, and microorganisms, whereas ascorbate peroxidase (APX; EC 1.11.1.11) is a plant-specific H2O2-scavenging enzyme. APX utilizes ascorbate as its specific electron donor to reduce H2O2 to water with the concomitant generation of mono-dehydroascorbate. APX isoforms have been described in various plant cell compartments including cytosol, chloroplasts, mitochondria, and peroxisomes. These isoforms result both from differential expression of the various members of a multigene family and from posttranscriptional regulation such as alternative splicing involved in the production of chloroplast APX isozymes (for review, see Shigeoka et al., 2002.). APX gene regulation in response to biotic and abiotic stresses has received increased attention these last years. In particular, the expression of cytosolic APX has been shown to be regulated in response to several oxidative stress factors, revealing the presence of some functional cis-regulatory elements in its promoter (Shigeoka et al., 2002). For example, a heat shock response element has been described in the cytosolic APX genes (cAPX) of all plant species scrutinized so far, and it has been shown to be required in the in vivo heat shock induction of the AtAPX1 gene from Arabidopsis (Storozhenko et al., 1998). In addition, Gadea et al. (1999) have demonstrated that the leader intron observed in the 5′-untranslated region (UTR) of a tomato (Lycopersicon esculentum) cAPX gene is required to confer constitutive gene expression in leaves but not in other plant organs.
Metal ion overload can lead to intracellular concentrations beyond the storage and detoxification capacities of plant cells, and such a displacement of the balance toward a pro-oxidant state activates antioxidant defenses. This direct link between metal ion toxicity and activation of responses against oxidative stress is poorly documented. It has been shown that artificial Fe overload of Nicotiana plumbaginifolia doubled APX and catalase activities in the leaves (Kampfenkel et al., 1995). Also, characterization of the tomato chloronerva mutant revealed abnormally high levels of Fe and Cu in leaves, an increase in oxygen radical production, and activation of antioxidant enzymes (Pich and Scholz, 1993; Pich et al., 1994). Among these enzymes, cytosolic APX and Cu/Zn superoxide dismutase are more abundant in this mutant (Herbik et al., 1996). In a previous work, we demonstrated that APX mRNA abundance increases in response to Fe overload in cotyledons of Brassica napus (Vansuyt et al., 1997). Fe-induced expression of APX was confirmed recently for the bean (Phaseolus vulgaris) APX gene encoding a cytosolic form (Pekker et al., 2002). These authors showed that such a regulation operates through an Fe-mediated production of ROS, in a pathway involving gluthatione (GSH) as an intermediate signal.
In this work, we demonstrate that Fe overload specifically induces the expression of a cytosolic APX gene in Arabidopsis leaves, and we analyze the role of 5′-upstream sequences, including the first intron, in the control of the expression of this gene. The integration of ROS and GSH into the Fe response pathway is also discussed.
RESULTS
Tissue-Specific Expression and Fe-Induced Expression of the AtAPX1 Gene
We have demonstrated previously that APX mRNA abundance increased in response to Fe overload in cotyledons of B. napus (Vansuyt et al., 1997). These data were obtained using a nonspecific probe, making us unable to show precisely which member(s) of the APX family was turned on in response to the Fe treatment.
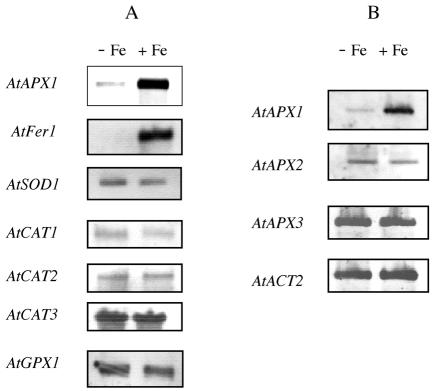
Using a 3′-UTR probe specific for the AtAPX1 gene from Arabidopsis, we observed that the corresponding transcript abundance was increased in response to Fe treatment of Arabidopsis cells (Fig. 1A). This response was specific because none of the three Arabidopsis catalase transcripts (AtCAT1–3), superoxide dismutase mRNA encoded by the AtSOD1 gene, or gluthatione peroxidase (AtGPX1) transcript had their abundance increased in response to Fe overload (Fig. 1A). In contrast and as a positive control, the AtFer1 gene, encoding one of the four Arabidopsis ferritins (Petit et al., 2001a), had its expression induced in response to the Fe treatment (Fig. 1A). Using derooted Arabidopsis plants to facilitate Fe loading (Lobréaux et al., 1995), it was also observed by semiquantitative RT-PCR that AtAPX1 transcript abundance increased in leaves in response to the treatment, as observed for cultured cells (Fig. 1B). Specificity of this response was confirmed within the APX family. Only AtAPX1 (GenBank accession no. X59600) was induced by Fe, whereas AtAPX2 (GenBank accession no. X98275), a soluble cytosolic APX involved in the photooxidative response (Karpinski et al., 1997), and AtAPX3 (GenBank accession no. X98003), a microsomal isoform of APX, were unaffected by the Fe treatment (Fig. 1B). As a control, the actin mRNA (AtACT2, GenBank accession no. U41998) abundance was unchanged whatever the Fe conditions used (Fig. 1B).
Figure 1.
Specific increased abundance of AtAPX1 mRNA in response to Fe treatment. A, Northern blot of cytosolic APX (AtAPX1), catalase 1 to 3, (AtCAT1–3), ferritin (AtFer1), superoxide dismutase (AtSOD1), and gluthatione peroxidase (AtGPX1) from cultured Arabidopsis cells treated or not with 500 μm Fe-citrate for 6 h. B, Reverse transcriptase (RT)-PCR of APX (AtAPX1–3) and of actin (ACT2) using RNA from Arabidopsis cotyledons treated or not with 500 μm Fe-citrate for 6 h.
Reexamination of the Transcriptional Start Site of the AtAPX1 Gene
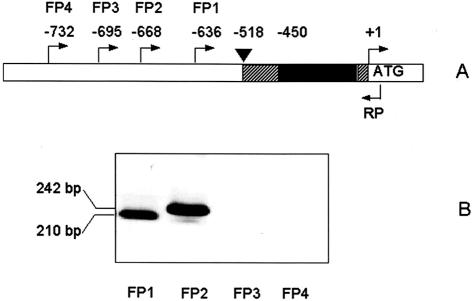
Transcription start site of the AtAPX1 gene was published to occur at position –518 in Figure 2 (Storozhenko et al., 1998), using the A of start codon as +1, in agreement with data reported by Kubo et al. (1993). Both of these mappings were quoted as data not shown in the corresponding publications. The occurrence in the Arabidopsis database of an expressed sequence tag (EST; GenBank accession no. R89992) that is longer in its 5′ region than what could be predicted from the published transcription initiation site (Fig. 2) caused a need for reinvestigation of this point. To analyze leaf AtAPX1 transcripts by RT-PCR, we designed oligonucleotides covering this region of the promoter sequence (FP1–4 in Fig. 3) and a common reverse primer (RP in Fig. 3) anchored within the AtAPX1 coding sequence. These RT-PCR reactions revealed amplified DNA fragments of 210 and of 242 bp when oligos FP1 and FP2 were used, respectively, as forward primers (Fig. 3). No amplified DNA fragments were observed when using FP3 and FP4. Sizes of the amplified DNA fragments when using FP1 and FP2 are consistent with amplification from a spliced AtAPX1 transcript lacking the 5′-UTR intron sequence. The sequence of a fragment obtained by PCR using the RP and FP2 primers showed that the intron sequence was absent in this processed transcript (data not shown). These data indicate that AtAPX1 transcript can extend 5′ up to the sequence region covered by oligo FP 2 (Fig. 3) but cannot reach the sequence region covered by oligo FP3. Thirty-three base pairs upstream of the oligo FP 2 sequence a putative TATA box (positions –703 to –708 in Fig. 2) can be observed, preceded by a CAAT box between positions –761 and –765 (Fig. 2). It indicates that transcription initiation of AtAPX1 occurs about 155 bp upstream from what has been reported previously (Storozhenko et al., 1998), and the three As found at position –669/–671 are good candidates to be initiator nucleotides. The same result has been obtained by using root RNA as template (data not shown).
Figure 2.
Nucleotide sequence upstream of the ATG of the AtAPX1 gene. One thousand kilobase pairs upstream of the ATG are shown. ▴, 5′ Positions of the Δ5 to Δ7 deletions used to drive β-glucuronidase (GUS) expression (see Figs. 6 and 7). ▾, Initiation start point defined by Storozhenko et al., 1998. ♦,5′ Extremity of the longest AtAPX1 EST recovered from the database (R89992). ▿, Putative initiation transcription sites define from the RT-PCR experiments described in Figure 3; they are found downstream TATA and CAT boxes (bold and underlined). The first intron sequence located in the 5′-UTR is indicated in italic and bold. In bold and underlined with stars is shown the heat shock cis-element (HSE). The nucleotide positions (in base pairs) are referred to the A of the ATG translation initiation codon as +1.
Figure 3.
RT-PCR analysis of the transcription initiation region of the AtAPX1 gene. A, Schematic representation of the AtAPX1 gene upstream region. ▪, First 5′-UTR intron. ▾, Position of the initiation transcription site according to (Storozhenko et al., 1998). FP 1 to 4, Forward primers used for RT-PCR. The numbers above indicate the position of the first nucleotide of the oligos, considering the A of the ATG translation initiation codon as +1. RP, Position of the common reverse primer located within the AtAPX1 coding sequence. B, RT-PCR performed with FP1 to 4 as forward primers and RP as reverse primer, using AtAPX1 leaf RNAs as template.
The First AtAPX1 Intron Contains Root-Specific Enhancer Element(s)
The AtAPX1 gene contains a 437-bp intron in its 5′-UTR (Fig. 2), reminiscent of the 718-bp leader intron found in the 5′-UTR of the tomato cytosolic APX20 gene (Gadea et al., 1999). In tomato, it was shown that this leader intron was required to confer constitutive gene expression in leaves but not in other organs of the plant (Gadea et al., 1999).
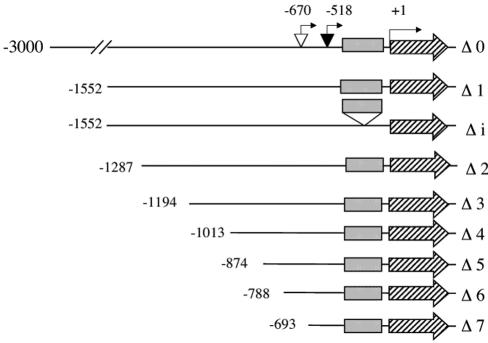
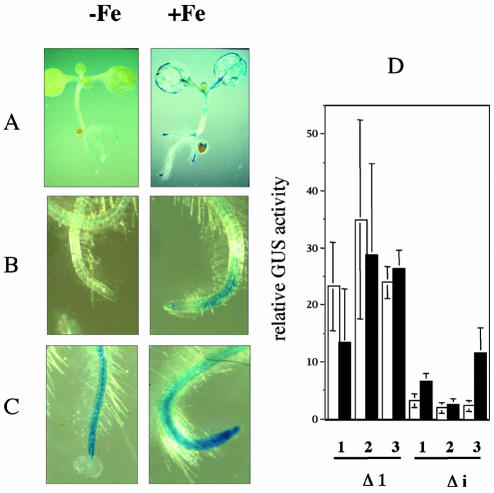
To demonstrate an eventual regulatory role of the first intron in the expression of the AtApx1 gene, we translationally fused the GUS reported gene to a 1,552-bp AtAPX1 DNA fragment located upstream from the ATG (Δ1 in Fig. 4) and to the same fragment deleted from the 5′-UTR intron (Δi in Fig. 4). As shown in Figures 5 and 6, young Arabidopsis plantlets transformed with the Δ1 construct exhibited a strong constitutive GUS staining in the roots whatever the Fe nutrition conditions, whereas the cotyledons were GUS stained only in the case of the Fe overload treatment. Constitutive expression of cAPX1 in Arabidopsis roots was confirmed by northern analysis (data not shown). In contrast, deletion of the leader intron (Fig. 5, A and B) resulted in an important loss of the constitutive root GUS staining, whereas the Fe induction was still observed in the cotyledons (Fig. 5A). However, the root zone just above the elongation tip became GUS stained in response to the Fe treatment (Fig. 5B), indicating that the AtAPX1 gene expression was induced by Fe in this peculiar part of the root and that this induction did not require the presence of the first intron. Quantification of the GUS activity in roots of three independent lines transformed with constructs containing or not containing the 5′-UTR intron confirmed the staining data (Fig. 5D). Constitutive GUS activity was, on average, 10-fold lower in absence of the intron. In presence of the intron, no induction of the GUS activity in response to Fe was observed. However, intron deletion enabled a low Fe-induced GUS activity in the root to be revealed, in agreement with the staining observed in response to Fe treatment in the zone just above the root tip (Fig. 5B).
Figure 4.
Scheme representing the various AtAPX1::GUS constructs used to determine cis-region(s) required for Fe-induced expression of the AtAPX1 gene. Δ0 is a 3-kb DNA fragment upstream of the AtAPX1 gene, fused to the GUS reporter gene. Δ1 to 7, 5′ Serial deletions generated from Δ0 (for details, see “Materials and Methods”). Δi, Δ1 with the 5′-UTR intron deleted. ▾, Position of the initiation transcription site according to Storozhenko et al., 1998. ▿, Position of the transcription initiation region as defined by experiments reported in Figure 3.
Figure 5.
Evidence for a root-specific enhancer sequence within the 5′-UTR intron. A, GUS staining of Arabidopsis plantlets transformed with the Δi construct (see Fig. 4) and grown with (+Fe) or without (–Fe) Fe excess. B, Root tip enlargement from a Δi plantlet grown with or without Fe excess. C, Same as B from Δ1 plantlets containing the 5′-UTR intron (see Fig. 4). D, Root GUS activity quantification of three independent lines transformed either by Δ1 or by Δi and grown without (white) or with (black) Fe excess. Each value is the mean of three independent measurements. Relative GUS activity is expressed as micrograms of Glc per hour per milligram of protein.
Figure 6.
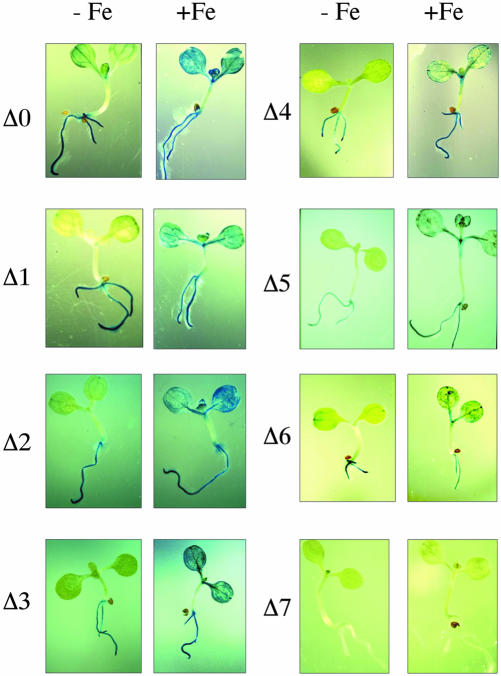
Histochemical GUS staining of 1-week-old Arabidopsis plantlets. transformed with the various AtAPX1::GUS constructs (except Δi) described in Figure 4 and treated (+ Fe) or not (–Fe) with 500 μm Fe-citrate
This result indicates that the 5′-UTR intron of the AtAPX1 gene is required to confer constitutive gene expression in Arabidopsis roots and is not necessary for its Fe overload-mediated expression. In contrast, elements upstream of the ATG codon, independent of the intron sequence, are necessary to promote reporter gene activity in response to the Fe treatment. This prompted us to perform a promoter sequence serial deletion analysis to define the DNA region responsible for the Fe response.
Characterization of a Minimal AtAPX1 Promoter Responsive to Fe Treatment
In addition to the Δ0 construct containing 3 kb of upstream AtAPX1 sequence fused to the GUS reporter gene, seven DNA fragments were fused to the GUS reporter gene of 1,552, 1,287, 1,194, 1,013, 874, 788, and 693 bp, respectively, upstream from the AtAPX1 ATG codon; these constructs were named Δ1 (–1,552) to Δ7 (–693; Fig. 4). The 5′ ends of the AtAPX1 DNA fragments corresponding to Δ5 to Δ7 are also indicated on the sequence presented in Figure 2. They all contain the 670 bp of the AtAPX1 5′-UTR including the leader intron and upstream promoter sequence of decreasing length, from 882 (Δ1) to 23 (Δ7) bp. GUS staining of the plantlets transformed with these constructs is presented in Figure 6, where it can be observed that the Fe induction of the GUS expression in cotyledons remains until deletion Δ6. Construct Δ7 has lost its ability to drive GUS expression, whatever the Fe nutrition conditions used. This is consistent with the fact that the putative TATA and CAAT boxes (Fig. 2) have been deleted and that only 23 bp upstream from the new initiation site described in Figure 4 are kept in the Δ7 construct.
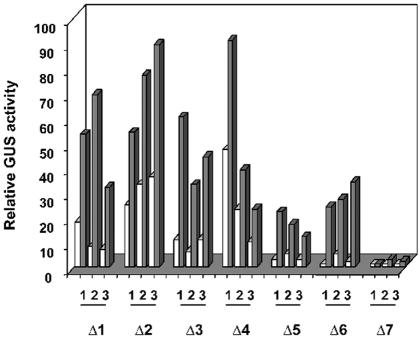
Quantification of these GUS activities has been performed on three independent transformed line plants for each construct, with or without Fe overload, and are presented in Figure 7. Δ1 to Δ6 constructs exhibited a marked Fe-induced GUS activity, whereas Δ7 construct, which lacks the putative TATA box, had almost no significant GUS activity. The Δ6 construct, which contains 118 bp of promoter sequence defines, therefore, a minimal AtAPX1 promoter able to induce the GUS activity by more than 10-fold.
Figure 7.
GUS activity quantification in Arabidopsis cotyledons of three independent transformed lines for each AtAPX1::GUS constructs described in Figure 4 (except Δi) and treated (black bars) or not (white bars) with 500 μm Fe-citrate. Relative GUS activity is expressed as micrograms of Glc per hour per milligram of protein. Each value is the mean of three independent measurements.
The Minimal AtAPX1 Promoter Responsive to Fe Treatment Contains a Nonfunctional Fe-Dependent Regulatory Sequence (IDRS)
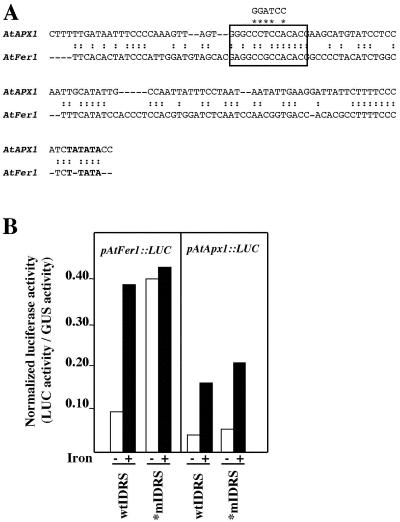
The IDRS is a 15-bp cis-element found within the upstream promoter sequence of the ZmFer1 and At-Fer1 maize (Zea mays) and Arabidopsis ferritin genes, respectively, and necessary for the repression of their expression under Fe-deficient conditions; Fe overload has been shown to derepress the expression of these genes (Petit et al., 2001b, Murgia et al., 2002). Comparison of the AtFer1 promoter sequence with the 5′ upstream sequence of the AtAPX1 gene reveals a partially conserved IDRS between positions –793 and –779 of the AtAPX1 gene (Figs. 2 and 8A). To test whether this sequence was required or not for the Fe-dependent expression of the AtAPX1 gene expression, it was mutated as indicated in Figure 8A. The minimal AtAPX1 promoter mutated within the IDRS was fused to the LUC reporter gene. This construct was transiently expressed in Arabidopsis cells Fe overloaded or not, in comparison with a construct containing the wild-type AtAPX1 minimal promoter. This procedure was previously successful to demonstrate the functionality of the ferritin IDRS (Petit et al., 2001b; Fig. 8B). Site-directed mutagenesis of the IDRS within the promoter sequence of AtAPX1 gene fused upstream of the LUC gene did not abolish the Fe-dependent activation of the reporter gene in such a transient expression assay, whereas mutagenesis of the AtFer1 IDRS confirmed the derepression of the expression of the LUC gene under Fe-deficient conditions (Fig. 8B). This indicates that the IDRS-like element observed within the AtAPX1 promoter is likely not involved in the Fe-regulated expression of the AtAPX1 gene.
Figure 8.
AtAPX1 IDRS-like analysis. A, Nucleotide sequence alignment of the promoter region from the Arabidopsis ferritin Fer1 gene (AtFer1) containing an IDRS (black framed box) with a similar region of the Arabidopsis APX1 gene (AtAPX1). Nucleotides that have been mutagenized within the AtAPX1 IDRS like to test its functionality in an Arabidopsis cell transient assay are indicated above and marked by stars. B, Effect of IDRS mutations (*mIDRS), comparatively with wild type IDRS (wtIDRS), on the Fe-induced expression of AtFer1::LUC and AtApx1::LUC fusions in a transient assay in the absence (–) or presence (+) of 500 μm Fe-citrate. Eight-day-old Arabidopsis cells were cotransformed with the pAtFer1::LUC or the pAtApx1::LUC plasmids and with the pRD109 plasmid expressing GUS under the control of a histone promoter as internal control and treated as previously described (Petit et al., 2001b). Values are the mean of three independent transformations from one representative experiment.
Is Fe Activation of AtAPX1 Gene Expression Mediated by an Oxidative Stress Pathway?
It has been recently demonstrated that induced expression of bean cAPX by high intracellular levels of free Fe requires oxygen and could use GSH as an intermediate signal (Pekker et al., 2002). It led these authors to conclude that ROS regulate the expression of cytosolic APX in bean, in response to an Fe-mediated oxidative stress, although H2O2 application to derooted bean plants did not affect Fe-mediated cAPX expression (Pekker et al., 2002). To investigate this point in Arabidopsis, we performed two types of experiments. First, various concentration of H2O2 (5, 10, and 20 mm) were added in presence of 50 μm Fe(III)-EDTA to the culture medium of Arabidopsis cell suspension cultures to promote more Fenton reaction. The abundance of AtAPX1 transcript was measured under such conditions and was found unchanged when compared with control cells untreated by H2O2 (data not shown). This result is in perfect agreement with our previous observation that the expression of B. napus APX gene was not induced by various concentrations of H2O2 in presence of 100 μm Fe(III)-EDTA (Vansuyt et al., 1997) and with the result of Pekker et al. (2002) showing that H2O2 did not affect Fe-mediated bean cAPX expression. This is an important difference with the maize Zmfer1 and Arabidopsis AtFer1 ferritin gene expression, which has been shown to be induced by exogenous H2O2 application (Savino et al., 1997; Petit et al., 2001a).
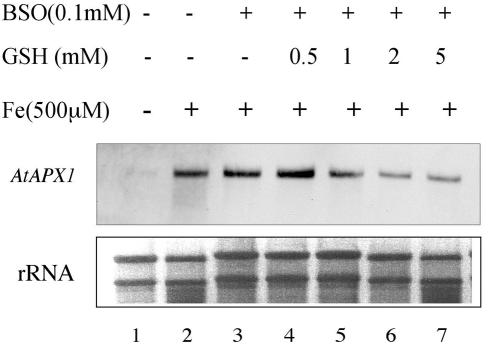
GSH has been shown to antagonize the Fe-induced ferritin expression in maize (Savino et al., 1997) and, in contrast, to increase bean cAPX transcript abundance, at least at a low (10 μm) Fe concentration in the culture medium (Pekker et al., 2002). We have shown previously that the Fe-mediated B. napus APX gene expression was insensitive to a pretreatment with the antioxidant molecule N-acetyl Cys, which in contrast inhibits the Fe induction of ferritin gene expression (Vansuyt et al., 1997). Also, Karpinski et al. (1997) have reported that excess light stress activated the expression of AtAPX1 and that this response was abolished by treating Arabidopsis leaves with GSH. To clarify these apparently contradictory results, we investigated the effect of GSH on AtAPX1 gene expression in Arabidopsis seedlings treated or not with an Fe excess (Fig. 9). Increase of AtAPX1 transcript abundance in Arabidopsis cotyledons in response to a treatment with 500 μm Fe(III)-EDTA for 6 h was unaffected by a 15-h pretreatment with BSO, a GSH synthesis inhibitor (Fig. 9, lanes 1–3). Treatment of the seedlings with various concentrations of GSH (0.5, 1, 2, and 5 mm) revealed a dose-dependent inhibition of AtAPX1 expression in response to the Fe excess treatment (Fig. 9, lanes 4–7).
Figure 9.
Effect of GSH on the Fe-mediated expression of AtAPX1. Fifteen-day-old Arabidopsis seedlings grown on agar plates were treated for 6 h with water (–Fe; lane 1) or 500 μm Fe (III)-EDTA (+Fe, lanes 2–7). Before this treatment, some plants were pretreated with 0.1 mm buthionine-sulfoximine (BSO) for 15 h (lanes 3–7) and for an additional hour with 0, 1, 2, or 5 mm GSH (lanes 4–7). RNA was prepared from frozen samples and analyzed by northern blots using an ATAPX1-specific 3′-UTR probe. SYBR gold (Molecular Probes, Eugene, OR) staining of rRNA was used as a control for RNA loading.
Taken together, these results all agree that exogenous H2O2 application did not affect Fe-mediated APX expression in B. napus (Vansuyt et al., 1997), in bean (Pekker et al., 2002), or in Arabidopsis (this work). It is also clear that exogenously applied GSH has an inhibitory effect on the activation of expression of AtAPX1 in response to excess light stress (Karpinski et al., 1997) or excess Fe stress (this work). This last point is in contrast with the effect of GSH on cAPX expression in bean, which increases the Fe-mediated expression of this gene at 10 μm Fe and has no effect at higher Fe concentrations (Pekker et al., 2002).
DISCUSSION
Fe overload ultimately can lead to an oxidative stress and, as a consequence, to activation of some of the resistance mechanisms to this stress (for review, see Briat, 2002). APX is one of the enzymes involved in such a response and has been shown previously to be induced by Fe overload (Vansuyt et al., 1997). We show here that the Arabidopsis AtAPX1 gene encoding a cytosolic form of APX has its expression specifically induced in response to Fe overload (Fig. 1). This result is in agreement with a report of Pekker et al. (2002) showing that a cAPX from bean is up regulated by Fe overload. The AtAPX1 gene, as with all cAPX genes reported so far, contains a large intron (437 bp) in its 5′-UTR. Deletion of this intron abolishes the strong constitutive expression of the GUS reporter gene in the roots, affecting only slightly the Fe-induced GUS activity in the cotyledons (Fig. 5). In Arabidopsis, therefore, the first intron of AtAPX1 behaves as a root-specific enhancer, raising the question of the function of this high level of cAPX expressed in this organ. The high level of constitutive expression of AtAPX1 measured through fusion of its promoter with the GUS reporter gene is consistent with the high abundance of the AtAPX1 mRNA detected by northern in Arabidopsis roots (data not shown). In tomato plants, the leader intron of the APX20 gene also has been described as containing an enhancer element for high constitutive expression but in the leaves, not in the roots (Gadea et al., 1999); such a difference between the two plant species is unexplained so far and would require additional experiments to be understood.
The AtAPX1 transcription initiation site has been reported as occurring at position –518 in Figure 2 (Storozhenko et al., 1998), in agreement with a previous report of Kubo et al. (1993). However, presence in the Arabidopsis database of the EST R89992 (GenBank Accession no.) revealed the existence of a longer transcript than predicted from the primer extension data previously reported. Furthermore, in The Arabidopsis Information Resource release of April 14, 2003, an even longer AtAPX1 cDNA has been reported (AY086425), its first nucleotide being located within the HSE sequence. RT-PCR experiments using oligonucleotides within this region of the AtAPX1 5′ upstream sequence (Figs. 2 and 3) enabled us to show that transcription initiation is likely to occur at position –669/–671 of the sequence presented in Figure 2, that is 154/156 bp upstream from the previously reported start point (Storozhenko et al., 1998).This result is in agreement with the fact that deletion Δ7 (Fig. 4) of the AtAPX1 upstream sequence, which removes the putative TATA and CAAT boxes indicated in Figure 2, abolishes completely the GUS activity in transgenic plantlets (Figs. 6 and 7), although the TATA box defined by Storozhenko et al. (1998) is still present in this construct. Although we observed the same position for transcription initiation both in leaves and roots (Fig. 3; data not shown), it cannot be excluded that elements defined by Storozhenko et al. (1998) could serve to define an alternative transcription start point for AtAPX1 transcription under particular conditions. One of the consequence of the AtAPX1 transcript being around 200 bp longer than the one previously described is to position the heat shock-responsive element previously described (Storozhenko et al., 1998) within the 5′ leader non-coding sequence of the AtAPX1mRNA. The occurrence of transcriptional cis-regulatory elements in the 5′-UTR region of plant genes has been described previously, although the role of regulatory elements within this region is more often related to regulation of translational efficiency, sometimes by increasing mRNA stability (Bolle et al., 1994; Dickey et al., 1998; Hua et al., 2001). One of the best documented 5′-UTR translational control element is the Fe-responsive element present in ferritin mRNA from animals but not from plants (Jacobson, 1996).
It is possible to delete DNA sequence up to position –788 from the AtAPX1 ATG initiation codon without loosing the Fe-induced activity of the GUS reporter gene in transgenic Arabidopsis plants (Figs. 6 and 7). Within this minimal promoter, an IDRS-like element can be observed (Fig. 8). Such an element is known to be necessary to repress ferritin gene expression under Fe deficiency condition. However, point mutations within this regulatory element (Fig. 8) did not affect LUC expression driven by such a modified promoter sequence, suggesting it plays no role in the Fe regulation of the AtAPX1 gene expression, whereas similar mutations within the AtFer1 IDRS abolished the Fe response (Fig. 8B; Petit et al., 2001b). Pekker et al. (2002) have demonstrated recently that high intracellular levels of free Fe in plants lead to the enhanced production of ROS, which in turn induces the expression of bean cAPX, possibly using GSH as an intermediate signal. Activation of maize ferritin gene expression by Fe overload also has been shown to be a complex response, part of it likely involving ROS, based on inhibition of the Fe response by GSH or N-acetyl cysteine treatments (Savino et al., 1997). However, Petit et al. (2001b) have reported that the IDRS was not required for the H2O2 induction of the ZmFer1 ferritin gene expression but was specific for the metal response, not related to the catalytic effect of this metal on ROS production. In other words, and as schematized in an hypothetical model presented in Figure 10, Fe overload has a specific regulatory effect, mediated by a functional IDRS, which leads to expression of at least some of the ferritin genes, but is not involved in cAPX expression. In contrast, Fe overload-mediated ROS production is responsible for cAPX gene expression and in part for ferritin gene expression through an IDRS-independent pathway requiring still-uncharacterized regulatory cis-elements. Whether this pathway is the same for the two genes is questionable because ferritin AtFer1 gene expression is activated by application of exogenous H2O2, whereas AtAPX1 is not (data not shown), in agreement with results published by Vansuyt et al. (1997) with B. napus APX and by Pekker et al. (2002) for bean cAPX. H2O2 compartmentation could be an important parameter for these responses to take place. Karpinski et al. (1997) have reported that high light stress, leading to photo-inhibition and, therefore, to a transient increase in H2O2 concentration within chloroplasts, was responsible for activation of the cytosolic AtAPX1 gene expression and that GSH had an inhibitory effect of this response. We did observe such a GSH effect in the case of the Fe-mediated expression of AtAPX1 (Fig. 9). To avoid interference with the preexistent GSH, we first depleted GSH pool by pretreatment with BSO. Therefore, it could be speculated that ROS production within the chloroplast, either in response to high light or to Fe stresses, is involved in the regulatory mechanisms controlling AtAPX1 expression. On the other hand, the fact that the AtFer1 ferritin gene expression is induced by extracellular application of H2O2 indicates that ROS implicated in this pathway act at another level than the one operating for AtAPX1 regulation. Whether GSH could be a modulator of this integrated response remains to be demonstrated. It is an interesting working hypothesis because GSH has been suggested to be an intermediate in the bean cAPX gene activation in response to Fe overload (Pekker et al., 2002). However, this antioxidant molecule has been shown to antagonize the Fe-induced ferritin expression (Savino et al., 1997) and to inhibit the Fe (this work; Fig. 9) or the high light (Karpinski et al., 1997) expression of the Arabidopsis AtAPX1 gene. Although there is a difference in GSH action between bean and Arabidopsis, possibly because of the fact that homogluthatione replaces GSH in bean (Wingate et al., 1988), such an antioxidant molecule clearly interferes with stress-mediated expression of cytosolic APX gene expression. In the case of ROS accumulation, the level of GSH is expected to decrease (Karpinski et al., 1997), possibly leading to the derepression of AtFer1 and AtAPX1.
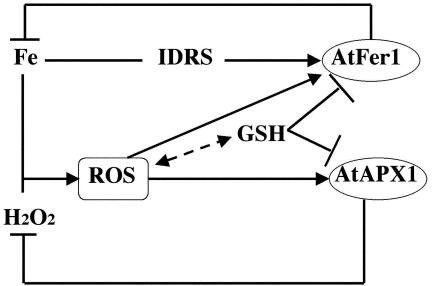
Figure 10.
Model integrating the Fe and oxidative stress responses regulating ferritin (AtFer1) and APX (AtAPX1) gene expression in Arabidopsis. Fe regulates AtFer1 gene expression through the IDRS. Ferritin in turn will store free Fe, decreasing its level and having a negative feedback effect (Petit et al., 2001b). H2O2 can react with free Fe, leading to the production of ROS, that can also activate ferritin gene expression through an IDRS-independent pathway (Lobréaux et al., 1995; Savino et al., 1997; Petit et al., 2001b). AtAPX1expression is also activated by ROS produced in case of Fe excess, and this pathway is inhibited by GSH treatment and is IDRS independent (this work). APX expression will have in turn a negative feedback effect by leading to H2O2 detoxification. Also, extracellular GSH treatment is known to antagonize the Fe induction of ferritin synthesis (Lobreaux et al., 1995; Savino et al., 1997). The dotted double arrow line indicate the balance between ROS and GSH levels.
There are several reports showing that Fe and oxygen metabolism are closely linked. In yeast (Saccharomyces cerevisiae), a cytosolic catalase and a Fe3+ reductase are both regulated by the transcription factor MAC1 (Jungmann et al., 1993). In Pseudomonas aeruginosa, Fe overload induces the expression of three oxidative stress defense genes: catalase, alkyl hydrogen peroxidase, and Fe-SOD (Vasil and Ochsner, 1999). Interestingly, a recent work from Pnueli et al. (2003) shows that ferritin expression is enhanced in an Arabidopsis knockout cApx1 mutant.
CONCLUSION
Taken together, our results indicate that the co-regulation of APX and ferritin gene expression by Fe excess could provide a synergistic way to protect plants from ROS formation because APX scavenges H2O2, whereas ferritin stores excess free Fe, preventing the generation of the highly toxic hydroxyl radical via the Fenton reaction.
MATERIALS AND METHODS
Plant and Cell Cultures
Arabidopsis cells were cultivated at 24°C with constant shaking in 100 mL of Murashige and Skoog medium (Sigma, St. Louis) containing 50 μm Fe-EDTA and supplemented with 0.2 g of Gln, 30 g of Suc and 2 mg of 2,4-dichlorophenoxyacetic acid per liter. Subcultures were made every 14 d using 10 mL of cell suspension added to 90 mL of fresh medium. For Fe overload experiments, 10-d-old cell cultures were treated with 500 μm Fe-citrate. Arabidopsis seeds were surface-sterilized and grown in vitro in Hoagland solid medium containing 2% (w/v) Suc and 10 μm Fe-EDTA.
AtAPX1 Promoter::GUS Constructs and Plant Transformation
A 3-kb DNA fragment upstream of the AtAPX 1 gene was fused to the GUS coding sequence at the ATG start codon and cloned into pGSV6 vector. Several 5′ deletions were generated by PCR and introduced into pGSV6 vector. Transgenic Arabidopsis lines harboring the different constructs were selected on the basis of their kanamycin resistance. A minimum of three independent lines was selected for each of the construct.
GUS Assay and Staining
Ten-day-old Arabidopsis were grown in vitro and used to prepare extracts for GUS assay. Ten cotyledon pairs were sonicated in 150 μL of 100 mm phosphate buffer (pH 7.8) containing 0.2% (v/v) Triton X-100 and 1 mm dithiothreitol. The extracts were centrifuged at 14,000g for 30 min at 4°C, and the supernatant was saved for analysis. For GUS activity, 25 μL of supernatant was added to 75 μL of 100 mm phosphate buffer (pH 7) containing 4-nitrophenyl β-d-glucuronide (Sigma) in 96-well plates. Extracts were incubated at 37°C, and the OD at 414 nm was recorded after 0, 10, 20, and 30 min. Protein concentration was determined according to Bradford (1976). In situ GUS activity was detected essentially according to Jefferson et al. (1987). Pigments were removed from the plantlets by incubation in 70% (v/v) ethanol. The samples were photographed with a stereo-microscope (BX51, Olympus, Tokyo)
AtAPX1 Promoter::Luciferase Constructs and Luciferase Assays
An 824-bp DNA fragment from the 5′-upstream sequence of the AtAPX1 gene, containing the IDRS (Fig. 8), was PCR amplified using oligos CCATCGATCCCAAAGTTAGTGGGCCCTCCACACGAAGCATGTorCCATCGATCCCAAAGTTAGTGGGCGGATCCCACGAAGCATGT as forward primers and oligo CTTCGCCATGGTAGCTAAGCTCTGGAACAAATACAC as reverse primer to generate wild-type or IDRS mutated promoter sequence, respectively. These DNA fragments were cloned by translational fusion with luciferase coding sequence at the ATG, using the pSLluc+de plasmid previously described (Petit et al., 2001b). Arabidopsis cell transformations by particle bombardment, transient luciferase assay, and internal standardization using the pRD109 construct were done as previously reported (Savino et al., 1997; Petit et al., 2001b).
RNA Analysis by Northern and RT-PCR
RNA was extracted from cell cultures, roots, and leaves of Arabidopsis. For northern analysis, equal amounts (15 μg) of RNA were denatured in 20 mm Tris-acetate (pH 7.0) containing 50% (v/v) dimethyl sulfoxide and 1 m glyoxal for 20 min at 50°C. The RNA samples were then fractionated on 1.2% (w/v) agarose gels and transferred to nylon membranes (Positive, Qbiogene, Carlsbad, CA) according to the recommendations of the manufacturer. Fluoresceine-labeled probes were generated by PCR reactions using cloned cDNAs to APX (3′-UTR sequence), ferritin, SOD, and catalases from Arabidopsis, with the appropriate oligonucleotide primers. Hybridizations were carried out as previously described (Vansuyt et al., 1997). The blots were then washed in 2× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) and 0.1% (w/v) SDS at 65°C two times for 15 min and in 0.2× SSC 0.1% (w/v) SDS at 60°C two times 15 min. Blots were incubated with antifluoresceine antibodies coupled to alcaline phosphatase and finally with chemiluminescent substrate (CDP-star, Boehringer Mannheim/Roche, Basel).
For RT-PCR, first strand cDNA synthesis was carried out with Superscript (Invitrogen, Carlsbad, CA) using 5 μg of RNA. Specific PCR amplification of APX1, 2, and 3 was carried out using oligos described by Panchuk et al. (2002). Primers used to amplify the AtAPX1 5′ leader sequence were: RP (5′GTTCTTCGTCATCTTAGCTAAGCTC 3′), FP1 (5′TGGCAAATTTGACCACACGATATC 3′), FP2 (5′ GCCGTGCGTTTAATCATGATCG 3′), FP3 (5′ CCCTAAGATCCGAACGTCCATTT 3′), and FP4 (5′ TTGAAGGATTATTCTTTTCCCATC 3′).
Acknowledgments
We acknowledge Prof. Robertson McClung (Dartmouth College, Hanover, NH) for the gift of catalase cDNA clones. We thank Dr. Jean-Michel Petit (Institut National de la Recherche Agronomique, Montpellier, France) for his help in AtAPX1:: LUC transient expression assay with Arabidopsis cells, and Dr. Marcel Salanoubat (Centre National de Séquençage, Evry, France) for providing the root Arabidopsis cDNA library.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.029876.
References
- Bolle C, Sopory S, Lubberstedt T, Herrmann RG, Oelmuller R (1994) Segments encoding 5′-untranslated leaders of genes for thylakoid proteins contain cis-elements essential for transcription. Plant J 6: 513–523 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Briat JF (2002) Metal-ion-mediated oxidative stress and its control. In M Montagu, D Inzé, eds, Oxidative Stress in Plants. Taylor and Francis Publishers, London, pp 171–189
- Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF (1998) Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell 10: 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea J, Conejero V, Vera P (1999) Developmental regulation of a cytosolic ascorbate peroxidase gene from tomato plants. Mol Gen Genet 262: 212–219 [DOI] [PubMed] [Google Scholar]
- Herbik A, Giritch A, Horstmann C, Becker R, Balzer HJ, Baumlein H, Stephan UW (1996) Iron and copper nutrition-dependent changes in protein expression in a tomato wild type and the nicotianamine-free mutant chloronerva. Plant Physiol 111: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XJ, Van de Cotte B, Van Montagu M, Verbruggen N (2001) The 5′ untranslated region of the At-P5R gene is involved in both transcriptional and post-transcriptional regulation. Plant J 26: 157–169 [DOI] [PubMed] [Google Scholar]
- Jacobson A (1996) Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem 65: 693–739 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Reins HA, Lee J, Romeo A, Hassett R, Kosman D, Jentsch S (1993) MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J 12: 5051–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfenkel K, Van Montagu M, Inze D (1995) Effects of iron excess on Nicotiana plumbaginifolia plants (implications to oxidative stress). Plant Physiol 107: 725–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Saji H, Tanaka K, Kondo N (1993) Genomic DNA structure of a gene encoding cytosolic ascorbate peroxidase from Arabidopsis thaliana. FEBS Lett 315: 313–317 [DOI] [PubMed] [Google Scholar]
- Lobréaux S, Thoiron S, Briat JF (1995) Induction of ferritin synthesis in maize leaves by an iron-mediated oxidative stress. Plant J 8: 443–449 [Google Scholar]
- Murgia I, Delledonne M, Soave C (2002) Nitric oxide mediates iron-induced ferritin accumulation in Arabidopsis. Plant J 30: 521–528 [DOI] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Tel-Or E, Mittler R (2002) Reactive oxygen intermediates and glutathione regulate the expression of cytosolic ascorbate peroxidase during iron-mediated oxidative stress in bean. Plant Mol Biol 49: 429–438 [DOI] [PubMed] [Google Scholar]
- Petit JM, Briat JF, Lobreaux S (2001a) Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem J 359: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, van Wuytswinkel O, Briat JF, Lobreaux S (2001b) Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J Biol Chem 276: 5584–5590 [DOI] [PubMed] [Google Scholar]
- Pich A, Scholz G (1993) The relationship between the activity of various iron-containing and iron-free enzymes and the presence of nicotiamine in tomato seedlings. Physiol Plant 88: 172–178 [Google Scholar]
- Pich A, Scholz G, Stephan UW (1994) Iron dependant changes of heavy metals, nicotianamine and citrate in different plant organs and in the xylem exudate of two tomato genotypes. Nicotianamine as possible copper translocator. Plant Soil 165: 189–196 [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203 [DOI] [PubMed] [Google Scholar]
- Savino G, Briat JF, Lobreaux S (1997) Inhibition of the iron-induced ZmFer1 maize ferritin gene expression by antioxidants and serine/threonine phosphatase inhibitors. J Biol Chem 272: 33319–33326 [DOI] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53: 1305–1319 [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Van Montagu M, Inze D, Kushnir S (1998) The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol 118: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansuyt G, Lopez F, Inze D, Briat JF, Fourcroy P (1997) Iron triggers a rapid induction of ascorbate peroxidase gene expression in Brassica napus. FEBS Lett 410: 195–200 [DOI] [PubMed] [Google Scholar]
- Vasil ML, Ochsner UA (1999) The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol 34: 399–413 [DOI] [PubMed] [Google Scholar]
- Wingate WC, Lawton MA, Lamb CJ (1988) Gluthatione causes a massive and selective induction of plant defense genes. Plant Physiol 87: 206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]